Abstract
Simulating catalytic reactivity under operative conditions poses a significant challenge due to the dynamic nature of the catalysts and the high computational cost of electronic structure calculations. Machine learning potentials offer a promising avenue to simulate dynamics at a fraction of the cost, but they require datasets containing all relevant configurations, particularly reactive ones. Here, we present a scheme to construct reactive potentials in a data-efficient manner. This is achieved by combining enhanced sampling methods first with Gaussian processes to discover transition paths and then with graph neural networks to obtain a uniformly accurate description. The necessary configurations are extracted via a Data-Efficient Active Learning (DEAL) procedure based on local environment uncertainty. We validated our approach by studying several reactions related to the decomposition of ammonia on iron-cobalt alloy catalysts. Our scheme proved to be efficient, requiring only ~1000 DFT calculations per reaction, and robust, sampling reactive configurations from the different accessible pathways. Using this potential, we calculated free energy profiles and characterized reaction mechanisms, showing the ability to provide microscopic insights into complex processes under dynamic conditions.
Similar content being viewed by others
Introduction
Dynamics has long been recognized as a key ingredient in chemical reactivity, particularly in heterogeneous catalysis where active sites continuously evolve according to reaction conditions1,2. High temperatures and pressures induce intricate transformations in the microscopic structure of the catalyst, ranging from surface diffusivity3 to reactant- or adsorbate-induced4 reconstructions. If capturing these dynamic phenomena experimentally is a formidable challenge, their computational modeling is no less so. Atomistic simulations, particularly molecular dynamics (MD), are an ideal candidate for providing microscopic insights into their workings5,6,7. However, to fully describe these dynamical effects, we need both accurate quantum-mechanical (QM) models of the potential energy surface and extended time- and length-scale simulations6, two often conflicting requirements.
Machine learning (ML) potentials8 have emerged in recent years as promising tools to address the accuracy-efficiency trade-off. They are optimized to reproduce energies and forces from a dataset of reference calculations, typically performed at the Density Functional Theory (DFT) level. Hence, their effectiveness depends on the quality of the training dataset, which must include not only the equilibrium structures but also the relevant high-energy ones. This is especially crucial for transition state (TS) geometries, whose energies are connected to reaction rates through an exponential relationship. However, the identification of these structures in a complex and dynamic environment remains elusive, especially under operating conditions where an ensemble of TS configurations often exists9,10,11. The quest is made even more challenging by the cost of ab initio calculations, which can rapidly become prohibitive for large systems or when an accurate electronic description is required. Therefore, the ability to efficiently construct reactive potentials with a minimum number of QM calculations is critical to enable their widespread use. In this regard, recent developments in data-efficient architectures can help reduce the number of points needed to train a robust model. These advances include equivariant graph neural networks12 or transfer learning approaches13 leveraging foundational models14,15. However, these techniques still do not solve the crucial issue of identifying the few relevant configurations to be included in the reference dataset set.
The training set construction typically involves active learning procedures16,17,18, in which an ML model is trained on an initial dataset and used to generate new structures (e.g., via molecular dynamics). A subset of these configurations is then labeled with single-point DFT calculations and added to the training set, proceeding iteratively until convergence. In the field of computational catalysis, such schemes have also been employed in combination with nudged elastic band19 and minima hopping20 to accelerate the calculation of energetic barriers and adsorbate geometries. However, we cannot rely on these static approaches to simulate the dynamics under operating conditions (e.g., T = 600–900 K) since the mechanism and the relevant environments may deviate significantly from the calculations performed at T = 0 K. In fact, the effects of dynamics under operating conditions can manifest themselves in many different ways9,21,22, including the dynamic change of the surface23 and the emergence of a broad ensemble of reactive pathways11, which would be impossible to capture by sampling the potential energy landscape with static calculations. Adaptive enhanced sampling techniques, such as metadynamics24, are better suited to sample the reactive landscape at finite temperatures and to collect a diverse set of atomic environments25,26. These methods introduce an external bias potential that increases the fluctuations of selected collective variables (CVs), allowing to sample the transitions between metastable states27. By integrating these sampling techniques with active learning strategies, it has been possible to construct ML potentials for a wide variety of rare events, from phase transitions28,29,30 to chemical reactions31,32,33,34,35,36,37,38,39,40, and catalytic processes11,23,41,42,43. Recently, these techniques have also been used to enhance the model uncertainty to explore high-uncertainty configurations44,45,46,47. Despite the success of enhanced sampling-based active learning, these studies often required many iterations and numerous DFT calculations, leaving room for improvement. Two aspects should be considered to make the process more efficient. At first, when there is little data available on the reactive regions, incremental learning48 approaches should be used in which the potential is frequently updated. Otherwise, by pushing the system out of equilibrium with enhanced sampling, we risk extrapolating poorly and sampling the wrong configurational regions. On the other hand, we also need an efficient criterion for selecting structures for DFT calculations from those obtained in the active learning process.
In this manuscript, we introduce a new scheme to efficiently construct reactive potentials, leveraging both enhanced sampling methods and on-the-fly selection of relevant structures. This is achieved following a two-stage protocol: an exploratory phase to harvest an initial pool of reactive configurations and a second stage in which we obtain a uniformly accurate description of the transition pathways (see Fig. 1 for the diagram of the protocol). For the second step, we developed a new active learning scheme named Data-Efficient Active Learning (DEAL), which allows us to identify a non-redundant set of structures to be added to the training set. As an enhanced sampling method, we used OPES49, a recent evolution of metadynamics that offers greater flexibility and comes with different variants that can be used to explore or converge the free energy landscape. Furthermore, another important ingredient is Gaussian processes (GPs)50, which we used first to learn the potential energy surface on-the-fly and then to identify novel local environments to build a minimal data set within our active learning scheme.
We illustrate these methodological advances by studying several reactions related to ammonia decomposition over iron-cobalt (FeCo) alloy catalysts. This process is the reverse of ammonia synthesis, carried out through the famous Haber-Bosch process, and is crucial to enable ammonia to be used as a hydrogen carrier51. On metal surfaces, cracking of ammonia typically occurs through three steps of dehydrogenation followed by nitrogen desorption via recombination52. In addition, the ferromagnetic and metallic properties of the surface make the description of the electronic structure particularly computationally expensive. Experimentally, an improvement in catalytic performance over pure Fe has been observed53,54, but a microscopic understanding of the mechanism is still lacking. In ref. 53, although experiments were conducted on Fe-Co alloys, the computational analysis was limited to pure Fe and Co. The authors suggested that the higher activity of FeCo with respect to monometallic catalysts could be attributed to a change in the free energy barriers (particularly in a reduction of the hydrogen formation), which would likely resemble the Co ones. However, no modeling of the alloy was performed because of its complexity. In ref. 54, Fe-Co alloys supported on MgO (with FeCo identified as having optimal catalytic activity among FexCo1−x studied) have been investigated, suggesting the metallic FeCo(110) as the active phase based on XPS data. However, their DFT calculations focused solely on nitrogen adsorption energies at T = 0 K without addressing the reaction mechanism or energy barriers. From the modeling perspective, even if this is a new system that has never been studied, it has the same crystal structure of α-Fe (bcc), on which we have extensively investigated both ammonia synthesis23,42 and decomposition11,55. The relevance of the system, coupled with the challenges behind its modeling, makes it an ideal candidate for demonstrating our data-efficient protocol in a real-world scenario.
Results
Stage 0: Preliminary construction of reactant potentials
In our pursuit to model the catalytic process of ammonia decomposition FeCo surfaces, we initiated our investigation by constructing ML potentials for the reactants. This involved gathering configurations to characterize the pristine 110 surface (see Supplementary Fig. 1) and the different intermediate species adsorbed on it (NH3, NH2, NH, N, N2, H,…). To efficiently accomplish this task, we employed GPs to learn on-the-fly the potential energy surface, with the sparse implementation of FLARE56 using the Atomic Cluster Expansion (ACE) descriptors57. Recognizing the limitations of GPs with large training datasets50, we trained separate models for each intermediate species.
Initially, we generated a dataset by conducting a set of uncertainty-aware molecular dynamics (MD) simulations based on GPs at the operando temperature of T = 700 K. Subsequently, we performed simulations at higher temperatures to diversify our configurations and capture surface dynamics. In addition, to obtain an exhaustive coverage of the reactant space, we carried out enhanced sampling simulations to explore the various adsorption sites and the diffusion of the molecules on the surface. This preparatory stage produced about 2500 configurations for all the different intermediates.
