Abstract
Harmful cyanobacterial blooms pose severe threats to aquatic ecosystems. Bloom-forming cyanobacteria form cyanobacterial aggregates (CAs) that create a phycosphere supporting diverse microbial interactions. Here, longitudinal metagenomics and metatranscriptomics were employed to explore the temporal variation of CA-attached viral communities throughout cyanobacterial blooms in Lake Taihu. Viral communities, represented by 5613 viral operational taxonomic units, showed increased relative abundance (RPKM) with the expansion of bloom areas. Among 1791 virus‒host linkages, host shifts followed the succession of two dominant cyanobacterial genera, Microcystis and Dolichospermum. Viruses demonstrated high virus‒host abundance ratios within all host genera and showed elevated transcriptional activities infecting Dolichospermum during the late bloom stage. Viruses featured high microdiversity and positively selected replication-associated genes in response to abundant host genera and variable trophic status. This study uncovered diverse active viral auxiliary metabolism associated with photosynthesis, biochemical cycling, and DNA biosynthesis, and highlighted the significant role of phycosphere-associated viruses during cyanobacterial blooms.
Similar content being viewed by others
Introduction
The widespread occurrence of cyanobacterial blooms has led to a vicious cycle of water-quality deterioration in natural aquatic ecosystems1. The uncontrolled proliferation of cyanobacterial biomass accelerates oxygen depletion, leading to hypoxic conditions that are unfavorable for the growth of submerged aquatic organisms2. Harmful blooming cyanobacteria also produce diverse toxic metabolites such as microcystins3. These toxins can cause severe digestive, neurological, and chronic kidney diseases in humans through contaminated drinking water or the food chain4.
Blooming cyanobacteria typically exist in the form of cyanobacterial aggregates (CAs), where diverse microbes are crowded in cyanobacteria-produced extracellular polysaccharides. This environment, known as the phycosphere, facilitates frequent and active biotic interactions during the whole cycle of cyanobacterial blooms5. CA-attached bacterial communities show the potential in catalyzing the turnover of complex organic matter during cyanobacterial blooms. A previous multi-omics study has also explored the taxonomic diversity and functional responses of bacterial communities in CAs to different stages of cyanobacterial blooms in Lake Taihu6. Functional linkages between cyanobacteria and CA-attached heterotrophic bacteria offered valuable insights into the ecological control and management of eutrophication. In addition to bacterial communities, CAs may harbor ubiquitous and diverse viral communities that potentially play pivotal roles in influencing microbial loop and regulating ecosystem-scale biochemical cycles. However, previous studies mainly focused on the dynamics of free-living viruses during algae blooms at the inter-population level. For example, Peng et al.7 suggested that increased viral lysis in eukaryotic populations and lysogeny in cyanobacteria played crucial roles in driving phytoplankton succession and Microcystis bloom formation in a large reservoir. Yuan et al.8 demonstrated that free-living viruses in a eutrophic lake encoded AMGs involved in central carbon, nitrogen, and phosphate metabolism that influenced nutrient cycling through host infection. Morimoto et al.9 revealed the suppression effect of antiviral gene expression on viral proliferation by exploring the infection dynamics of Microcystis-interacting viruses. As the primary structural units in eutrophic lakes, CAs created a unique habitat characterized by the enrichment of cyanobacteria and their viral counterparts, different from the spatially dispersed and taxonomically differentiated free-living biomass. However, limited information was available associated with the diversity and ecological roles of ubiquitous phycosphere-associated viral communities, creating a critical knowledge gap in understanding the mechanisms driving bloom formation and persistence.
Viruses are the most abundant biological entities in aquatic ecosystems10. As natural predators of cellular microorganisms, such as cyanobacteria, viruses can exert top-down control on host dynamics through lytic infections to cause cell death, or integrate their genomes into host chromosomes to replicate alongside host DNA during lysogenic infections11,12. Variations in aquatic nutrient concentrations may alter the “bloom-and-burst” cycles of microbes, subsequently affecting the inter-population diversity of viral communities through influencing host availability13. Different trophic status may also induce viruses to engage in diversified competitive and cooperative relationships with their microbial hosts14,15. For instance, the “kill the winner” theory suggests that increased host productivity favors the enhanced viral infections, which suppress the dominance of the hosts among microbial communities16. The “Red Queen” theory indicates that viruses and their hosts undergo long-term evolutionary arm races through bacterial resistance to viruses and viral counter-defense against their hosts17. Hence, viral predation might serve as a persistent bio-prevention strategy to eliminate highly abundant cyanobacterial genera, such as Microcystis and Dolichospermum, in bloom-forming CAs18. During the infection phase, viruses encode a large repertoire of auxiliary metabolic genes (AMGs) to reprogram the metabolic state of host cells for self-adaptation19. Previous studies have revealed that viruses infecting cyanobacteria could augment photosynthesis by expressing core photosystem II proteins psbA and psbD20, driving the photosynthetic carbon cycle in aquatic ecosystems. Due to the high turnover rates and evolutionary potentials21, viruses could drive intra-population genetic variations (known as microdiversity) to adapt to host dynamics and maintain stability under variable environmental conditions. Recent investigations have revealed distinct microdiversity patterns and evolutionary trajectories among viruses residing in habitat-specific environments, such as polar glaciers, the Mediterranean, and cold springs22,23,24. The integrated exploration of viral diversity and virus‒host interactions in CAs will deepen our understanding of the algae bloom-forming biosphere and its environmental implications.
As an important water supply source in the Yangtze River Delta and one of the five largest freshwater lakes in China, Lake Taihu has been suffering from extensive cyanobacterial blooms accompanied by the increasing occurrence of toxic cyanotoxins due to the massive discharge of industrial or agricultural wastewater25,26. Unlike other eutrophic lakes, the broad and shallow basin in Lake Taihu, coupled with favorable meteorological conditions, contribute to recurrent algal proliferation events that span several months and affect extensive surface areas of the lake27,28. The complex interplay of nutrient pollution and distinct hydrological environment makes Lake Taihu a typical and ideal case study for exploring viral dynamics throughout long-term cyanobacterial blooms29. Based on the paired time-series metagenomes and meta-transcriptomes, we conducted a longitudinal investigation on CA-attached viral communities during the whole cycle of cyanobacterial blooms in Lake Taihu, with the aim of addressing: (1) temporal variation of viral communities and its ecological drivers at different cyanobacterial blooming stages; (2) taxonomic variation of viruses and their hosts as well as the infection dynamics of virus‒host linkages; (3) genome-wide microdiversity patterns of viral populations during the whole cycle of cyanobacterial blooms; (4) functional potentials of viral communities in hosts and ecosystems. This study showed an integrated profile of temporal dynamics, microdiversity, and ecological functions of CA-attached viruses throughout the cyanobacterial blooms, offering valuable perspectives for eutrophication progression and national water quality management.
Results
Temporal variation of viral communities and its ecological drivers
A large-scale virus identification analysis of 16 shotgun metagenomes gave rise to 5613 nonredundant vOTUs with an average length of 7.2 kb. Viral genomes were assigned to four quality levels, including complete (1.5% of the vOTUs), high-quality (2.2%), medium-quality (4.5%), and low-quality (91.8%) (Supplementary Table 1). Viral communities showed a significantly higher Shannon index at the middle stage than at the late stage of cyanobacterial blooms, whereas no distinct differences in viral α-diversity were detected between the early and the other two stages (Supplementary Fig. 1). Linear regression analysis demonstrated a significantly positive correlation between viral and bacterial α-diversity (Supplementary Fig. 2, r = 0.78, p < 0.001). Principal co-ordinates analysis (PCoA) demonstrated clear variations of viral β-diversity corresponding to early and late stages (p < 0.001), and viral communities sampled at close periods were prone to cluster together (Fig. 1a). Linear regression analysis indicated a potential association between viral abundance and cyanobacterial bloom areas (Fig. 1b, r = 0.47, p < 0.05). Host-associated viral abundance peaked in sample M8, coinciding with the maximum extent of cyanobacterial bloom areas at 481.1 km2 (Supplementary Table 2).
a Principal co-ordinates analysis (PCoA) of viral β-diversity based on Bray-Curtis dissimilarity of viral communities. b Variation in average abundance (RPKM) of viral communities with sampling time. The fitted line is visualized using the locally estimated scatterplot smoothing (LOESS) nonparametric regression model. The subplot shows the correlation between viral abundance and cyanobacterial bloom areas (km2, log2). c Co-occurrence network of environmental factors and vOTUs. The size of each dot which represents environmental factor is proportional to the network degree. Pie charts demonstrate the proportion of the connected vOTUs in a single network module for each environmental factor. d Procrustes analysis based on Bray-Curtis distances of viral and bacterial communities.
