Abstract
Studying neuronal cells in space reveals how microgravity affects brain function, gene expression, and cellular processes. This study details the preparation and validation of a 3D neuronal electrophysiology (EPHYS) sensing microfluidic biodevice used during a suborbital space flight. Initially, the device’s function was tested with rat hippocampal neurons using EPHYS data collected via a microelectrode array (MEA). This system was later applied to human glutamatergic (Glu) neurons for eight days preceding a suborbital flight. A live-dead assay confirmed cell viability, and the system was integrated into a CubeLab to maintain a controlled environment. Two biological samples were flown, along with two control samples, to validate the EPHYS system. Results showed that human Glu-neurons exposed to microgravity exhibited altered expression of vesicular glutamate transporters (VGLUTs) while maintaining neuronal differentiation markers. The findings contribute to understanding neurological disorders, neuro-inflammation, and cognitive impacts of space travel, with broader applications for brain health research on Earth.
Similar content being viewed by others
Introduction
Long-duration space missions give rise to various risk factors that significantly impact astronaut brain structure and function, with potential long-term effects still being studied1. These effects encompass changes in brain morphology and cognitive abilities, including attention and decision-making1,2. Acute exposure to microgravity is known to affect spatial orientation and sensorimotor coordination3; whereas chronic exposure is believed to damage sensitive neuronal structures and induce epigenetic changes in their DNA, leading to alteration of cognition and behavioral functions. Cosmic radiation, on the other hand, may lead to amyloid-β accumulation, neuro-inflammation, and cognitive alterations4. Evidence suggests that spaceflight accelerates brain aging, paralleling effects observed in cardiovascular and musculoskeletal systems4, which is the International Space Station Research Laboratory's (ISS-NLs) primary focus in developing effective countermeasures. However, there have not been many studies conducted on the role of microgravity in age-related neurodegenerative diseases, such as Alzheimer’s disease (AD) or Parkinson’s disease (PD). Yet, it is recognized empirically that dementia in elderly patients may accelerate after becoming bedridden. If skeletal unloading in bedridden patients is partially analogous to the effect of microgravity in space, then there exists a strong possibility that neurodegenerative diseases may be accelerated in space. In addition, microgravity-induced neurodegeneration may be amplified by cosmic rays and other factors. Based on such a premise, in a recent suborbital flight study, we assessed the electrophysiological (EPHYS) behavior of human central nervous system (CNS) neurons when encapsulated within a microfluidic biodevice. It is broadly agreed that cytoskeletal rearrangements can occur in cells during a flight-based study, which can uniquely affect neuronal function and EPHYS5. Thus, studying how neuronal cells behave upon exposure to microgravity (in a low Earth orbit (LEO)) is a vital first step in understanding the effects of microgravity on brain function.
This research involves developing and validating a 3D neuronal EPHYS sensing microfluidic biodevice during a recent suborbital spaceflight. Initially, rat hippocampal neurons were used to characterize the device’s function, collecting EPHYS data via a microelectrode array (MEA) probe interfacing with the neuronal cell networks. The system was then applied to human glutamatergic (Glu) neurons, with EPHYS data collected over eight days leading up to the spaceflight launch. To maintain the viability of the human neuronal cells during the suborbital flight, the system was integrated into a CubeLab, which provided a closed environment maintained at 37 °C and facilitated automated media exchanges. Designed specifically for this project, the CubeLab housed and stabilized four microfluidic devices, supplemental medium and waste storage, heaters to maintain active temperature control of each sample chamber, OneBox data acquisition systems to enable for EPHYS monitoring, and a LattePanda, which allowed for automation of each of the components without compromising the sterility of the CubeLab upon closure. The CubeLab was purged with 5% CO2 and 20% O2 following the integration of microfluidic devices and was then sealed to maintain this environment passively. The fluid flow was fixed at a constant speed of 0.4 mL/min via the pumps attached to the device. A schematic showing the layout of components within the CubeLab is shown in Supplementary Fig. 1. Medium exchanges were performed immediately before locker handover to Blue Origin, and upon payload return to ground after launch. Due to spatial limitations within the flight locker and CubeLab, only four microfluidic devices could be sustained. For this reason, two biological samples were selected for the flight based on healthy cell morphology and EPHYS activity. Two control samples containing only Matrigel were also included to validate the EPHYS recording system, thereby allowing for the verification of microfluidic device function and assessment of baseline interference levels captured by the MEA due to external electric fields and flight vehicle vibrations. Matrigel was selected as the primary substrate for 3D cell culture based on its ability to promote neuronal network formation and spontaneous electrical activity of iCell glutamatergic neurons6, thus providing a framework for sustaining spontaneously firing 3D neuronal cultures within our device. This study lays the groundwork for understanding the impact of microgravity on neuronal function and provides valuable insights for space exploration and neurobiology. It also offers potential applications in studying neurological disorders on Earth, such as Alzheimer’s and Parkinson’s disease, and contributes to ensuring astronaut well-being during long missions.