Given the complexity of the reaction pathways and the potential existence of multiple channels, we approached the collection of reactive configurations through a two-step process. In the first, the reactive pathways are discovered, while in the second, the description of these pathways is improved until high accuracy is achieved.
Stage 1: Reactive pathways discovery via uncertainty-aware flooding simulations
The initial step in harvesting reactive configurations is the discovery of reaction pathways and transition state structures. While for gas phase reactions, a simple interpolation between reactants and products can provide a reasonable guess, for catalyzed reactions, this is not possible, especially at high temperatures. Indeed, the active site is not known beforehand or may change due to dynamics23 or, again, there may be multiple reaction pathways that need to be sampled11. To address this challenge, we used OPES to perform a set of “flooding-like"58 simulations together with uncertainty-aware MD (see Methods). OPES-flooding introduces an external potential to fill the reactant basin and then let the reactive event occur as spontaneously as possible along the low free-energy pathways. This allowed us to sample reactive processes with minimal knowledge of the reaction mechanism. The only requirement for these methods is the definition of collective variables (CVs) that can distinguish between reactant and product states. We note that, in the case where the products of the reaction to be studied are not known, generic CVs could be used to discover the possible products59. In addition, the integration with uncertainty-aware MD brings two significant advantages: it allows for an efficient selection of reactive configurations based on the uncertainty of the local environments, and it updates the potential energy surface model in an incremental manner, correcting wrong extrapolations to nonphysical regions of phase space.
To illustrate our workflow and demonstrate its effectiveness in sampling reactive pathways, we start with the case of N2 dissociation, a process that we have extensively examined on Fe surfaces23,42. Nitrogen molecules can adsorb onto the (110) surface in two main configurations, either parallel (α) or perpendicular (γ) to the surface (see Fig. 2a). A priori, we do not know from which adsorption site the molecule may break. To find out, we performed a set of iterative OPES-flooding simulations, using the same CVs of ref. 23, namely the N-N distance dN,N and the coordination of N with surface atoms CN,Fe∣Co. As can be seen in Fig. 2b, the bias potential promoted the exploration of new regions of the potential energy surface. Whenever the uncertainty of the local environments (Fig. 2c) exceeded a predefined threshold (i.e., it was structurally different from the environments in the training set), a new DFT calculation was performed, and the GP model was updated. The reactive pathway of N2 dissociation can be effectively visualized in the plane defined by the two CVs (thick orange curve in Fig. 2d), illustrating the progression from the gas phase through the adsorption sites γ and α and finally to the transition state region. Whenever the system reached the product state (vertical dashed line), the simulation was stopped, and a new one started. In this way, an ensemble of reactive pathways was iteratively collected (Fig. 2d), ensuring continuous refinement of the potential energy surface and inclusion of a diverse set of reactive environments. Indeed, each pathway exhibited geometric diversity covering a wide range of coordination with surface atoms while originating from the same horizontal adsorption site α.
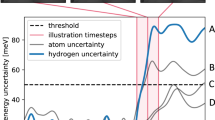
We report a schematic presentation for the case of N2 dissociation. a The reactant free energy landscape constructed at the end of the preliminary phase, highlighting the metastable minima (gas phase and two adsorption states). Arrows indicate that the reaction paths toward the product state are unknown a priori. Several iterations are then initiated from the reactant basin. The time evolution of the distance between N atoms dN,N (b) and the GP maximum uncertainty on the local environments (c) are shown for a specific iteration. Every time the uncertainty of the GP exceeds the threshold of 0.1 (red dotted line), energy and forces are recomputed at the DFT level (red dots), and the GP is updated. Panel d shows all iterations in the 2D space defined by dN,N and the coordination number between N and surface atoms CN,Fe∣Co, with the trajectory shown in b, c highlighted. Above the top axis, we report the distribution of points where DFT calculations were performed, projected along dN,N. Snapshots and graphs were made with Ovito86 and Matplotlib87, respectively.
The need for a flexible and adaptive approach is even more important for modeling the three dehydrogenation steps from ammonia to atomic nitrogen. To harvest such reactive structures, we followed the same protocol, using uncertainty-aware flooding simulations starting from the GP potentials optimized for each reactant. In previous work on the Fe surfaces11, despite being three steps of the same chemical process (dehydrogenation), the different interactions of the adsorbates with surface atoms had necessitated the use of different CVs to converge the free energy calculations. Since we are in an exploratory phase here, we wanted to use instead a single generic CV (i.e., the planar distance between N and H) for all reaction steps to demonstrate our approach’s ability to find the accessible pathways with minimal knowledge. The resulting trajectories are visualized in the plane defined by nitrogen-hydrogen and nitrogen-cobalt coordination (Fig. 3), whose distribution revealed a broad spectrum indicative of the existence of at least two distinct reaction channels. To put our results into context, we superimposed the reactive pathways on the free energy surface derived later from our study (analogous to Fig. 6). The remarkable alignment between the sampled and minimum free energy pathways confirmed the effectiveness of our methodology in accurately sampling the crucial reactive configurations without prior assumptions on the adsorption sites or the transition states. Although each dehydrogenation step occurred from different adsorption sites characterized by increasing coordination with the surface, thanks to the flooding scheme, we were able to find reactive pathways using a generic CV that only distinguished reactants from products. Furthermore, an analysis of the evolution of the sampled paths as a function of iterations demonstrated the importance of an incremental approach: the transition pathways initially pass through high free-energy regions and then converge to low free-energy regions as new data are added (Supplementary Fig. 2). In total, about 400 reactive configurations were collected in this exploratory phase, demonstrating that our approach enables systematic and efficient sampling of reaction pathways under operando conditions, even when multiple ones are available.
The trajectories are visualized in the space defined by CN,H and CN,Co. To facilitate the understanding of the sampled paths, we have shown in the background the free energy surface obtained from the final simulations, which are shown up to ~ 2 kBT above the barriers, highlighting the minimum free energy paths for each reaction (see Fig. 6 for the full colored version). The dots represent the structures recalculated with DFT, whose distributions along CN,H are projected above the upper axis.
Stage 2: Uniform accuracy along reactive paths through GNNs and Data-Efficient Active Learning (DEAL)
Having collected the first dataset of reactive structures, we moved on to the second phase of our approach, aiming to converge the accuracy along the reactive paths. For this purpose, we used equivariant graph neural networks (GNNs)12 to represent the potential energy surface. These architectures are more flexible than GPs60 and have demonstrated remarkable data efficiency, enabling robust simulations as early as hundreds/thousands of training points61. In particular, we used MACE62, which integrates the descriptors used in our previous approach (i.e., ACE) in a message-passing scheme. Resorting to neural networks allowed us to consolidate all previously collected data into a single potential, overcoming the limitations imposed by having separate GPs for each intermediate. In addition, this allowed us to conduct long (~ ns) enhanced sampling simulations instead of the many short (~ ps) simulations of the exploratory phase. This way, we sampled hundreds of different reactive events, providing a more thorough sampling of reaction pathways under dynamic conditions.
To enrich the training dataset further and improve the accuracy of our model, we turned our attention to selecting additional structures from the new MD trajectories. As before, we illustrate our method while focusing on the nitrogen recombination and dissociation process. Our first step is to examine the uncertainty of the MACE model, evaluated using the maximum standard deviation of a committee of models (see Fig. 4a). The distribution of the maximum uncertainty per configuration peaks around 80 meV Å−1. We note that this value is high due to the large magnitudes of the N forces involved in the reaction: 90 percent of these forces have a relative uncertainty of less than 20 percent, see Supplementary Fig. 4. To assess the quality of the potential in describing the reactive event, it’s revealing to analyze the average uncertainty as a function of the collective variable, which describes the progress of the reaction (Supplementary Fig. 3). Indeed, this quantity showed a strongly nonuniform behavior, with significantly higher uncertainty in the region between the reactant and product states (i.e., 1.5 < dN,N < 2.2). Furthermore, by comparing the mean uncertainty with the distribution of the selected configurations (Fig. 4b) we realized that the query-by-committee criterion would lead to an imbalance between the region of the space which is most sampled (2.2 < dN,N < 2.7), but whose configurations are only slightly above the threshold, and the one that really needs to be improved. In previous works, it was necessary to manually select multiple configurations with high uncertainty41 or from a specific region in CV space32 to accurately model reactive events.