The β-diversity of viral communities was influenced by multiple environmental factors, such as water temperature (WT), nutrient concentrations, and β-diversity of bacterial communities. Canonical correspondence analysis (CCA) revealed significant effects of WT, dissolved oxygen (DO), chemical oxygen demand (COD), biological oxygen demand (BOD), total nitrogen (TN), total phosphorus (TP), and ammonium nitrogen (NH4+-N) on viral communities (Supplementary Table 3, p < 0.05). Within the co-occurrence network between environmental factors and viral communities, WT showed the most correlation linkages to viruses and was clustered with 23.6% of the vOTUs into a single network module, followed by TN (19.8%), COD (15.7%), and NH4+-N (14.9%) (Fig. 1c). Procrustes analysis (Fig. 1d, M2 = 0.13, p < 0.001) and mantel test (Supplementary Fig. 3, p < 0.001) further demonstrated a significant correlation between viral and bacterial communities.
Temporal variation of virus‒host taxonomy and infection dynamics
Over 20% of the 5613 vOTUs were assigned to 15 phyla, 18 classes, and 67 families (Supplementary Table 4). During the whole cycle of cyanobacterial blooms, tailed double-stranded DNA phages belonging to the Caudoviricetes class, Uroviricota phylum, showed the highest relative abundances among taxonomically classified viruses (51.1–92.1%) (Supplementary Fig. 4). Inoviridae were also ubiquitously detected in CAs, with their relative abundance ranging from 3.1% to 20.9%. In silico approaches predicted 1,084 vOTUs linked to putative bacterial hosts (Supplementary Table 5 and Fig. 2a), including 12.9% vOTUs with known taxonomy. Given that viral replication is highly dependent on host availability, viral abundances showed significantly positive correlations with predicted host abundances in each sample (Supplementary Fig. 5, p < 0.001). Variance partitioning analysis demonstrated over 50% variance contribution of viral communities to their hosts, suggesting the significant influence of viruses on the host community structure in the phycosphere (Supplementary Fig. 6). At the family level, the Peduoviridae_like phage prevailed at the early and middle stage of cyanobacterial blooms, reaching a maximum relative abundance of 64.2% in the sample M3 (Fig. 2b). Two other families belonging to the Uroviricota phylum, Autographiviridae and Mesyanzhinovviridae, primarily dominated at the late period of the middle stage. The presence of other viral populations, such as Lipothrixviridae and Cressdnaviricota, may reflect their potential interactions with archaea, eukaryotic phytoplankton, or protists that coexisted with cyanobacteria, though their ecological roles in CAs remained speculative and warranted further validation. Among the predicted 1791 virus‒host pairs, bacterial hosts spanned 14 phyla, 20 classes, 55 families, and 68 genera (Supplementary Table 5). Notably, two Cyanobacteriota genera, Microcystis (190) and Pseudnanabaena (47), and one Pseudomonadota genus, Rubrivivax (75), ranked as the top three host genera with the most linkages to viruses (Fig. 2a). Cyanobacteriota and Pseudomonadota occupied an average relative abundance of 52.3% and 34.5% among all host phyla, respectively (Fig. 2c). At the early stage of cyanobacterial blooms, viruses were linked to highly abundant species affiliated with Pseudomonadota and Bacteroidota, which accounted for two-thirds of relative abundance among viral hosts. Microcystis was the most dominant host genus with an extremely high average relative abundance of 44.4% at the middle stage, which served as a major attack target for the most highly expressed viruses (105.9 TPM) (Fig. 3a). At the late stage of cyanobacterial blooms, Dolichospermum appeared as a new dominant host genus with an average relative abundance up to 52.9% (Fig. 2c), while the average relative abundance of viral hosts within non-cyanobacterial phyla dropped to 13.2%. The virus‒host abundance ratio (VHR) served as an estimate of viral replication strength30. Lineage-specific VHR for each genus was higher than one, suggesting the active host infection and viral proliferation in CAs (Supplementary Table 6). The high VHRs during the whole cycle of cyanobacterial blooms (Fig. 3b) indicated the top-down control of viral predations on microbial communities in the phycosphere. VHRs demonstrated a significantly negative correlation with host abundance for the majority of the top 20 genera with the highest abundance, as opposed to Dolichospermum (Supplementary Fig. 7).
a Virus‒host association network based on in silico host prediction. The bottom-right inset indicates the number of vOTUs linked to the specific bacterial genus in the network. Two representative network modules are highlighted by circles. b Relative abundance of viral families with host assignment at different samples. Viral families with relative abundance < 1% are assigned to a separate group. c Relative abundance of host genera at different samples. Host genera with relative abundance < 1% are assigned to a separate group.
a Transcriptomic expression of viruses infecting different host genera. Boxplots show the expression level of viruses linked to three representative Cyanobacteriota genera at early, middle, and late stages of cyanobacterial blooms. b Changes in virus–host abundance ratios (VHRs) for different genera across 16 samples. The significance of p value is marked by asterisks (***p < 0.001; **p < 0.01; ns: p > 0.05).
Microdiversity pattern of viral populations
By tracking viral nucleotide diversity, single nucleotide variant (SNV), and ratio of non-synonymous to synonymous mutations (pN/pS), we uncovered the microdiversity pattern of viral communities during the whole cyanobacterial bloom cycle. Viruses showed relatively higher nucleotide diversity and SNV frequencies at the middle stage than at the early or late stages (Fig. 4a). Nucleotide diversity and SNV frequencies displayed positive Spearman’s correlations with WT and pH and negative correlations with TN and COD (Fig. 4b). Host proliferation may serve as a driving force for genome-wide evolutionary dynamics of viruses, as evidenced by the increased trend of viral nucleotide diversity with the host abundance (Fig. 4c). For example, viruses infecting Microcystis, Pseudanabaena, Rubrivivax, and Roseomonas_B, which showed high abundance among host communities, exhibited high-level nucleotide diversity over the 95% quantile threshold. At the gene level, the majority of the viral genes were subject to purifying selection, while ~3.0% of the genes with pN/pS > 1 underwent positive selection (Fig. 4d) and exhibited significantly higher expression levels at the middle stage than at the late-stage (Supplementary Fig. 8, p < 0.05). The highest proportion of positively selected genes were involved in DNA replication, recombination, and repair (Fig. 4d). In addition, a large number of viral genes with pN/pS > 1 engaged in carbohydrate transport and metabolism, transcription, cell wall/membrane/envelope biogenesis, and posttranslational modification, protein turnover, chaperones at the middle stage (Supplementary Fig. 9). Microcystis genus was linked to viruses carrying the most genes under positive selection, followed by Rubrivivax and Pseudanabaena (Fig. 4e).
a Nucleotide diversity and average SNV frequencies (per 1000 base pairs) of viruses at different cyanobacterial blooming stages. b Spearman correlation between viral nucleotide diversity or SNV and environmental factors. Asterisks indicate statistically significant differences between sampling stages (*p < 0.05; **p < 0.01; ***p < 0.001; ns: p > 0.05). c Linear regression between viral nucleotide diversity and host abundance. Host genera infected by viruses with nucleotide diversity above the 95% quantile threshold are specifically labeled. d Proportion of viral genes under positive selection (pN/pS > 1) or under purifying selection (pN/pS < 1). e Number of positively selected genes encoded by viruses that infect specific bacterial genera.