Extended space travel in low Earth orbit (LEO) exposes the neurons in the central nervous system (CNS) to various stressors such as microgravity and cosmic radiation. These stressors can harm delicate cell structures and cause epigenetic changes in their DNA, which can lead to alterations in functions such as cognition and behavior7. Thus, understanding the effects of long-term spaceflight on electrically active neurons, while also considering their benefits to life on Earth, is an immediate necessity for the space research community8. However, present techniques to study such a phenomenon rely on invasive whole-cell recording at intermittent time points, resulting in limited data acquisition and even reduced insight into cell behavior. Currently, there exists no validated technology for continuous electrophysiology recording and real-time monitoring of cells in physiological systems exposed to long-duration spaceflight missions. Utilizing Glutamatergic (Glu)-Neurons, derived from human induced pluripotent stem cells, we made a significant advancement in studying human neuronal physiology compared to traditional rodent models in a flight-ready platform9. We have utilized a disruptive combination of tools, including a microfluidic tissue-on-a-chip biodevice with an integrated 3D probe, to quantify the behavior of functional neuronal networks which are critical to understanding cell function in extreme environments like spaceflight and microgravity9. This model also offers other advantages over rodent cultures, including greater relevance to human performance, reproducibility, and scalability.
The intellectual merits of this study lie in addressing the urgent need to develop a 3D neuronal electrophysiology sensing microfluidic device for space applications. Furthermore, our study offers a radically different approach or disruptive innovation that may significantly enhance or enable new human exploration missions. Such a platform can be especially helpful in understanding the effects of long-term space travel on CNS neurons, particularly considering stressors like microgravity and cosmic radiation. Utilizing advanced techniques such as a microfluidic tissue-on-a-chip biodevice with an integrated 3D microelectrode array for electrophysiology, the study aimed to develop a platform for continuous and real-time monitoring of electrically active neurons, providing insights into their behavior in the extreme environmental conditions imposed by a spaceflight. The outcomes from the suborbital flight study validate the functionality of our biodevice and offer a promising platform for studying neurological disorders on Earth.
Results
Pre-Flight
Blue Origin’s New Shepard NS-24 mission launched from the West Texas site in Van Horn on Dec 19th, 2023, and carried many scientific payloads, including our own, on a suborbital flight to the edge of space, reaching above the Kármán line10. The vehicle, powered by its BE-3 engine, provided 3.43 min of microgravity for experiments before the booster and capsule began their descent, with a total flight duration of 16.11 min. The engine and crew capsule experienced average G-forces of 1.2249 G, 0.0274 G, and 1.3124 G for launch, microgravity, and landing phases, respectively, with maximum G-forces during booster separation just before microgravity and upon reentry into Earth’s atmosphere. The complete flight log containing the elapsed time, acceleration magnitude, and measured G-force can be found in the supplementary data. The booster executed a controlled, vertical landing, while the capsule descended separately, using parachutes and retro-thrust for a soft landing in the desert. This mission demonstrated reusable technology to reduce space access costs and supported microgravity research, enhancing our understanding of spaceflight effects on various systems. The results obtained from our payload aboard this suborbital flight are reported in two stages, namely, pre-flight and post-flight, respectively.
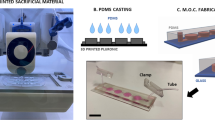
Multiple iterations of a microfluidic device were tested to create a device capable of sustaining and collecting EPHYS data acquisitions from 3D in-vitro cultures for at least one week of in-vitro culture. The initial device design featured a central hydrogel chamber (1 × 1 × 10 mm, h x w x l) with medium channels on either side (1st Gen: Fig. 1). Our design was inspired by published design ideas of microfluidic devices with pillars and posts to integrate electrodes for studying cell behavior from others11. A half-wall separating the medium channels and central hydrogel chamber allowed for the flow of media to the hydrogel during culture. Medium exchange is facilitated via two inlets and outlets at the top and bottom of each channel. The Neuropixels probe is seated at an opening at the base of the device, which is connected directly to the hydrogel chamber. This design was later altered to facilitate simplified medium exchanges via a single inlet and outlet in 2nd Gen devices employing the same hydrogel–Neuropixels interface, with a medium path adjusted to a more streamlined closed-loop design. Finalized adaptations of this device (3rd Gen) utilize the core structure of the second generation, with the incorporation of two ports centered over the medium channels to serve as a means to ground the medium when conducting EPHYS measurements using an external reference. See Supplementary Fig. 2 for the process used in the fabrication of the microfluidic device.
Shown are the iterations of microfluidic devices which were designed to accommodate hydrogels containing cell cultures, media, and a Neuropixels probe. The 1st Gen design was changed to allow for automated media exchanges via a single inlet and outlet (2nd Gen), and ultimately the addition of two inlets to provide easier access to grounding to obtain reduced noise measurements (3rd Gen).