We illustrate the GP-based active learning of the MACE structures for the N recombination and dissociation process. a Distribution of the maximum uncertainty on the forces \({\sigma }_{max}^{(MACE)}\) (calculated from a committee model) on the simulation performed with MACE before the active learning. The gray dotted line at 90 meV Å−1, represents the chosen threshold for the query-by-committee selection. In the inset (a1), we report the distribution of the same configurations along the collective variable dN,N. b Distribution of the DFT single point performed with DEAL (red bars), among the ones pre-selected via query-by-committee (blue bars). The number of configurations in each bin is reported on top of the bars. c Distribution of the maximum uncertainty on the forces \({\sigma }_{max}^{(MACE)}\) for initial model (blue histogram) and after DEAL (red histogram). The second distribution is obtained from a new MD with identical parameters performed with the MACE model after DEAL. In the inset (c1), we report the average uncertainty along the reaction coordinate dN,N for the two simulations generated respectively with the initial MACE model (blu line) and after DEAL (red line).
Our objective is to avoid these issues and systematically achieve uniform accuracy along the entire reaction path with a minimal number of DFT calculations. To this end, we propose an active learning procedure that uses the uncertainty of local environments to select a minimal set of representative configurations. In the following, we refer to this scheme as Data-Efficient Active Learning (DEAL). DEAL consists of two parts: first, a pre-selection of structures with high uncertainty is made using the query-by-committee criterion (see Fig. 4a). Next, a representative subset of non-redundant configurations is extracted using Gaussian processes. Specifically, we use the GP models trained in the previous stage to decide which configurations to recalculate at the DFT level by measuring the uncertainty of the local environments of the pre-selected structures. In this way, the uncertainty estimate is updated on-the-fly after each single-point calculation, allowing us to filter out all the redundant configurations. In the case of the N2⇌2N reaction, this approach allowed us to perform only ~ 100 DFT calculations, which corresponds to about 5% of those selected using the query-by-committee criterion (Fig. 4b). Moreover, this percentage varies significantly as a function of CV, ranging from 2 to 3% in the most sampled region to 12−25% in the TS region, demonstrating how our algorithm can automatically target the relevant configurations without requiring any input on the reaction coordinate.
The addition of the configurations selected by DEAL to the MACE model effectively lowered the uncertainty of the configurations generated in the MD simulations (Fig. 4c). In fact, already after only one cycle of active learning, the uncertainty drops below the chosen threshold. Otherwise, we would have performed another cycle of active learning with the same protocol. Similar results were obtained for all the other three dehydrogenation steps; see Supplementary Fig. 6.
Finally, before moving to the validation of the potential, we note that in this scheme, the DFT calculations are performed serially, unlike in the query-by-committee selection. It is worth mentioning that our goal is to extract a representative set of configurations from those flagged with high uncertainty. We found that this can be accomplished, even in the lack of a pre-trained model, by optimizing a GP model from scratch, using the energy and forces predicted by the ML potential as labels (see Methods). This is possible because the uncertainty provided by the GP depends on the spatial relationship between the inputs (i.e., the similarity between local environments as measured by the kernel function) and not on the labels used56. In this way, once the selection of structures has been performed with DEAL, the DFT calculations can be performed in an embarrassingly parallel way, making our approach even more practical. We validated this approach by comparing the distribution of the selected structures with that obtained previously with the pre-trained GP models (Supplementary Fig. 7). In addition, we trained a MACE potential using the newly collected structures and verified that the description along the entire reaction pathway was equally accurate (Supplementary Fig. 8).
Validation of the workflow
In this section, we assess the quality of the final potential, as well as investigate its evolution through the different stages. The root mean square error (RMSE) of the final MACE model on the validation dataset was 0.4 meV/atom for energies and 17 meV Å−1 for forces (corresponding to a relative error of less than 3 percent). Supplementary Fig. 9 reports the accuracy of force predictions divided per chemical species, from which we see that the adsorbate atoms (N, H) have the largest error, also due to the higher magnitude of the forces.
To confirm the accuracy and reliability of our potential in describing reactive processes and to better understand the importance of each stage, we conducted an extensive post-hoc analysis. To this end, we trained three different MACE models using the configurations collected at the end of each of the three stages (stage 2 thus represents the final potential). We constructed an additional test set composed of configurations extracted from the production simulations and recalculated at the DFT level. Specifically, we selected 200 structures for the N2⇌2N reaction and 150 for each dehydrogenation step, uniformly distributed along the CVs. We then evaluated how the accuracy of the predictions of the energies and forces (of the adsorbates) changes when expanding the training set as the workflow progresses. This analysis shows that our procedure is able to achieve uniform accuracy along the reaction paths for both energies (Supplementary Fig. 10) and forces (Fig. 5 and Supplementary Fig. 11). It also clarifies the relevance of each phase. In fact, focusing on the force predictions (Fig. 5, top panel), we can observe that, at the end of the preliminary step, a significant fraction of the configurations have errors greater than 0.5 eV Å−1. Therefore, using directly the MACE potential to perform reactions by skipping the exploratory phase could be dangerous, as we risk extrapolating to unphysical configurations.
(top) Force accuracy for the N forces along the CV (N-N distance), with the violin plot denoting the distribution within each bin and the solid lines representing the average along the CV. Each line/violin plot corresponds to the accuracy of the MACE model optimized using as a training set the configurations collected until a given stage (bottom). Number of configurations in the reactive range collected at each stage. For each stage, it is specified the type of simulation, while the number and length of the simulations are reported in the Methods section. The results for the other dehydrogenation steps are reported in Supplementary Fig. 11.
We also observed how the relationship between the uncertainty \({\sigma }_{max}^{(MACE)}\) and the error on the forces changes in the different stages (Supplementary Fig. 12). Interestingly, in the initial stage, the committee-based uncertainty strongly underestimates the error in the transition region, while this gap decreases as more points are added. Additionally, we also investigated the possibility of using a standard feed-forward neural network potential such as DeePMD63,64 instead of MACE, using the same parameters of ref. 11. However, Supplementary Fig. 13 shows that, after the exploratory phase, the DeepMD errors are still large up to more than 1 eV Å−1, which leads the simulations to be unstable. This implies that DeepMD requires larger training datasets to obtain stable simulations, in agreement with what was observed in ref. 61, and confirms the importance of using data-efficient architectures in our scheme. At the same time, this analysis shows that using the dataset of configurations collected with our workflow with other architectures still yields uniform (though higher) accuracy along the entire reactive path, further confirming the quality of the data selection scheme.
Finally, we conclude this section with a comment on the number and distribution of the structures used in the training set. The number of configurations collected in each stage is reported in Supplementary Table 1, while the distributions along the CVs at different stages are reported in the bottom panels of Fig. 5 and Supplementary Fig. 11. It is worth noting that these distributions are complementary and collectively result in uniform coverage of the reactive path. Remarkably, this uniform distribution was achieved without using the CVs in the selection process, but simply by providing our active learning schemes with reactive trajectories. If we focus on nitrogen dissociation/recombination, our approach produces a robust potential with less than 1k configurations (958). Considering the entire catalytic cycle, which consists of 6 different intermediates, a total of 5k DFT calculations were used. Compared to the number of configurations used in previous studies11,23 on Fe surfaces (30k for N2 dissociation23 and 110k for ammonia decomposition11), we obtained a more than 20-fold improvement in data efficiency. This was possible not only because of data-efficient architectures but, more importantly, due to an efficient protocol for sampling reactive configurations and selecting the most relevant ones.
Unraveling the free energy landscape and dehydrogenation mechanism
Thanks to the potential we have constructed, we can study the mechanism of ammonia decomposition on FeCo alloys in detail. As a representative example, we focus on the dehydrogenation steps of ammonia to atomic nitrogen, showing how the same set of simulations used to construct the potential also allowed us to reconstruct the free energy profiles and unravel the reaction mechanism.
We first performed a set of 25 ns-long OPES simulations, which allowed us to calculate the free energy profiles of the three dehydrogenation steps (as depicted in Fig. 6). The resulting free energy barriers exhibited varying heights, with NH2 → NH+H being the lowest (0.7 eV), followed by NH3 → NH2+H (1 eV), and finally NH → N+H (1.2 eV). The free energies are accurately reconstructed, with a sampling error smaller than 20 meV. Analyzing the 2D free energy surface as a function of the maximum distance between N and H atoms, \(\max \left({d}_{N,H}\right)\), and the coordination number between N and Co atoms, CN,Co, revealed further insights on the reaction mechanism. Multiple pathways are clearly present in all three reaction steps, as already observed in the exploration phase. Notably, these paths exhibit very similar barrier heights but distinct geometric configurations, underscoring the necessity of considering all possible routes. In the case of NH3 dehydrogenation, we observed two predominant paths characterized by the nitrogen-cobalt coordinations being around 0.5 or 1, with a preference for the latter. Similarly, for NH2, the reaction occurred at either coordination 1 or 2, albeit with a slight favor towards the former. Finally, for NH, there was substantial parity between the two paths at coordination values 1 and 2. If we look instead from the perspective of hydrogen, its total coordination with the surface in the TS region is always around 1, but the reaction occurs preferentially when the reactive hydrogen is in contact with cobalt atoms (Supplementary Fig. 14).