Transcriptomic expression of virus-encoded AMGs
Transcriptomic analyses provided gene expression profiles of viruses during the whole cycle of cyanobacterial blooms. The number of the up-regulated viral genes was approximately 1 ~ 3-fold higher than that of the down-regulated genes in comparison groups between middle stage and early or late stages (Supplementary Fig. 10), suggesting enhanced activities of viral functional genes during the large-scale cyanobacterial blooming period. Specifically, viruses possess the capacity to alter the metabolic state of their hosts through the function of AMGs31. Using DRAM-v, we identified 292 AMG proteins assigned to 78 gene ontologies, primarily involved in carbon metabolism, photosynthesis, genome replication, amino acid metabolism, and modification/regulation (Supplementary Table 7). AMGs associated with photosynthetic reaction center protein (PF00124/K02703, psbA) demonstrated the highest average expression level of 104.8 TPM (Fig. 5a), almost 15-fold of the total expression of other AMGs. These photosynthesis-related AMGs showed an up-regulation pattern at the late stage of cyanobacterial blooms (Supplementary Fig. 11), significantly affecting the energetic efficiency of the photosynthetic carbon cycle in CAs. Virus-encoded phosphate starvation-inducible AMGs (phoH) exhibited higher expression levels than other nutrient metabolic AMGs and showed active up-regulation in an earlier period of cyanobacterial blooms under a relatively low phosphorus supply. Mannosyl-glycoprotein endo-β-N-acetylglucosaminidase (PF01832) ranked as the third most transcriptionally expressed AMG and displayed higher expression at the middle stage (Fig. 5a). In the early-to-middle period of cyanobacterial blooms, we discovered highly expressed and up-regulated AMGs associated with key nucleotide metabolism pathways, including ribonucleotide reductase (PF02867/PF00317) ribonucleoside-diphosphate reductase (K00525/K00526), and dUTP pyrophosphatase (K01520) (Fig. 5a and Supplementary Fig. 11). These AMGs were predominantly encoded by lytic viruses (Supplementary Table 7). Lysogenic viruses possessed AMGs linked to central carbon metabolism, specifically aconitate hydratase and isocitrate dehydrogenase, which functioned as key enzymes in the tricarboxylic acid (TCA) cycle. Some other differentially expressed AMGs in the middle stage participated in carbohydrate metabolism, such as concanavalin A-like lectin (PF13385), UDP-glucose/GDP-mannose dehydrogenase family (PF00984/PF03721), and UDPglucose 6-dehydrogenase (K00012). These representative differentially expressed AMGs were positioned within internal regions of viral contigs and were located adjacent to viral-like genes based on genomic context visualization (Supplementary Fig. 12). Their functions were further validated through tertiary protein structure analysis. The number of actively expressed AMGs showed a significantly positive correlation with WT and negative correlations with COD, TN, and NH4+-N, while the total expression levels of AMGs exhibited positive correlations with NH4+-N, TN, COD, and TP (Fig. 5b).
a Expression distribution of AMGs at different samples. The expression level is Z-score normalized by samples. Differentially expressed AMGs are indicated by asterisks. A bar plot demonstrates the average expression level of AMGs. b Correlation between environmental factors and the number of actively expressed AMGs, as well as the total expression level of AMGs. The significant differences of spearman’s correlation are denoted by asterisks (***p < 0.001; **p < 0.01; *p < 0.05; ns: p > 0.05).
Discussion
Viruses are ubiquitously distributed across aquatic ecosystems and serve as important bioindicators of water quality, yet limited studies have unveiled the diversity and distribution pattern of viral communities within the phycosphere during cyanobacterial blooms. According to a long-span estimation on the relationship between nutrients and chlorophyll a32, high-level nutrient concentrations contributed to the extensive outbreak of cyanobacteria in Lake Taihu, which served as a representative case for studying viral dynamics during cyanobacterial blooms6. In the present study, viral communities represented by the identified 5613 vOTUs showed sensitive responses to bacterial communities, nutrient concentrations, and water temperature (Fig. 1). Since viral replication highly depends on bacterial hosts33, viral and bacterial diversity inclined to convergent dynamics and environment-driven changes in bacterial distribution simultaneously affect viral communities. For example, nitrogen and phosphorus are key growth-limiting nutrients for autotrophic cyanobacteria, and regulate heterotrophic bacteria and viral populations through the microbial loop34 within the phycosphere. The increased COD supply provides CA-attached heterotrophic microbes with the necessary carbon and energy, fostering the growth and proliferation of viral communities. From the early to middle stages of cyanobacterial blooms, increased nutrient influx created a favorable condition for the rapid growth of cyanobacteria in Lake Taihu6, thereby supporting viral proliferation reflected by the observed increases in viral α-diversity and abundance. Water temperature as a thermodynamic limiting factor has been demonstrated to show a strong correlation with viral communities in marine environments21,35. At the late stage of cyanobacterial blooms, low temperature may influence enzymatic activities, kinetics, and DNA replication performance of viral communities36 which led to their lower viral diversity.
The Uroviricota phylum showed the highest abundance and the most connections to bacterial hosts among the CA-attached viral communities (Fig. 2b and Supplementary Table 5). Multiple studies have observed large members of cyanophages in Uroviricota and highlighted their great ecological contributions to marine cyanobacteria37,38. A recent study suggested that free-living viruses in Lake Taihu were primarily linked to Proteobacteria, Actinobacteriota, Bacteroidota, and Planctomycetota8, while CA-attached viruses showed infection preferences toward two dominant cyanobacterial genera, Microcystis and Dolichospermum, with no detectable associations with Actinobacteriota. Parveen et al.39 indicated that the phycosphere was characterized by the notable absence of Actinobacteria, which prevailed in the free-living communities in a French eutrophic lake. Viral host taxonomy displayed distinct temporal succession patterns across different bloom stages. At the early stage of cyanobacterial blooms, frequent material exchange and intercellular signal transduction facilitated the proliferation of cyanobacteria-interacting microbial consortia6, comprising diverse species affiliated with Pseudomonadota, Bacteroidota, and other phyla, such as JALHMQ01, ELB16-189, and Rubrivivax (Fig. 2c). Especially, highly expressed viruses were predicted to infect Rubrivivax (Fig. 3a), which acts as an intimate partner of CA-attached microbes by supplying necessary carotenoids and metabolite products for the turnover of heterotrophic bacteria40. Following the progression of cyanobacterial blooms, Microcystis occupied the dominant niche, accompanied by a decline in the relative abundance of other attached bacteria within non-cyanobacterial phyla. Previous studies have demonstrated that Microcystis occurred as the predominant cyanobacterial genus within the phycosphere at the middle stage of cyanobacterial blooms18. Bloom-forming Microcystis can produce hazardous microcystins that affect aquatic organisms and cause severe human diseases through the food chain41,42. High Microcystis biomass served as a major host source for the most abundant and transcriptionally active viruses (Fig. 3a). At the late bloom stage, variations in water temperature and nitrogen concentration drove an alternate succession of Dolichospermum as the predominant viral host genus, accompanied by a significant reduction in the relative abundance of other associated microbial hosts. According to the significant positive correlation between VHR and Dolichospermum abundance (Supplementary Fig. 7), viruses likely followed the “kill the winner” dynamics16,43 that underwent lytic infection cycles and rapid proliferation during periods of high host productivity. However, the occurrence of viral replication during bloom development indicated that virus–host dynamics in bloom formation may involve more complex mechanisms than simple density-dependent relationships. A study on Emiliania huxleyi suggested that viruses maintained prolonged lysogenic states in healthy host cells but rapidly transitioned to lytic infection as host cells faced physiological pressure44, which may help explain why viruses persisted with their hosts during long-term cyanobacterial blooms and showed high-level lysis cycles at the late bloom stage when CAs began to break down. Our findings suggested that the active CA-attached viruses potentially functioned as natural bio-antimicrobial agents of the dominant culprit of cyanobacterial blooms from the perspective of virus–host interactions. Comparatively, free-living viruses may divert their infection pressure toward dominant non-cyanobacterial taxa, thereby indirectly conferring a competitive advantage to cyanobacteria and contributing to bloom succession7. Recently, the application prospects of viruses in managing harmful algal blooms have received increasing attention45. Future investigations can focus on developing bacteriophage therapies by isolating specific viruses capable of attacking bloom-forming cyanobacterial species, which may offer a promising approach to mitigating the detrimental effects of cyanobacterial blooms on aquatic ecosystems.