The uncertainties on the 1D free energy profiles (shadow region, ~ 0.01 eV) are calculated with a weighted block average49.
To gain a deeper understanding of the reaction paths, we conducted a new set of flooding simulations, yielding an ensemble of 100 reactive trajectories per step (Fig. 7). From the distributions reported on the right of the figure, we can learn a qualitatively different behavior between NH3 and NH2/NH. Notably, NH3 trajectories exhibited a broadened distribution in the coordination between H and Co, CH,Co, reacting uniformly in contact with either iron (CH,Co = 0) and cobalt (CH,Co = 1). Conversely, NH2/NH trajectories displayed a greater flux over higher coordination values, indicating a propensity for dehydrogenation when hydrogen is cobalt-coordinated. See also Supplementary Fig. 15 for a detailed analysis of TS configurations in terms of N and H coordination. This comprehensive analysis underscores the multiplicity of reaction pathways facilitated by high-temperature dynamic conditions. It also demonstrates the effectiveness of our approach, not only in parsimoniously constructing the potential but also in investigating the process under operative conditions with the same simulations, thus elucidating the reaction mechanism and reconstructing the free energy profile of the different catalytic steps. Although the main focus of this manuscript has been on developing methodologies for constructing reactive ML potentials in a data-efficient manner, we can already gain some interesting insights into this process. In particular, our results indicate that the reaction mechanism and free energy barriers on FeCo are similar to those on Fe11, challenging the hypothesis that Co induces a radically different mechanism53. Moreover, our findings are in line with what has been reported in ref. 54, where the authors measured similar apparent activation energies between Fe and FeCo, suggesting that the reason for the better catalytic performance might be related rather to FeCo’s resistance to nitride formation.
An ensemble of 100 trajectories is calculated with the OPES-flooding scheme and visualized in the 2D plane defined by max(dN,H) and CH,Co. Only the 40 fs around the reactive event (determined by max(dN,H) crossing 1.45 Å, denoted by a vertical line) are reported for each trajectory. For convenience, we report in the background the free energy surfaces projected on the same coordinates obtained from the OPES simulations (Supplementary Fig. 14), which are shown up to ~ 2 kBT above the barriers for each reaction. On the right axis, we projected the distribution of the coordination number CH,Co when crossing the line at max(dN,H) = 1.45 Å (black dashes).
Discussion
The development of methods that require few QM calculations is essential to enable widespread use of ML-based potentials. In this manuscript, we introduced a data-efficient scheme for constructing reactive potentials. Applied to the study of ammonia decomposition on iron-cobalt alloy catalysts, our protocol effectively captured complex reaction mechanisms composed of multiple channels without prior knowledge of the adsorption sites or the transition states. Remarkably, the number of needed training points (approximately one thousand DFT calculations for each catalytic step) is at least one order of magnitude less than previous work11,23. These advances can facilitate the simulation of more realistic systems, both in complexity and underlying level of theory. Our approach achieved this result by relying on both data-efficient architectures and a well-designed protocol for sampling and selecting relevant configurations. Because we do not know in advance which configurations are relevant, especially at high temperatures, we divided the protocol into two phases: first exploring reactive pathways and then obtaining uniform accuracy along them.
The exploratory phase is carried out using flooding simulations that push the system out of the reactants, requiring only collective variables that distinguish reactants and products. To avoid extrapolating into non-physical regions, we performed uncertainty-aware MD based on GPs, which are optimized on-the-fly whenever a new local environment is found. This provided an initial set of reactive structures, most of which lie on the minimum free energy pathways. In the convergence step, we used graph neural networks to extensively sample the reactions with reversible enhanced sampling simulations. Since the query-by-committee criterion was found to undersample the transition state region, we proposed a novel active learning strategy (DEAL) based on the local environment uncertainty provided by GPs. This way, we extracted a small set of nonredundant configurations from the configurations with high-uncertainty generated by the GNN. The result is a uniformly accurate potential along the entire reaction path, achieved with a parsimonious number of DFT calculations.
Also, it is important to note that the proposed approach is highly modular. For example, if one has already collected reactive configurations, it is possible to move directly to the second stage of active learning with DEAL. Or, one could leverage foundational models instead of GPs to collect the initial set of reactive configurations. In addition, it will be interesting to test the parallel variant of DEAL as a tool to reduce the size of existing databases. These features make our approach a general and versatile framework for constructing reactive potentials, applicable not only to catalytic processes but also to the broader field of chemical reactions and materials. Indeed, we believe that having a data-efficient way to construct ML potentials is critical to enable the study of increasingly complex systems, both in terms of the environments (such as reactions in the presence of solvent or surface dynamics) and the level of theory of the reference calculations. Finally, this workflow could be further integrated with ML techniques to design collective variables65 in a data-driven away, thereby reducing the need to identify process-specific CVs. Similarly, path sampling methods could be integrated into the active learning scheme to investigate the mechanism in a more unbiased way33. For all these reasons, we believe that our scheme can be effectively used to construct reactive potentials for a wide variety of processes occurring under dynamic conditions, gaining insights into their workings at the atomistic level.
Methods
Machine learning frameworks
Here, we briefly describe the two main architectures that we used in the different stages of our approach to represent the potential energy surface: Gaussian Processes and graph neural networks.
Gaussian Processes (GPs) are kernel-based methods that can be used for regression and probabilistic classification tasks. Starting with the Gaussian Approximation Potential (GAP) method66, GPs have been successfully used for representing the potential energy surface of extended systems. Here, we used the sparse GP implementation of FLARE (Fast Learning of Atomistic Rare Events)56,67, where the energy is modeled as a sum over atomic contributions that depend on all atom-centered local environments of the system: E = ∑iϵi. These environments are characterized by a set of invariant descriptors obtained as the product of equivariant ones56 based on the atomic cluster expansion (ACE)57. What distinguishes GPs from other ML methods is their ability to provide confidence intervals on the predictions. Because of the energy decomposition, we can also measure the variance associated with each atomic contribution ϵi, obtaining a measure of the uncertainty of local environments, i.e., how different they are with respect to the training ones. In particular, per each configuration, we monitor the maximum local uncertainty defined as:
where \({\tilde{\sigma }}_{{\epsilon }_{i}}\) is the variance on the local environment rescaled by the kernel hyperparameter to obtain a dimensionless quantity that ranges between 0 and 1 (see ref. 56 for the details). The uncertainty estimate can be used to guide the active learning process by selecting the new configurations to be labeled with DFT single point calculations. In particular, this can be done on-the-fly by running “uncertainty-aware” MD simulations, in which the GP model is used to perform the next MD step only if the uncertainty is low; otherwise, a DFT calculation is run, and the GP is updated67. The same property can be used to screen an existing dataset of configurations, using the uncertainty estimate to monitor the presence of new local environments against those in the training set. On the other hand, GPs scale unfavorably with the number of points in the training dataset, making them feasible only for small datasets (≲ thousands of atomic configurations).
Compared with kernel-based methods, the parameterization of the potential energy surface by neural networks allows for greater expressiveness60 and eliminates the scaling with the number of training data points. In the case of Graph Neural Networks (GNNs), the description of local environments (the “message") is propagated across neighbors, resulting in a larger effective receptive field than with kernel methods or standard neural networks. In particular, we used MACE62, one of the state-of-the-art architectures, excelling in both in-domain and out-domain predictions and demonstrating remarkable data efficiency68. MACE accomplishes this through a higher-order equivariant message-passing scheme related to ACE, which enables accurate modeling of complex atomic interactions while maintaining efficiency. On the other hand, GNNs do not have a direct estimate of the uncertainty; this can be approximated by the standard deviation of the predictions of a committee of models trained on different partitions of the training dataset. In particular, we focus on the standard deviation of the components α of the forces acting on the i-th atom over a committee of N different models: \({\sigma }_{i,\alpha }=\sqrt{\frac{{\sum }_{k}{({f}_{i,\alpha }-{\bar{f}}_{i,\alpha })}^{2}}{N}}\) and monitor the maximum value per each atomic configuration:
This quantity is typically used in active learning schemes to select new configurations to be recalculated at the DFT level (query-by-committee selection)17.
Data-Efficient Active Learning (DEAL)
DEAL is an active learning scheme that aims to improve the accuracy of an ML potential in a data-efficient way. Given a set of configurations (typically obtained from MD simulations using the potential itself), we want to identify a non-redundant subset to be recomputed at the DFT level and added to the training set of the potential. DEAL is composed of two steps: I) pre-filtering via query-by-committee and II) non-redundant selection of the local environments using the uncertainty provided by GPs.
In the first step, we select a set of configurations that are not correctly described by the ML potential, as measured by the uncertainty computed by a committee of NNs. That is, we select all the configurations whose uncertainty is above a given threshold. We suggest choosing the threshold not based on the absolute value of uncertainty but in such a way as to select the subset of configurations that are described worse, e.g., slightly above the peak of the uncertainty distribution41. In fact, especially in the early stages, the uncertainty could significantly underestimate the error (see Supplementary Fig. 12), just as the absolute value depends greatly on the architecture of the NN (see Supplementary Fig. 13). Also, it is appropriate to set a limit on the maximum uncertainty to avoid including non-physical configurations.
The second step is the identification of a non-redundant subset from those with high uncertainty. To achieve this goal, we train a GP model on the filtered configurations, using the maximum uncertainty on the local environments \({\sigma }_{max}^{(GP)}\). This allows the uncertainty estimate to be updated on-the-fly and, in turn, selects a non-redundant set of configurations to be calculated at the DFT level. In practice, this can be accomplished in two different ways: (a) using the pre-trained GP models on the previously collected configurations and performing a DFT calculation before updating the GP or (b) training a GP model from scratch only on the filtered configurations and using the MD energy/forces as labels (selection only). This is possible because the uncertainty provided by the GP is based on the similarity of the local environments and does not depend on the actual labels used for training. We note that the second strategy has several practical advantages. First, the training time for the GP is greatly reduced because the size of the training set remains small. More importantly, DFT calculations can be performed in parallel after selection. The only downside is the fact that it might select structures that are already in the ML potential’s training set, but the query-by-committee pre-selection ensures that all these structures are not described well by the ML potential. Another element to consider is that, when building a sparse GP model from scratch, more of the first parsed structures will be selected. For this reason, we tested two different schemes: shuffling the trajectory or sorting the configurations by their uncertainty (Supplementary Fig. 7).
Finally, we note that while this scheme was designed to achieve uniform accuracy along reactive paths (by applying it to enhanced sampling simulations), it does not require any knowledge of the reaction coordinate and is thus applicable as a general active learning scheme also to non-reactive simulations.
Enhanced sampling simulations
Even with machine learning potentials, many processes, such as chemical reactions, occur on time scales that are too long to be simulated with standard MD. For this reason, several enhanced sampling methods69 have been developed. In particular, a widely used family is based on the identification of selected collective variables (CVs), which describe the most important degrees of freedom involved in the process. Introducing an external potential as a function of these CVs enhances their fluctuations and accelerates the sampling of the transition. Notable examples are Umbrella Sampling70 and Metadynamics24. In this work we used On-the-fly Probability Enhanced Sampling (OPES)49, a recent evolution of Metadynamics, which converges faster and requires fewer hyperparameters. Given a collective variable s, OPES first reconstructs its probability distribution at equilibrium P(s), using a Kernel Density Estimate (KDE). Then, it introduces a potential to guide the system to sample a given target distribution ptg(s):
A typical choice of the target distribution is the well-tempered one: \({p}^{tg}(s)\propto P{(s)}^{\frac{1}{\gamma }}\), which lowers the free energy barriers by a factor γ, and correspond to the same distribution sampled in Well-Tempered Metadynamics71. Since the target is not known a priori, the probability and, thus, the bias are constructed iteratively during the simulation. Furthermore, a useful feature is the ability to limit the amount of bias deposited. In case one does not know the height of the barrier to be overcome, an exploratory variant is also available72, which can be used to quickly explore the free energy landscape. When the OPES bias is converged to a quasi-static value, the free energy along the biased CV or any other variables can be recovered from a simple Umbrella Sampling-like reweighting scheme49.
In addition, we also employed OPES-flooding, is a variant of the OPES method in which the potential is applied only in the reactant basin. This can be ensured by the barrier hyperparameter that sets the maximum amount of deposited bias. In addition, this can be enforced by restricting the application of the potential within a certain region of the CV space that bounds the reactants58. In this way, free energy paths that are sampled are more likely to be close to the lowest free energy ones. Once the system has overcome the free energy barrier and reached the product basin, the simulation is terminated. By performing a set of flooding simulations, we can also obtain statistics on the reactive pathways and characterize them.
Construction of the reactive ML potential
Here, we describe the protocol used for the collection of the configurations for training the reactive potential, focusing on the active learning policies used in the different stages to select the relevant samples.
In the preliminary step, we collected configurations from the reactants states. We ran uncertainty-aware MD through the on-the-fly code implemented in FLARE73, using LAMMPS74 as MD engine. For each intermediate (NH3, NH2, NH, N/N2, H), we trained a different GP model. These models were used in parallel to perform a series of simulations starting with the adsorbate in different initial conditions to study the interaction of the intermediates with the surface. The criterion for selecting the configurations is based on the maximum uncertainty over the local environments predicted by FLARE. Whenever this exceeds a given threshold, a DFT calculation is performed, and a (sparse) set of local environments is added to the training dataset. In the initial phase, a small threshold of 0.02 was used to avoid extrapolating into unphysical regions. We ran about 10 iterations for each reactant until the simulations proceeded for about 1 ps without DFT calculations. Furthermore, to achieve a more extensive sampling, we performed OPES simulations to sample the different adsorption sites and the diffusion on the surface by enhancing the coordination of nitrogen with the surface and the x,y coordinates of the center of mass, respectively; see below for the definition of the CVs. At the beginning of the preliminary stage, one short AIMD (400 fs), which was used to initialize the GP model, was performed.
For the reactive pathway discovery step (Stage 1), we performed a set of OPES flooding-like simulations with uncertainty-aware MD, biasing collective variables capable of distinguishing reactants from products (e.g., N-N distance for N2 dissociation; see below). For each reaction step (N2 → 2N, NH3 → NH2+H, NH2 → NH+H, NH → N+H), we performed 30 iterations, stopping the simulation each time the molecule reacted and starting again to sample a new reactive event. As a threshold for the maximum local uncertainty, we used a value of 0.1 for the first 20 iterations, then lowered it to 0.075 for the remaining ones in order to continue labeling a few structures from each path.
In Stage 2, we trained a single model with MACE75 using all the configurations collected earlier. With this model, we ran long MD simulations (3 ns) for each reaction step, sampling tens of reactive events within each. We then calculated the MACE uncertainty on the predictions \({\sigma }_{max}^{(MACE)}\) through a committee of 4 models. The MACE uncertainty is used for the first step of DEAL, that is, to select the configurations whose uncertainty was above a given threshold. The threshold (chosen based on the peak of the maximum uncertainty distribution) is equal to 90 meV Å−1 for N2 → 2N and 50 meV Å−1 for NHx → NHx−1+H. Then, we use the GP to screen the high-uncertainty configurations and select a non-redundant set to be recalculated at the DFT level and added to the MACE training set. We used the pre-trained models used in stage 1 and a GP threshold equal to 0.075. In the case of the N2 ↔ 2N reaction, the preliminary potential (stage 0) contained configurations of both reactants and products. In contrast, in the case of the dehydrogenation reactions, the products (NHx + H) had not been sampled. Therefore, we added a set of product configurations selected via the query-by-committee criterion.