CA-attached viruses drove active microdiversity for persistent adaptation in response to host dynamics and trophic status. The average nucleotide diversity of viral populations reached 8.4 × 10−3, significantly surpassing that of viruses found in global oceans (GOV2), cold seep sediments, and soil environments22, indicating the high-level intra-population genetic variation of viruses in CAs. Viral nucleotide diversity and SNV frequencies were positively affected by water temperature (Fig. 4b), which has been shown as a significant factor influencing genetic selection and enzyme kinetics of cold-stressed viruses36. Comparatively, TN and COD showed a negative correlation with viral nucleotide diversity and SNV frequencies, since lower nutrient supply may exert evolutionary pressures on viral communities. Host-associated viruses with higher nucleotide diversity are inclined to infect host genera with high abundance, such as Microcystis, Pseudanabaena, and Rubrivivax (Fig. 4c). Such viral microdiversity patterns might be associated with the “Red Queen” dynamics as the increased host abundance induced intensified frequency of virus‒host interactions, necessitating that viruses should drive inter-population genetic variation to maintain constant co-existence with host communities17,46. Viruses carried highly expressed and positively selected viral genes linked to DNA replication, repair, and recombination at the middle stage (Fig. 4d and Supplementary Fig. 9), facilitating rapid genomic adaptation to variable host dynamics and environmental stimuli. Our findings provided novel insights into viral adaptive strategy during cyanobacterial blooms from the perspective of intra-population genetic variation beyond the inter-population level.
CA-attached viruses carried transcriptionally expressed AMGs involved in diverse functions, actively participating in biochemical processes during the whole cycle of cyanobacterial blooms. The most highly expressed AMGs (psbA) engaged in the primary photochemistry of photosynthetic reaction centers (PSII) (Fig. 5a). Previous studies have revealed that marine cyanophages can improve the photosynthetic productivity of host cells through horizontal gene transfer of psbA, accelerating the sequestration of dissolved organic matter pool in the ocean47. The expression of viral psbA may also confer photoprotection by mitigating light stress-induced reactive oxygen species in infected hosts48. The phosphate starvation-inducible phoH gene helped to transfer free phosphate to ATP synthase for energy production, suggesting the enhanced environmental adaptation of viruses for phosphorus acquisition49. The high expression of mannosyl-glycoprotein endo-β-N-acetylglucosaminidase (PF01832) at the middle stage (Fig. 5a) facilitated bacterial flagellar synthesis for improved host mobility and more frequent community interactions. The actively expressed AMGs linked to DNA biosynthesis and replication showed the capability of viruses to shift host metabolism towards nucleotide metabolism for increased host availability and viral self-production50. Viruses adopting different lifestyles showed different functional roles in regulating microbial metabolic activities and biochemical cycles throughout the cyanobacterial blooms. Lytic viruses tended to encode more AMGs involved in nucleotide metabolism, such as ribonucleotide reductase, dCTP deaminase, and dUTP pyrophosphatase, which can hijack cellular genome resources for the rapid production of next-generation virions (Supplementary Table 7). Lysogenic viruses harbor AMGs associated with central metabolic processes like the TCA cycle, which were absent in lytic viruses. These AMGs supported fundamental biochemical pathways that promoted both host fitness and viral persistence. Carbon and nitrogen levels showed negative correlations with the number of expressed AMGs and positive correlations with the total expression levels of AMGs (Fig. 5b). In environments with relatively lower nutrient availability, viruses may carry a larger number of AMGs associated with diverse functions such as nutrient acquisition, carbohydrate metabolism, and DNA replication, to confer fitness advantages for host survivability and self-production, while in the late period of cyanobacterial blooms characterized by a nutrient-rich condition, viruses tended to increase AMG expression to enhance specialized functional potential of hosts in nutrient utilization (e.g., photosynthesis) for co-prosperity of viruses and hosts. In all, CA-attached viruses showed nutrient-driven and viral lifestyle-dependent functional potentials, which significantly affected microbial metabolism and biochemical cycles during cyanobacterial blooms.
Harmful cyanobacterial blooms are a significant worldwide environmental issue that necessitates urgent attention. Taking Lake Taihu as a typical and pivotal case, we employed multi-omics approaches to unveil the distribution pattern of viral communities in the cyanobacterial phycosphere and their ecological role in regulating microbial dynamics throughout the annual cycle of cyanobacterial blooms. However, the limited number of samples and lack of laboratory validation may represent some limitations that should be acknowledged. Firstly, the present study was based on 16 CA samples due to climate impacts and gaps in monitoring data, which may not well represent the whole cycle of cyanobacterial blooms in Lake Taihu. In addition, a small sample size may not accurately characterize the variation trends in microbial dynamics. A well-designed sampling campaign with consistent sampling frequencies would help better elucidate the microbial dynamics during cyanobacterial blooms. Secondly, our findings were mainly based on macro-omics analyses. More studies should integrate multi-omics technologies and experimental validation methods to decipher the virus‒host interactions and their potential applications in managing cyanobacterial blooms. In particular, enormous efforts have been made to mitigate ecological risks posed by harmful cyanobacterial blooms and the production of toxic cyanotoxins through various treatment technologies and management strategies51. A key challenge is to explore alternative and cost-effective strategies by using biological forces to address the problem. Since viral predations exert top-down control on bacterial hosts, multiple studies have tried to use viruses as antibacterial agents to treat pathogenic infections in humans52. Analogies to targeted phage therapy in the medical field53, developing strategies for applying active cyanophages may help alleviate the environmental risks posed by harmful cyanobacterial blooms. Based on longitudinal monitoring of a representative natural phycosphere, our study presented valuable insights into viral dynamics and virus‒host interactions during cyanobacterial blooms, providing guidance for managing eutrophication with a designed viral therapy strategy.