Computational details
DFT calculations
All DFT calculations used to build the reference database were performed using the PWscf code of Quantum ESPRESSO76,77,78. Exchange-correlation effects were treated using the generalized-gradient approximation with the Perdew-Burke-Ernzerhof (PBE) functional79. We employed ultrasoft pseudopotentials selected from the SSSP PBE Precision v1.3.0 library80 with 1, 5, 16, and 17 valence electrons for H, N, Fe, and Co, respectively. The wave function and the charge density cutoff were set at 90 and 1080 Ry. The Marzari-Vanderbilt cold smearing technique81 with a Gaussian spreading of 0.04 Ry was used to treat the collinear spin-polarized electronic occupations. Initial spin polarizations were set to 0.6 for Fe∣Co. To ensure the consistency of the dataset, spurious high-energy non-ferromagnetic configurations were removed from the dataset. A vacuum layer of at least 12 Å was included in the z-direction of all the slab models to prevent self-interaction effects. The Brillouin zone was sampled using a Monkhorst-Pack k-point grid82 with a maximum k-spacing of 0.25 Å−1, and the SCF convergence threshold was set to 10−6 Ry.
FLARE
The local environments are described through the ACE B2 descriptors, using a basis expansion with Nmax = 8, and lmax = 3. We used a cutoff equal to 5.5 Å for Fe and Co and 4 Å for the interaction with the other species. The kernel used to compare the environments was a squared normalized dot product.
MACE
All the GNN models in this work were constructed using MACE version 0.3.075. The models employed a cutoff radius of 6 Å for the atomic environments with 256 channels and L = 0, finding a balance between accuracy and computational efficiency. The dataset was split into training/validation subsets with a ratio of 95:5, and the model was optimized with AMSGrad using a learning rate of 0.01 and a batch size of 4. The performance was evaluated on energy and forces with a weighted root mean square error (RMSE) loss function. The weights of the energy and forces were initially set to 1 and 100, respectively. In the last ~25% epochs, the weight of energy has been increased by a factor of 10 three times. The optimization was interrupted via an early stopping criterion with a patience of 200 epochs.
Molecular dynamics
MD simulations were performed using the Large-scale Atomic/Molecular Massively Parallel Simulator (LAMMPS) software74, supplemented by FLARE83, MACE v0.3.075, and PLUMED v2.984. The canonical sampling through velocity rescaling thermostat85 with a coupling constant of 50 fs was employed to control the temperature in all the simulations. During the construction of the preliminary reactants potentials, we simulated FeCo slabs cut along the 110 surface containing 60 (3 × 4 × 5) and 144 (4 × 6 × 6) atoms. A vacuum region of at least 12 Å was added in all configurations to avoid self-interaction effects. The slab was simulated in both bulk-terminated (by fixing the two bottom layers) and double-interface (by fixing a central layer) conditions. In the latter, adsorbate molecules were placed on both sides. The surface dynamics was simulated at 700 K and then 800 K, while the interaction with the different reaction intermediates (NH3, NH2, NH, N/N2, H) was simulated at 700 K. At this stage, atomic interactions were described by FLARE, and the timestep was set to 1 fs. In the reaction discovery stage, as well as during the GNN active learning and the final simulations, we simulated 120 atoms (4 × 6 × 5) bulk-terminated (2-fixed layers) FeCo(110) slabs at T = 700 K. For the final simulations, we repeated the simulations with a larger system (800 atoms) to rule out possible finite-size effects. Periodic boundary conditions were applied in the x and y directions, while a reflecting wall was implemented over the surface in the z-direction.
Enhanced sampling
Here, we first describe the different CVs used and, later, the details of the OPES simulations employed in the different stages. In addition to the distance between different atoms, we focused on the coordination numbers to measure both the interaction between the adsorbate and the substrate atoms and to distinguish the different NHx species. The coordination is defined in a continuous and differentiable way as:
where rij is the distance between the i-th atoms of species X and the j-th atoms of species Y, and r0, d0, n, and m, are tunable parameters. For the coordination between N and Fe, Co, and Fe∣Co, we set the parameters d0 = 1 Å and r0 = 1.5 Å n = 6, m = 12, while for the coordination between H and Fe, Co, and Fe∣Co, we used d0 = 0.8 Å r0 = 1.0 Å. Finally, for the coordination between N and H, we used d0 = 0.7 Å r0 = 0.8 Å n = 5, m = 7.
In the preliminary step, we used OPES to enhance a) the exploration of the different adsorption states by using as CV the coordination between N and Fe/Co atoms and b) the diffusion of the adsorbed molecule by using the x and y components of its center of mass.
In the exploratory stage, we iteratively performed OPES-flooding simulations to harvest an ensemble of reactive pathways. For N2 dissociation, we used the distance between the two N atoms dN,N and the coordination CN,Fe∣Co as CVs, while for the hydrogenation steps, we used the inverse of the planar distance between N and one H atom. The probability estimate was updated every 50 steps, and the the barrier was chosen to be 50 kJ mol−1, in order to apply the bias only in the reactant region. Simulations were interrupted after the occurrence of the reaction, namely when dN,N exceeded 2.5 Å or when the coordination CN,H was decreased by 1 with respect to the equilibrium value.
During the active learning stage with MACE and the final production simulations, we used OPES with the standard well-tempered target distribution. To study nitrogen recombination, we use the distance dN,N as CVs. The barrier parameter was set to 200 kJ mol−1, and the bias was updated every 200 steps. For the first dehydrogenation step (NH3 → NH2+H), we biased the simulation along three CVs: CN−H, CN,Fe∣Co, and the angular variable Θ defined in Ref. 11. For the second reaction (NH2 → NH+H), we used CN,H and CN,Fe∣Co, while for the last step (NH → N+H), we employed the distance dN,H and CN,Fe∣Co. The barrier parameters were set equal to 80, 60, and 100 kJ mol−1, respectively, and the bias was updated every 200 steps. Harmonic restraints were applied in all reactions to facilitate reversible sampling. For the 2N → N2 process, the restraint was applied if dN,N ≤1.2 or dN,N ≥3.2 Å, while for the dehydrogenation steps if dN,H ≤2.5 Å, with an elastic constant of 5000 kJ mol−1 Å−2.
Finally, to study the reactive pathways in detail, we perform a set of 100 OPES flooding simulations for each dehydrogenation step. The CV used for all the steps was the coordination CN,H. To avoid biasing the TS region, we exclude from bias deposition the regions where CN,H is lower than 2.82, 1.85, and 0.9 for the three hydrogenation steps, using an OPES barrier parameter equal to 70, 30, and 80 kJ mol−1, respectively. All simulations were stopped once the reaction occurred, terminating the simulation when the maximum NH distance exceeded 1.9 Å.
Data availability
Input files of the enhanced sampling simulations are available on the PLUMED-NEST repository with plumID:24.035. The outputs of the simulations are available from the corresponding author on reasonable request.
Code availability
The code to reproduce the workflow for the construction of ML potentials is available at https://github.com/luigibonati/DEAL/. LAMMPS (www.lammps.org), Quantum ESPRESSO (www.quantum-espresso.org), PLUMED (www.plumed.org), FLARE (https://github.com/mir-group/flare) and MACE (https://github.com/ACEsuit/mace) are open-source codes.
References
Schlögl, R. Heterogeneous Catalysis. Angew. Chem. Int. Ed. 54, 3465–3520 (2015).
Shi, X. et al. Dynamics of heterogeneous catalytic processes at operando conditions. JACS Au 1, 2100–2120 (2021).
Spencer, M. S. Stable and metastable metal surfaces in heterogeneous catalysis. Nature 323, 685–687 (1986).
Eren, B. et al. Activation of Cu(111) surface by decomposition into nanoclusters driven by CO adsorption. Science 351, 475–478 (2016).
Grajciar, L. et al. Towards operando computational modeling in heterogeneous catalysis. Chem. Soc. Rev. 47, 8307–8348 (2018).
Piccini, G. et al. Ab initio molecular dynamics with enhanced sampling in heterogeneous catalysis. Catal. Sci. Technol. 12, 12–37 (2022).
Van Speybroeck, V., Bocus, M., Cnudde, P. & Vanduyfhuys, L. Operando modeling of zeolite-catalyzed reactions using first-principles molecular dynamics simulations. ACS Catal. 13, 11455–11493 (2023).
Unke, O. T. et al. Machine Learning Force Fields. Chem. Rev. 121, 10142–10186 (2021).
Sun, G. & Sautet, P. Metastable Structures in Cluster Catalysis from First-Principles: Structural Ensemble in Reaction Conditions and Metastability Triggered Reactivity. J. Am. Chem. Soc. 140, 2812–2820 (2018).
Zhang, Z., Zandkarimi, B. & Alexandrova, A. N. Ensembles of Metastable States Govern Heterogeneous Catalysis on Dynamic Interfaces. Acc. Chem. Res. 53, 447–458 (2020).