Methods
Data collection and reading processing
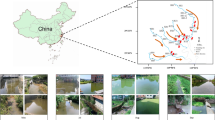
The sixteen paired metagenomic and metatranscriptomic datasets were acquired and sequenced as previously reported6, which were deposited at the NCBI BioProject database under accession numbers PRJNA664299 and PRJNA664620, respectively. Briefly, a sampling campaign was conducted weekly or monthly from 2nd April 2015 to 27th January 2016 in the northern bays of Lake Taihu where large-size CAs (>50 μm) have been observed based on previous monitoring54. The cyanobacterial bloom areas (covering areas of cyanobacteria in the lake) were monitored using a remote sensing technology and analyzed using Environment for Visualizing Images v4.255. Since real-time remote sensing data was missing for several samples due to cloudy days, the unavailable data was replaced with the data of the closest date from the sampling time. The remote sensing analysis revealed a significant expansion of the blooming areas from April/May (~4.5 km2) to June (~129.1 km2), which remained relatively stable until January before declining (~1.70 km2). Based on the observation of variation trend in cyanobacterial bloom areas, the sampling time was divided into three stages (Early: E1~E3; Mid: M1~M10; Late: L1~L3), which was consistent with Zhu et al.’s6 definition of time intervals during the annual cycle of cyanobacterial blooms. The sample details are listed in Supplementary Table 2. Water sample of 5 L each was collected at 0.5 m depth on each sampling date and then filtered through 40 μm Cell Strainers (BD Falcon) to obtain CAs. CAs that floated to the surface were collected by micro-pipetting into tubes. Free-living bacteria were removed using a vortex and washing method56. During the sampling process, eight representative environmental factors were measured, including pH, water temperature (WT), dissolved oxygen (DO), chemical oxygen demand (COD), biological oxygen demand (BOD), ammonium nitrogen (NH4+-N), total nitrogen (TN), and total phosphorus (TP). Following DNA and mRNA extraction, metagenome and metatranscriptome libraries were constructed and sequenced on an Illumina HiSeq2500 platform.
The paired-end metagenomic reads were quality-controlled using TrimGalore v0.6.4 within metaWRAP v1.2.2 (https://github.com/bxlab/metaWRAP). De novo assembly was performed by metaSPAdes v3.13.057 with a minimum contig size of 1000 bp. Raw metatranscriptomic reads were also quality-checked using TrimGalore and then searched against the SILVA database to exclude rRNA sequences from bacterial, archaeal, and eukaryotic taxa58.
Binning, taxonomy, and abundance calculation of bacterial genomes
Contigs of each assembly were binned (binning module: --metabat2 --maxbin2 --run-checkm) and merged into metagenome-assembled genomes (MAGs) using metaWRAP v1.2.2. Genomes with >50% completeness and <10% contamination were selected to meet the standard threshold of medium-to-high quality genomes according to the Minimum Information about a Metagenome-Assembled Genome (MIMAG) criteria59. These filtered MAGs were dereplicated using dRep v3.4.260. Taxonomic classification was performed using GTDB-Tk based on the Genome Taxonomy Database (GTDB, http://gtdb.ecogenomic.org) Release 202. Read mapping was conducted using Bowtie2 v2.3.5 (http://bowtie-bio.sourceforge.net/bowtie2) against a pre-indexed database of MAGs by a bowtie2-build module. The bam files created by Bowtie2 and SAMtools v1.9 (http://samtools.sourceforge.net) were used as input for CoverM (https://github.com/wwood/CoverM) to calculate the abundance of MAGs (genome mode; -m rpkm; --min-read-percent-identity 95; --min-read-aligned-percent 90). The abundance value was measured in the form of reads per kilobase per million mapped reads (RPKM) values, calculated by the number of reads mapped to the targeted contig (×109) divided by the contig length and the total number of mapped reads.
Virus identification and taxonomic annotation
Viral contigs in each metagenomic assembly were recovered using viralVerify v1.161, VIBRANT v1.2.162, VirSorter2 v2.2.463, DeepVirFinder v1.064, PPR-Meta v1.165, geNomad v1.5.166, and Earth Virome Pipeline67. The completeness of the putative viral contigs was estimated using CheckV v1.0.168 with the minimum threshold of 1,500 bp for viruses with medium- to high-quality genomes or complete genomes and 3000 bp for viruses with low-quality genomes69. Lysogeny was determined using geNomad and CheckV. Identified viral contigs were de-replicated at 95% average nucleotide identity (ANI) and 85% alignment coverage using CD-HIT v4.7 (https://github.com/weizhongli/cd-hit), generating non-redundant viral operational taxonomic units (vOTUs). PhaGCN2 v2.170 used a deep learning classifier to taxonomically annotate vOTUs with genome length < 10 kbp based on the latest ICTV viral classification. vConTACT2 v0.9.1571 constructed a gene-sharing network with the Markov Cluster Algorithm for the taxonomic classification of vOTUs with over 10 kbp genome size. The abundance of vOTUs was calculated via the same pipeline of genome abundance estimation, except for utilizing the “contig” mode in CoverM.
Viral host prediction
Considering the challenge in exploring virus‒host relationships within the vast reservoir of newly discovered uncultured viral and bacterial genomes, we conducted viral host prediction through in silico metagenome-based approaches, including CRISPR (clustered regularly interspaced short palindromic repeat) spacer match, sequence similarity, and transfer RNA (tRNA) match. These methods have been widely applied in unveiling a diverse broad-spectrum of virus‒host linkages in marine, river, groundwater, and soil ecosystems72,73,74,75. CRISPR represents an acquired prokaryotic immune mechanism to recognize and memorize short segments from the genome of the viral invader76. If a virus matches the spacers from the CRISPR array, it can build a specific and reliable virus‒host linkage. The CRISPRs in MAGs were identified using MinCED77 and subsequently aligned against viral contigs using BLASTn with ≤1 mismatch and 100% coverage. As some viruses show adjustment of codon usage profiles to match tRNA genes to that of hosts under evolutionary stress, the tRNA match method could also be used to predict virus‒host linkages by searching shared tRNA genes between viruses and their hosts67. The tRNA genes from MAGs and vOTUs were identified using ARAGORN78 (‘-t’ option), followed by the subsequent alignment of tRNAs using BLASTn with stringent cut-off values of 100% sequence identity and 100% coverage. These two methods were implemented based on the “Earth Virome” pipeline by the Joint Genome Institute, which showed high taxonomic accuracy with 98.5% of spacer matches and 92.5% of tRNA matches against viral genomes agreeing with their known host taxonomy at the genus level67. Given that CRISPR systems occur in only 40~70% of prokaryotes and viral tRNA genes are not ubiquitous, we employed sequence similarity method by comparing the nucleotide homology between viral and bacterial genomes79 for supplement of diverse virus-host linkages. Specifically, the sequence alignment between vOTUs and MAGs was conducted by BLASTn based on the strict threshold of ≥ 90% minimum nucleotide identity and ≤0.001 e-value80. Edwards et al.81 evaluated the accuracy of sequence homology approaches based on a benchmarking dataset of virus host relationships. BLASTn-based sequence similarity method produced the highest host prediction accuracy at the genus level (62.32%). The results from the three methods were united to represent 1922 virus‒host linkages. To minimize potential false-positive errors during in silico predictions, we validated the predicted virus‒host linkages by comparing them with the Virus‒Host Database (https://www.genome.jp/virushostdb) to exclude 131 taxonomically unmatched virus‒host linkages. Viruses belonging to Lipothrixviridae, Cressdnaviricota, Kitrinoviricota, Taleaviricota, and Lenarviricota were removed from host prediction results, as these taxa were known to infect archaea or eukaryotes. The virus‒host association network was visualized in Gephi (https://gephi.org/) based on validated host prediction results (Supplementary Table 5).
Viral genes and AMGs annotation
Viral proteins predicted by Prodigal v2.6.382 were queried against the eggNOG database (http://eggnog6.embl.de/) using emapper.py v1.0.3. Prior to AMG screening, we used CheckV68 to identify prophage boundaries and remove host contamination. VirSorter2 v2.2.4 (--prep-for-dramv) provided the annotation files of virus-associated and viral hallmark genes for subsequent AMG identification using DRAM-v v1.4.683. The genes with auxiliary scores of 1 ~ 3 and M/F AMG flags were annotated as putative AMGs. Then we checked the genomic context of the AMG-containing viral contigs to remove the AMGs with no neighboring virus-like or viral hallmark genes and those located at the contig’s edge69. PROSITE84 helped the automated annotation of conserved domains and active sites in AMGs. Given that tertiary protein structures are more conserved than their protein sequences, Phyre285 was employed to predict AMG protein structures with confidence > 90% and coverage > 70%, thereby providing structural validation for the predicted biological functions of AMGs.
Nucleotide diversity, SNV calling, and pN/pS calculation
InStrain v1.7.586 used the BAM files generated by Bowtie2 to profile viral microdiversity. Viral populations with > 5x average read depth coverage were retained for downstream analysis. Nucleotide diversity (π) was used as a measurement of genetic diversity within viral populations and was calculated as the frequency of variations between two sequencing reads at a specific position, following the formula: 1 − ((frequency of A)2 + (frequency of C)2 + (frequency of G)2 + (frequency of T)2). A single nucleotide variant (SNV) was determined if a minimum frequency exceeded 0.05 and a false discovery rate met the threshold of 1e-06. These SNVs were then categorized as synonymous or nonsynonymous, depending on their alignment with gene annotations. The ratio of non-synonymous to synonymous mutations (pN/pS) implied whether genes were biased towards purifying selection to prevent mutation (<1) or positive selection to mutate actively (>1).
Transcriptomic analysis
Viral contigs and genes identified from metagenomic datasets were used as transcript references and were built into Bowtie2 indices. The non-rRNA reads were then mapped to the constructed indices using the rsem-calculate-expression module within RSEM v1.3.387. The expression levels of viral contigs and genes were represented by transcripts per million (TPM) values and Z-score normalized along the sample axis. The differentially expressed genes (DEGs) were identified by DEseq2 with |log2 (fold change)| ≥ 1 and p value < 0.05 between different cyanobacterial blooming stages88.
Statistical analyses
Statistical analyses were conducted in R 4.1.1. vOTU counts in respective samples were rarefied to the identical level using the “phyloseq” R package. The estimateR and diversity functions in the “vegan” R package were applied to calculate the Shannon-Wiener α-diversity index. Principal co-ordinates analysis (PCoA) showed the distribution of viral β-diversity using Bray-Curtis distances of viral communities. Permutational multivariate analysis of variance (PERMANOVA) was employed to estimate the significance of differences. Canonical correspondence analysis (CCA) and mantel test were conducted to reveal the relationship between viral communities and environmental factors/bacterial communities. The significance test was performed by the ANOVA function based on 999 permutations. The co-occurrence network was constructed according to Spearman’s correlation between vOTUs and environmental gradients. Benjamini-Hochberg Correction was applied to adjust p-values based on their ranks and the total number of comparisons. The associations with a correlation coefficient ≥ 0.7 and adjusted p value < 0.05 were retained after multiple comparison correction. Procrustes analysis was applied to investigate the relationship between bacterial and viral communities. Variance partitioning analysis (VPA) estimated the relative contribution of viral communities and environmental factors to the variance in host community structure.
Data availability
The used metagenome and meta-transcriptome datasets are acquired from the NCBI Sequence Read Archive under accession numbers SRR12678595-SRR12678610 and SRR12686983-SRR12686998, respectively. The authors declare that all other data supporting the findings of this study are available from the corresponding author on request.
Code availability
No custom codes were used in the present study.
References
Huang, J. et al. Effects of hydrological and climatic variables on cyanobacterial blooms in four large shallow lakes fed by the Yangtze River. Environ. Sci. Ecotechnol. 5, 100069. https://doi.org/10.1016/j.ese.2020.100069 (2021).
Huisman, J. et al. Cyanobacterial blooms. Nat. Rev. Microbiol. 16, 471–483, https://doi.org/10.1038/s41579-018-0040-1 (2018).
Wei, J. et al. Biodegradation of MC-LR and its key bioactive moiety Adda by Sphingopyxis sp. YF1: Comprehensive elucidation of the mechanisms and pathways. Water Res. 229, 119397. https://doi.org/10.1016/j.watres.2022.119397 (2023).
Feng, S. et al. Microcystin-LR combined with cadmium exposures and the risk of chronic kidney disease: A case–control study in central China. Environ. Sci. Technol. 56, 15818–15827, https://doi.org/10.1021/acs.est.2c02287 (2022).
Gao, D. et al. Determinants of total and active microbial communities associated with cyanobacterial aggregates in a eutrophic lake. mSystems 8, e00992–00922, https://doi.org/10.1128/msystems.00992-22 (2023).
Zhu, C. et al. Responses of cyanobacterial aggregate microbial communities to algal blooms. Water Res. 196, 117014. https://doi.org/10.1016/j.watres.2021.117014 (2021).
Peng, K. et al. Viruses may facilitate the cyanobacterial blooming during summer bloom succession in Xiangxi Bay of Three Gorges Reservoir, China. Front. Microbiol. 14, 1112590. https://doi.org/10.3389/fmicb.2023.1112590 (2023).
Yuan, L. et al. Seasonal succession, host associations, and biochemical roles of aquatic viruses in a eutrophic lake plagued by cyanobacterial blooms. Environ. Int. 193, 109125. https://doi.org/10.1016/j.envint.2024.109125 (2024).
Morimoto, D. et al. Cooccurrence of broad- and narrow-host-range viruses infecting the bloom-forming toxic cyanobacterium Microcystis aeruginosa. Appl. Environ. Microbiol. 85, e01170–01119, https://doi.org/10.1128/aem.01170-19 (2019).
Suttle, C. A. Marine viruses — major players in the global ecosystem. Nat. Rev. Microbiol. 5, 801–812, https://doi.org/10.1038/nrmicro1750 (2007).
Rohwer, F. & Thurber, R. V. Viruses manipulate the marine environment. Nature 459, 207–212, https://doi.org/10.1038/nature08060 (2009).
Silveira, C. B. & Rohwer, F. L. Piggyback-the-Winner in host-associated microbial communities. npj Biofilms Microbiomes2, 16010. https://doi.org/10.1038/npjbiofilms.2016.10 (2016).
Touchon, M., Bernheim, A. & Rocha, E. P. C. Genetic and life-history traits associated with the distribution of prophages in bacteria. ISME J. 10, 2744–2754, https://doi.org/10.1038/ismej.2016.47 (2016).
Rodriguez-Valera, F. et al. Explaining microbial population genomics through phage predation. Nat. Rev. Microbiol. 7, 828–836, https://doi.org/10.1038/nrmicro2235 (2009).
Fernández, L., Rodríguez, A. & García, P. Phage or foe: an insight into the impact of viral predation on microbial communities. ISME J. 12, 1171–1179, https://doi.org/10.1038/s41396-018-0049-5 (2018).
Knowles, B. et al. Lytic to temperate switching of viral communities. Nature 531, 466–470, https://doi.org/10.1038/nature17193 (2016).
Ignacio-Espinoza, J. C., Ahlgren, N. A. & Fuhrman, J. A. Long-term stability and Red Queen-like strain dynamics in marine viruses. Nat. Microbiol. 5, 265–271, https://doi.org/10.1038/s41564-019-0628-x (2020).
Zhu, C.-M. et al. Alternate succession of aggregate-forming cyanobacterial genera correlated with their attached bacteria by co-pathways. Sci. Total Environ. 688, 867–879, https://doi.org/10.1016/j.scitotenv.2019.06.150 (2019).
Ji, M. et al. Temporal turnover of viral biodiversity and functional potential in intertidal wetlands. npj Biofilms Microbiomes 10, 48, https://doi.org/10.1038/s41522-024-00522-8 (2024).
Ruiz-Perez, C. A., Tsementzi, D., Hatt, J. K., Sullivan, M. B. & Konstantinidis, K. T. Prevalence of viral photosynthesis genes along a freshwater to saltwater transect in Southeast USA. Environ. Microbiol. Rep. 11, 672–689, https://doi.org/10.1111/1758-2229.12780 (2019).
Sunagawa, S. et al. Structure and function of the global ocean microbiome. Science 348, 1261359. https://doi.org/10.1126/science.1261359 (2015).