Perego, S., Bonati, L., Tripathi, S. & Parrinello, M. How Dynamics Changes Ammonia Cracking on Iron Surfaces. ACS Catal. 14, 14652–14664 (2024).
Batatia, I. et al. The Design Space of E(3)-Equivariant Atom-Centered Interatomic Potentials. https://arxiv.org/abs/2205.06643v2 (2022).
Falk, J. I. et al. Transfer learning for atomistic simulations using GNNs and kernel mean embeddings. Adv. Neural Inf. Process. Syst. 36, 29783–29797 (2023).
Chanussot, L. et al. Open Catalyst 2020 (OC20) Dataset and Community Challenges. ACS Catal. 11, 6059–6072 (2021).
Batatia, I. et al. A foundation model for atomistic materials chemistry, https://arxiv.org/abs/2401.00096v2 (2023).
Smith, J. S., Nebgen, B., Lubbers, N., Isayev, O. & Roitberg, A. E. Less is more: Sampling chemical space with active learning. J. Chem. Phys. 148, 241733 (2018).
Schran, C., Brezina, K. & Marsalek, O. Committee neural network potentials control generalization errors and enable active learning. J. Chem. Phys. 153, 104105 (2020).
Ang, S. J., Wang, W., Schwalbe-Koda, D., Axelrod, S. & Gómez-Bombarelli, R. Active learning accelerates ab initio molecular dynamics on reactive energy surfaces. Chem 7, 738–751 (2021).
Schaaf, L. L., Fako, E., De, S., Schäfer, A. & Csányi, G. Accurate energy barriers for catalytic reaction pathways: an automatic training protocol for machine learning force fields. npj Computational Mater. 9, 1–10 (2023).
Jung, H., Sauerland, L., Stocker, S., Reuter, K. & Margraf, J. T. Machine-learning driven global optimization of surface adsorbate geometries. npj Computational Mater. 9, 1–8 (2023).
Wang, Y. G., Mei, D., Glezakou, V. A., Li, J. & Rousseau, R. Dynamic formation of single-atom catalytic active sites on ceria-supported gold nanoparticles. Nat. Commun. 6, 1–8 (2015).
Li, W. L. et al. Critical Role of Thermal Fluctuations for CO Binding on Electrocatalytic Metal Surfaces. JACS Au 1, 1708–1718 (2021).
Bonati, L. et al. The role of dynamics in heterogeneous catalysis: Surface diffusivity and N2 decomposition on Fe(111). Proc. Natl Acad. Sci. 120, e2313023120 (2023).
Laio, A. & Parrinello, M. Escaping free-energy minima. Proc. Natl Acad. Sci. 99, 12562–12566 (2002).
Yoo, D., Jung, J., Jeong, W. & Han, S. Metadynamics sampling in atomic environment space for collecting training data for machine learning potentials. npj Computational Mater. 7, 131 (2021).
Xu, J., Cao, X.-M. & Hu, P. Accelerating metadynamics-based free-energy calculations with adaptive machine learning potentials. J. Chem. theory Comput. 17, 4465–4476 (2021).
Valsson, O., Tiwary, P. & Parrinello, M. Enhancing Important Fluctuations: Rare Events and Metadynamics from a Conceptual Viewpoint. Annu. Rev. Phys. Chem. 67, 159–184 (2016).
Bonati, L. & Parrinello, M. Silicon Liquid Structure and Crystal Nucleation from Ab Initio Deep Metadynamics. Phys. Rev. Lett. 121, 265701 (2018).
Niu, H., Bonati, L., Piaggi, P. M. & Parrinello, M. Ab initio phase diagram and nucleation of gallium. Nat. Commun. 11, 1–9 (2020).
Abou El Kheir, O., Bonati, L., Parrinello, M. & Bernasconi, M. Unraveling the crystallization kinetics of the ge2sb2te5 phase change compound with a machine-learned interatomic potential. npj Computational Mater. 10, 33 (2024).
Galib, M. & Limmer, D. T. Reactive uptake of n2o5 by atmospheric aerosol is dominated by interfacial processes. Science 371, 921–925 (2021).
Yang, M., Bonati, L., Polino, D. & Parrinello, M. Using metadynamics to build neural network potentials for reactive events: the case of urea decomposition in water. Catal. Today 387, 143–149 (2022).
Zhang, J., Zhang, O., Bonati, L. & Hou, T. Combining transition path sampling with data-driven collective variables through a reactivity-biased shooting algorithm. J. Chem. Theory Comput. 20, 4523–4532 (2024).
Guan, X., Heindel, J. P., Ko, T., Yang, C. & Head-Gordon, T. Using machine learning to go beyond potential energy surface benchmarking for chemical reactivity. Nat. Computational Sci. 3, 965–974 (2023).
David, R. et al. ArcaNN: automated enhanced sampling generation of training sets for chemically reactive machine learning interatomic potentials. Digit. Discov. https://doi.org/10.1039/D4DD00209A (2025).
Zhang, H., Juraskova, V. & Duarte, F. Modelling chemical processes in explicit solvents with machine learning potentials. Nat. Commun. 15, 1–11 (2024).
Young, T. A., Johnston-Wood, T., Zhang, H. & Duarte, F. Reaction dynamics of Diels-Alder reactions from machine learned potentials. Phys. Chem. Chem. Phys. 24, 20820–20827 (2022).
Zeng, Z. et al. Mechanistic insight on water dissociation on pristine low-index TiO2 surfaces from machine learning molecular dynamics simulations. Nat. Commun. 14, 1–9 (2023).
David, R., Tuñón, I. & Laage, D. Competing Reaction Mechanisms of Peptide Bond Formation in Water Revealed by Deep Potential Molecular Dynamics and Path Sampling. J. Am. Chem. Soc. 146, 14213–14224 (2024).
Benayad, Z., David, R. & Stirnemann, G. Prebiotic chemical reactivity in solution with quantum accuracy and microsecond sampling using neural network potentials. Proc. Natl Acad. Sci. USA 121, e2322040121 (2024).
Yang, M., Raucci, U. & Parrinello, M. Reactant-induced dynamics of lithium imide surfaces during the ammonia decomposition process. Nat. Catal. 2023 6:9 6, 829–836 (2023).
Tripathi, S., Bonati, L., Perego, S. & Parrinello, M. How Poisoning Is Avoided in a Step of Relevance to the Haber-Bosch Catalysis. ACS Catalysis 4944–4950, https://pubs.acs.org/doi/abs/10.1021/acscatal.3c06201 (2024).
Mambretti, F., Raucci, U., Yang, M. & Parrinello, M. How Does Structural Disorder Impact Heterogeneous Catalysts? The Case of Ammonia Decomposition on Non-stoichiometric Lithium Imide. ACS Catal. 16, 1252–1256 (2024).
Kulichenko, M. et al. Uncertainty-driven dynamics for active learning of interatomic potentials. Nat. Computational Sci. 3, 230–239 (2023).
van der Oord, C., Sachs, M., Kovács, D. P., Ortner, C. & Csányi, G. Hyperactive learning for data-driven interatomic potentials. npj Computational Mater. 9, 1–14 (2023).
Tan, A. R., Dietschreit, J. C. B. & Gomez-Bombarelli, R. Enhanced sampling of robust molecular datasets with uncertainty-based collective variables. http://arxiv.org/abs/2402.03753 (2024).
Zaverkin, V. et al. Uncertainty-biased molecular dynamics for learning uniformly accurate interatomic potentials. npj Computational Mater. 10, 83 (2024).
Vandenhaute, S., Cools-Ceuppens, M., DeKeyser, S., Verstraelen, T. & Van Speybroeck, V. Machine learning potentials for metal-organic frameworks using an incremental learning approach. npj Computational Mater. 9, 1–8 (2023).
Invernizzi, M. & Parrinello, M. Rethinking Metadynamics: From Bias Potentials to Probability Distributions. J. Phys. Chem. Lett. 11, 2731–2736 (2020).
Rasmussen, C. E. Gaussian processes in machine learning. In Summer school on machine learning, 63–71 (Springer, 2003).
Lucentini, I., Garcia, X., Vendrell, X. & Llorca, J. Review of the Decomposition of Ammonia to Generate Hydrogen. Ind. Eng. Chem. Res. 60, 18560–18611 (2021).
Bell, T. E. & Torrente-Murciano, L. H2 Production via Ammonia Decomposition Using Non-Noble Metal Catalysts: A Review. Top. Catal. 59, 1438–1457 (2016).