Peng, Y. et al. Viruses in deep-sea cold seep sediments harbor diverse survival mechanisms and remain genetically conserved within species. ISME J. 17, 1774–1784, https://doi.org/10.1038/s41396-023-01491-0 (2023).
Zhong, Z.-P. et al. Lower viral evolutionary pressure under stable versus fluctuating conditions in subzero Arctic brines. Microbiome 11, 174. https://doi.org/10.1186/s40168-023-01619-6 (2023).
Coutinho, F. H., Rosselli, R. & Rodríguez-Valera, F. Trends of microdiversity reveal depth-dependent evolutionary strategies of viruses in the Mediterranean. mSystems 4, e00554–00519, https://doi.org/10.1128/msystems.00554-19 (2019).
Wang, J., Fu, Z., Qiao, H. & Liu, F. Assessment of eutrophication and water quality in the estuarine area of Lake Wuli, Lake Taihu, China. Sci. Total Environ. 650, 1392–1402, https://doi.org/10.1016/j.scitotenv.2018.09.137 (2019).
Qin, B., Xu, P., Wu, Q., Luo, L. & Zhang, Y. Environmental issues of Lake Taihu, China. Hydrobiologia 581, 3–14, https://doi.org/10.1007/s10750-006-0521-5 (2007).
Xu, H., Paerl, H. W., Qin, B., Zhu, G. & Gaoa, G. Nitrogen and phosphorus inputs control phytoplankton growth in eutrophic Lake Taihu, China. Limnol. Oceanogr. 55, 420–432, https://doi.org/10.4319/lo.2010.55.1.0420 (2010).
Chen, Y., Qin, B., Teubner, K. & Dokulil, M. T. Long-term dynamics of phytoplankton assemblages: Microcystis-domination in Lake Taihu, a large shallow lake in China. J. Plankton Res. 25, 445–453, https://doi.org/10.1093/plankt/25.4.445 (2003).
Bai, C. et al. Contrasting diversity patterns and community assembly mechanisms of bacterioplankton among different aquatic habitats in Lake Taihu, a large eutrophic shallow lake in China. Environ. Pollut. 315, 120342. https://doi.org/10.1016/j.envpol.2022.120342 (2022).
Parikka, K. J., Le Romancer, M., Wauters, N. & Jacquet, S. Deciphering the virus-to-prokaryote ratio (VPR): insights into virus–host relationships in a variety of ecosystems. Biol. Rev. 92, 1081–1100, https://doi.org/10.1111/brv.12271 (2017).
Kieft, K. et al. Ecology of inorganic sulfur auxiliary metabolism in widespread bacteriophages. Nat. Commun. 12, 3503. https://doi.org/10.1038/s41467-021-23698-5 (2021).
Zou, W. et al. Temporal dependence of chlorophyll a–nutrient relationships in Lake Taihu: Drivers and management implications. J. Environ. Manag. 306, 114476. https://doi.org/10.1016/j.jenvman.2022.114476 (2022).
Roossinck, M. J. & Bazán, E. R. Symbiosis: viruses as intimate partners. Annu. Rev. Virol. 4, 123–139, https://doi.org/10.1146/annurev-virology-110615-042323 (2017).
Mooshammer, M. et al. Decoupling of microbial carbon, nitrogen, and phosphorus cycling in response to extreme temperature events. Sci. Adv. 3, e1602781. https://doi.org/10.1126/sciadv.1602781 (2017).
Gregory, A. C. et al. Marine DNA viral macro- and microdiversity from pole to pole. Cell 177, 1109–1123.e1114, https://doi.org/10.1016/j.cell.2019.03.040 (2019).
Alarcón-Schumacher, T. et al. Ecogenomics and adaptation strategies of southern ocean viral communities. mSystems 6, e00396–00321, https://doi.org/10.1128/msystems.00396-21 (2021).
Zhou, K. et al. Genomic and transcriptomic insights into complex virus–prokaryote interactions in marine biofilms. ISME J. 17, 2303–2312, https://doi.org/10.1038/s41396-023-01546-2 (2023).
Doron, S. et al. Transcriptome dynamics of a broad host-range cyanophage and its hosts. ISME J. 10, 1437–1455, https://doi.org/10.1038/ismej.2015.210 (2015).
Parveen, B. et al. Bacterial communities associated with Microcystis colonies differ from free-living communities living in the same ecosystem. Environ. Microbiol. Rep. 5, 716–724, https://doi.org/10.1111/1758-2229.12071 (2013).
de Lima, L. K. F., Ponsano, E. H. G. & Pinto, M. F. Cultivation of Rubrivivax gelatinosus in fish industry effluent for depollution and biomass production. World J. Microb. Biot. 27, 2553–2558, https://doi.org/10.1007/s11274-011-0725-3 (2011).
Le, V. V., Srivastava, A., Ko, S.-R., Ahn, C.-Y. & Oh, H.-M. Microcystis colony formation: Extracellular polymeric substance, associated microorganisms, and its application. Bioresour. Technol. 360, 127610. https://doi.org/10.1016/j.biortech.2022.127610 (2022).
Ren, X. et al. Fate, abundance and ecological risks of microcystins in aquatic environment: The implication of microplastics. Water Res 251, 121121. https://doi.org/10.1016/j.watres.2024.121121 (2024).
Howard-Varona, C., Hargreaves, K. R., Abedon, S. T. & Sullivan, M. B. Lysogeny in nature: mechanisms, impact and ecology of temperate phages. ISME J. 11, 1511–1520, https://doi.org/10.1038/ismej.2017.16 (2017).
Knowles, B. et al. Temperate infection in a virus–host system previously known for virulent dynamics. Nat. Commun. 11, 4626. https://doi.org/10.1038/s41467-020-18078-4 (2020).
Zhu, X. et al. From natural to artificial cyanophages: Current progress and application prospects. Environ. Res. 223, 115428. https://doi.org/10.1016/j.envres.2023.115428 (2023).
Wang, J., Li, L. & Lin, S. Active viral infection during blooms of a dinoflagellate indicates dinoflagellate-viral co-adaptation. Appl. Environ. Microbiol. 89, e01156–01123, https://doi.org/10.1128/aem.01156-23 (2023).
Ignacio-Espinoza, J. C. & Sullivan, M. B. Phylogenomics of T4 cyanophages: lateral gene transfer in the ‘core’ and origins of host genes. Environ. Microbiol. 14, 2113–2126, https://doi.org/10.1111/j.1462-2920.2012.02704.x (2012).
Sander, J. et al. Functional characterization and quantification of the alternative PsbA copies in Thermosynechococcus elongatus and their role in photoprotection. J. Biol. Chem. 285, 29851–29856, https://doi.org/10.1074/jbc.m110.127142 (2010).
Breitbart, M., Bonnain, C., Malki, K. & Sawaya, N. A. Phage puppet masters of the marine microbial realm. Nat. Microbiol. 3, 754–766, https://doi.org/10.1038/s41564-018-0166-y (2018).
Enav, H., Mandel-Gutfreund, Y. & Béjà, O. Comparative metagenomic analyses reveal viral-induced shifts of host metabolism towards nucleotide biosynthesis. Microbiome 2, 9. https://doi.org/10.1186/2049-2618-2-9 (2014).
Song, L. et al. Harmful cyanobacterial blooms: Biological Traits, mechanisms, risks, and control strategies. Annu, Rev. Env. Resour. 48, 123–147, https://doi.org/10.1146/annurev-environ-112320-081653 (2023).
Rehman, S., Ali, Z., Khan, M., Bostan, N. & Naseem, S. The dawn of phage therapy. Rev. Med. Virol. 29, e2041. https://doi.org/10.1002/rmv.2041 (2019).
Little, J. S. et al. Bacteriophage treatment of disseminated cutaneous Mycobacterium chelonae infection. Nat. Commun. 13, 2313. https://doi.org/10.1038/s41467-022-29689-4 (2022).
Zhu, W. et al. High nutrient concentration and temperature alleviated formation of large colonies of Microcystis: Evidence from field investigations and laboratory experiments. Water Res. 101, 167–175, https://doi.org/10.1016/j.watres.2016.05.080 (2016).
Kiage, L. M. & Walker, N. D. Using NDVI from MODIS to Monitor Duckweed Bloom in Lake Maracaibo, Venezuela. Water Resour. Manag. 23, 1125–1135, https://doi.org/10.1007/s11269-008-9318-9 (2009).
Cai, H.-Y., Yan, Z.-S., Wang, A.-J., Krumholz, L. R. & Jiang, H.-L. Analysis of the attached microbial community on mucilaginous cyanobacterial aggregates in the eutrophic Lake Taihu reveals the importance of Planctomycetes. Microb. Ecol. 66, 73–83, https://doi.org/10.1007/s00248-013-0224-1 (2013).
Nurk, S., Meleshko, D., Korobeynikov, A. & Pevzner, P. A. metaSPAdes: a new versatile metagenomic assembler. Genome Res 27, 824–834, https://doi.org/10.1101/gr.213959.116 (2017).
Kopylova, E., Noé, L. & Touzet, H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28, 3211–3217, https://doi.org/10.1093/bioinformatics/bts611 (2012).
Bowers, R. M. et al. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat. Biotechnol. 35, 725–731, https://doi.org/10.1038/nbt.3893 (2017).
Olm, M. R., Brown, C. T., Brooks, B. & Banfield, J. F. dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J. 11, 2864–2868, https://doi.org/10.1038/ismej.2017.126 (2017).
Antipov, D., Raiko, M., Lapidus, A. & Pevzner, P. A. MetaviralSPAdes: assembly of viruses from metagenomic data. Bioinformatics 36, 4126–4129, https://doi.org/10.1093/bioinformatics/btaa490 (2020).
Kieft, K., Zhou, Z. C. & Anantharaman, K. VIBRANT: automated recovery, annotation and curation of microbial viruses, and evaluation of viral community function from genomic sequences. Microbiome 8, 90. https://doi.org/10.1186/s40168-020-00867-0 (2020).
Guo, J. et al. VirSorter2: a multi-classifier, expert-guided approach to detect diverse DNA and RNA viruses. Microbiome 9, 37. https://doi.org/10.1186/s40168-020-00990-y (2021).
Ren, J. et al. Identifying viruses from metagenomic data using deep learning. Quant. Biol. 8, 64–77, https://doi.org/10.1007/s40484-019-0187-4 (2020).
Fang, Z. C. et al. PPR-Meta: A tool for identifying phages and plasmids from metagenomic fragments using deep learning. Gigascience 8, giz066. https://doi.org/10.1093/gigascience/giz066 (2019).
Camargo, A. P. et al. IMG/VR v4: an expanded database of uncultivated virus genomes within a framework of extensive functional, taxonomic, and ecological metadata. Nucleic Acids Res. 51, D733–D743, https://doi.org/10.1093/nar/gkac1037 (2022).
Paez-Espino, D. et al. Uncovering Earth’s virome. Nature 536, 425–430, https://doi.org/10.1038/nature19094 (2016).
Nayfach, S. et al. CheckV assesses the quality and completeness of metagenome-assembled viral genomes. Nat. Biotechnol. 39, 578–585, https://doi.org/10.1038/s41587-020-00774-7 (2021).
Pratama, A. A. et al. Expanding standards in viromics: in silico evaluation of dsDNA viral genome identification, classification, and auxiliary metabolic gene curation. PeerJ 9, e11447. https://doi.org/10.7717/peerj.11447 (2021).
Jiang, J.-Z. et al. Virus classification for viral genomic fragments using PhaGCN2. Brief. Bioinform. 24, bbac505. https://doi.org/10.1093/bib/bbac505 (2022).
Jang, H. B. et al. Taxonomic assignment of uncultivated prokaryotic virus genomes is enabled by gene-sharing networks. Nat. Biotechnol. 37, 632–639, https://doi.org/10.1038/s41587-019-0100-8 (2019).
Emerson, J. B. et al. Host-linked soil viral ecology along a permafrost thaw gradient. Nat. Microbiol. 3, 870–880, https://doi.org/10.1038/s41564-018-0190-y (2018).
Li, Z. et al. Deep sea sediments associated with cold seeps are a subsurface reservoir of viral diversity. ISME J. 15, 2366–2378, https://doi.org/10.1038/s41396-021-00932-y (2021).
Luo, X.-Q. et al. Viral community-wide auxiliary metabolic genes differ by lifestyles, habitats, and hosts. Microbiome 10, 190. https://doi.org/10.1186/s40168-022-01384-y (2022).
Cheng, R. et al. Virus diversity and interactions with hosts in deep-sea hydrothermal vents. Microbiome 10, 235. https://doi.org/10.1186/s40168-022-01441-6 (2022).
Horvath, P. & Barrangou, R. CRISPR/Cas, the immune system of bacteria and archaea. Science 327, 167–170, https://doi.org/10.1126/science.1179555 (2010).
Bland, C. et al. CRISPR Recognition Tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinforma. 8, 209. https://doi.org/10.1186/1471-2105-8-209 (2007).
Laslett, D. & Canback, B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32, 11–16, https://doi.org/10.1093/nar/gkh152 (2004).
Dutilh, B. E. et al. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat. Commun. 5, 4498. https://doi.org/10.1038/ncomms5498 (2014).
Coutinho, F. H. et al. Marine viruses discovered via metagenomics shed light on viral strategies throughout the oceans. Nat. Commun. 8, 15955. https://doi.org/10.1038/ncomms15955 (2017).
Edwards, R. A., McNair, K., Faust, K., Raes, J. & Dutilh, B. E. Computational approaches to predict bacteriophage–host relationships. FEMS Microbiol. Rev. 40, 258–272, https://doi.org/10.1093/femsre/fuv048 (2015).
Hyatt, D. et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinforma. 11, 119. https://doi.org/10.1186/1471-2105-11-119 (2010).
Shaffer, M. et al. DRAM for distilling microbial metabolism to automate the curation of microbiome function. Nucleic Acids Res. 48, 8883–8900, https://doi.org/10.1093/nar/gkaa621 (2020).
Sigrist, C. J. A. et al. New and continuing developments at PROSITE. Nucleic Acids Res. 41, D344–D347, https://doi.org/10.1093/nar/gks1067 (2012).
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N. & Sternberg, M. J. E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858, https://doi.org/10.1038/nprot.2015.053 (2015).
Olm, M. R. et al. inStrain profiles population microdiversity from metagenomic data and sensitively detects shared microbial strains. Nat. Biotechnol. 39, 727–736, https://doi.org/10.1038/s41587-020-00797-0 (2021).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinforma. 12, 323. https://doi.org/10.1186/1471-2105-12-323 (2011).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. https://doi.org/10.1186/s13059-014-0550-8 (2014).
Acknowledgements
This work was supported by the Shenzhen Natural Science Fund (the Stable Support Plan Program 20220808011114001). Supports from the High-performance Computing Platform of Peking University are acknowledged.
Author information
Authors and Affiliations
Contributions
T.L. designed the research. T.Y.C. and Y.X.X. performed the work and wrote the manuscript with the help of J.H.Z., Q.E.Z., J.W., and N.X. T.L. edited the manuscript. All authors contributed ideas, read, and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chen, T., Xiong, Y., Zhang, J. et al. Temporal dynamics, microdiversity, and ecological functions of viral communities during cyanobacterial blooms in Lake Taihu. npj Biofilms Microbiomes 11, 178 (2025). https://doi.org/10.1038/s41522-025-00771-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41522-025-00771-1