Wang, Y. et al. Understanding Reaction Networks through Controlled Approach to Equilibrium Experiments Using Transient Methods. J. Am. Chem. Soc. 143, 10998–11006 (2021).
Chen, S. et al. Highly loaded bimetallic iron-cobalt catalysts for hydrogen release from ammonia. Nat. Commun. 15, 1–11 (2024).
Purcel, M. et al. Iron Nitride Formation and Decomposition during Ammonia Decomposition over a Wustite-Based Bulk Iron Catalyst. ACS Catalysis 13947–13957, https://pubs.acs.org/doi/full/10.1021/acscatal.4c04415 (2024).
Vandermause, J., Xie, Y., Lim, J. S., Owen, C. J. & Kozinsky, B. Active learning of reactive bayesian force fields applied to heterogeneous catalysis dynamics of h/pt. Nat. Commun. 13, 5183 (2022).
Drautz, R. Atomic cluster expansion for accurate and transferable interatomic potentials. Phys. Rev. B 99, 014104 (2019).
Ray, D., Ansari, N., Rizzi, V., Invernizzi, M. & Parrinello, M. Rare Event Kinetics from Adaptive Bias Enhanced Sampling. J. Chem. Theory Comput. 18, 6500–6509 (2022).
Raucci, U., Rizzi, V. & Parrinello, M. Discover, Sample, and Refine: Exploring Chemistry with Enhanced Sampling Techniques. J. Phys. Chem. Lett. 13, 1424–1430 (2022).
Owen, C. J. et al. Complexity of many-body interactions in transition metals via machine-learned force fields from the tm23 data set. npj Computational Mater. 10, 92 (2024).
Fu, X. et. al. Forces are not enough: Benchmark and critical evaluation for machine learning force fields with molecular simulations. Trans. Mach. Learn. Res. (2023).
Batatia, I., Kovacs, D. P., Simm, G., Ortner, C. & Csányi, G. Mace: Higher order equivariant message passing neural networks for fast and accurate force fields. Adv. Neural Inf. Process. Syst. 35, 11423–11436 (2022).
Zhang, L. et al. End-to-end Symmetry Preserving Inter-atomic Potential Energy Model for Finite and Extended Systems. Adv. Neural Inf. Processing Syst., 31, http://www.deepmd.org/database/deeppot-se-data/ (2018).
Zeng, J. et al. DeePMD-kit v2: A software package for deep potential models. J. Chem. Phys. 159, 54801 (2023).
Bonati, L., Trizio, E., Rizzi, A. & Parrinello, M. A unified framework for machine learning collective variables for enhanced sampling simulations: mlcolvar. J. Chem. Phys. 159, 014801 (2023).
Bartók, A. P., Payne, M. C., Kondor, R. & Csányi, G. Gaussian approximation potentials: The accuracy of quantum mechanics, without the electrons. Phys. Rev. Lett. 104, 136403 (2010).
Vandermause, J. et al. On-the-fly active learning of interpretable bayesian force fields for atomistic rare events. npj Computational Mater. 6, 20 (2020).
Kovacs, D. P., Batatia, I., Arany, E. S. & Csanyi, G. Evaluation of the MACE Force Field Architecture: from Medicinal Chemistry to Materials Science. J. Chem. Phys. 159, 44118 (2023).
Hénin, J., Lelièvre, T., Shirts, M. R., Valsson, O. & Delemotte, L. Enhanced Sampling Methods for Molecular Dynamics Simulations [Article v1.0]. Living J. Computational Mol. Sci. 4, 1583–1583 (2022).
Torrie, G. M. & Valleau, J. P. Nonphysical sampling distributions in monte carlo free-energy estimation: Umbrella sampling. J. Computational Phys. 23, 187–199 (1977).
Barducci, A., Bussi, G. & Parrinello, M. Well-tempered metadynamics: a smoothly converging and tunable free-energy method. Phys. Rev. Lett. 100, 020603 (2008).
Invernizzi, M. & Parrinello, M. Exploration vs Convergence Speed in Adaptive-Bias Enhanced Sampling. J. Chem. Theory Comput. 18, 3988–3996 (2022).
Vandermause, J. et al. On-the-fly active learning of interpretable Bayesian force fields for atomistic rare events. npj Computational Mater. 2020 6:1 6, 1–11 (2020).
Thompson, A. P. et al. LAMMPS - a flexible simulation tool for particle-based materials modeling at the atomic, meso, and continuum scales. Computer Phys. Commun. 271, 108171 (2022).
Batatia, I., Kovács, D. P., Simm, G. N. C., Ortner, C. & Csányi, G. MACE: Higher Order Equivariant Message Passing Neural Networks for Fast and Accurate Force Fields. Adv. Neural Inf. Process. Syst. 35, 11423–11436 (2022).
Giannozzi, P. et al. QUANTUM ESPRESSO: a modular and open-source software project for quantum simulations of materials. J. Phys.: Condens. Matter 21, 395502 (2009).
Giannozzi, P. et al. Advanced capabilities for materials modelling with Quantum ESPRESSO. J. Phys.: Condens. Matter 29, 465901 (2017).
Giannozzi, P. et al. Quantum ESPRESSO toward the exascale. J. Chem. Phys. 152, 154105 (2020).
Perdew, J. P., Burke, K. & Ernzerhof, M. Generalized Gradient Approximation Made Simple. Phys. Rev. Lett. 77, 3865 (1996).
Prandini, G., Marrazzo, A., Castelli, I. E., Mounet, N. & Marzari, N. Precision and efficiency in solid-state pseudopotential calculations. npj Computational Mater. 4, 1–13 (2018).
Marzari, N., Vanderbilt, D., De Vita, A. & Payne, M. C. Thermal Contraction and Disordering of the Al(110) Surface. Phys. Rev. Lett. 82, 3296 (1999).
Monkhorst, H. J. & Pack, J. D. Special points for Brillouin-zone integrations. Phys. Rev. B 13, 5188 (1976).
Xie, Y., Vandermause, J., Sun, L., Cepellotti, A. & Kozinsky, B. Bayesian force fields from active learning for simulation of inter-dimensional transformation of stanene. npj Computational Mater. 7, 1–10 (2021).
Tribello, G. A., Bonomi, M., Branduardi, D., Camilloni, C. & Bussi, G. PLUMED 2: New feathers for an old bird. Comput. Phys. Commun. 185, 604–613 (2014).
Bussi, G., Donadio, D. & Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 126, 14101 (2007).
Stukowski, A. Visualization and analysis of atomistic simulation data with OVITO-the Open Visualization Tool. MODEL. Simul. Mater. Sci. Eng. 18, 015012 (2010).
Hunter, J. D. Matplotlib: A 2d graphics environment. Comput. Sci. Eng. 9, 90–95 (2007).
Acknowledgements
We are grateful to Enrico Trizio, Umberto Raucci, Andrea Rizzi, and Michele Parrinello, for stimulating discussions and providing feedback on the manuscript. We thank the anonymous reviewers for their constructive feedback on the manuscript. We acknowledge support from the Data Science and Computation Facility and its Support Team at Fondazione Istituto Italiano di Tecnologia, the CINECA award under the ISCRA initiative (IscraB28_AmmoFeCo) and the Max Planck Computing and Data Facility; and the Federal Ministry of Education and Research, Germany (Bundesministerium für Bildung und Forschung, BMBF, Hydrogen flagship project: TransHyDE Forschungsverbund AmmoRef, FKZ 03HY203A) for funding.
Author information
Authors and Affiliations
Contributions
S.P. and L.B. performed the calculations, discussed the results, and wrote the manuscript; L.B. supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Perego, S., Bonati, L. Data efficient machine learning potentials for modeling catalytic reactivity via active learning and enhanced sampling. npj Comput Mater 10, 291 (2024). https://doi.org/10.1038/s41524-024-01481-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41524-024-01481-6
This article is cited by
-
Machine Learning-Enhanced Molecular Dynamics: Current State, Challenges and Perspectives
Archives of Computational Methods in Engineering (2026)
-
Fast and Fourier features for transfer learning of interatomic potentials
npj Computational Materials (2025)
-
Accelerating structure relaxation in chemically disordered materials with a chemistry-driven model
npj Computational Materials (2025)
-
Machine learning interatomic potentials in biomolecular modeling: principles, architectures, and applications
Biophysical Reviews (2025)
-
Managing the complexity of emerging contaminants in aquatic environments: exploring their ecotoxicological impacts, detection techniques, and the use of innovative technologies for their remediation
Discover Catalysis (2025)