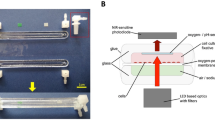
Data acquisitions were collected from cell-laden hydrogels, R-HI-501 or iCell Gluta-neurons, maintained in the microfluidic devices as early as 24 h in culture. Acquisitions were facilitated by a headstage, cable, and OneBox data collection system developed by imec. This system captured EPHYS activity from up to 300 low-impedance electrodes simultaneously, transmitting the data instantaneously to a PC. EPHYS data was collected from each microfluidic device for 60 sseconds using a Python-based algorithm (developed in collaboration with imec). This code possesses the ability to automatically alternate data acquisition from up to four probes for a set duration of time while maintaining flexibility with key parameters such as bank selection, gain, and tip or external reference points. Samples were maintained at 37 °C, 5% CO2 for the duration of each acquisition. Data was stored on a PC as a binary file. A schematic of this setup is shown in Fig. 2. In addition to this layout, an actual photograph of the biodevice integrated within the microfluidics circuit for the in-flight experiment is included in Supplementary Fig. 1.
Neural chips containing human glutamatergic neurons are maintained in a cube-lab which maintains adequate culture conditions. Recordings are collected automatically by a computer with technology specific software designed specifically for this project which employs the data collection system provided by imec.
In processing the collected data, initial sorting of the raw binary files collected from the Neuropixels probe was completed using Kilosort312, followed by manual curation of sorted units using the Phy software13,14. Kilosort3 matches neuronal spikes found in the raw EPHYS data with template waveforms of neuronal action potentials. Machine learning was then used to cluster similar waveforms into single units, which can be considered as spikes detected from individual neurons as described below.
EPHYS Data Acquisition and Processing was performed as follows: Data acquisitions began 24 h after the seeding of cell-laden hydrogels into the microfluidic devices. Acquisitions were facilitated via a headstage, cable, and data collection system designed and developed by imec. These components are connected to microfluidic devices and provide the Neuropixels probe with a power supply, bi-directional communication capabilities, and data transmission to a PC. EPHYS data is then collected from each microfluidic device for 60 s using a Python-based algorithm (developed in collaboration with imec) intended to automate the data collection process aboard a suborbital spaceflight. This code possesses the ability to automatically alternate data acquisition from up to four probes for a set duration of time while maintaining flexibility with key parameters such as bank selection and gain settings. Data is stored on a PC as a binary file. Data Processing was then performed as follows: Initial sorting of the raw binary files collected directly from the Neuropixels probe is completed using Kilosort3, followed by manual curation of sorted units using Phy. Kilosort3 works by matching spikes found in the raw EPHYS data with template waveforms of neuronal action potentials. Then, machine learning is used to cluster similar waveforms into single units, which can be considered as spikes detected from individual neurons. The resulting data is presented as a Python file, which can be opened in additional programs for viewing and elimination of false-positive spikes.
Parameters for Kilosort3 | ||
|---|---|---|
Parameter Name | Function | Settings used |
Threshold | [Th(1) Th(2)]. Sets the threshold of the template projections, which the raw data is required to match to be considered a spike. Th(1) sets the first pass and should be high to reduce noise contamination. Th(2) sets the second pass and is low to detect all spikes found within a unit. | [10 4] |
Number of Blocks for Rigid Registration | Indicates whether drift correction is turned on or off. Drift correction is used to account for movement of neurons relative to the probe due to tissue relaxation in vivo. | 0 |
Lambda | Determines the level of matching of spike amplitudes with the mean in a cluster. | 25 |
AUC Split | Threshold on the area under the curve (AUC) criterion for performing a split in the final step. | 0.9 |
The number of blocks for rigid registration, for which the default is 5, has been set to 0 (off). This parameter is used to correct drift in recordings, which may occur as a result of tissue relaxation or movement of the probe during in vivo acquisitions. Since the probe is fixed in the device, and cells are seeded around the probe rather than inserted for recording, drift correction is not critically necessary in processing or analyzing our data. The lambda parameter, default 10, has also been set to 25 to increase the level of matching between spike amplitudes found within a sorted unit. Sorted units are viewed in Phy, and the total number of spikes identified in each unit is manually extracted. Data is represented as the sum of the total number of spikes identified in each device. The total number of spikes identified from the same bank in a microfluidic device without cells has been subtracted to correct false-positive spikes in the sample data.
The resulting data is stored as a Python file, which can be opened in additional programs for viewing and elimination of false positive spikes (Supplementary Fig. 3). Sorted Neuropixel data was manually curated using Phy, which allows for the detection and quantitative assessment of neuronal activity. This provides us with insight into the quality of our recordings. A normal distribution of spikes tells us that all spikes for this unit were detected within the set threshold (Kilosort) and that the spikes for this unit are likely coming from an actual neuron. This can also be observed in the even distribution of spikes throughout the entire recording. Noise may present itself as isolated spikes of the same amplitude across the recording.
Our team was able to record EPHYS data from the human Glu neurons pre- and post-flight to study their viability during the entire suborbital flight study duration. Developing technologies are categorized by NASA into specific technology readiness levels (TRLs) to assess the progress towards incorporation into actual missions. The TRL categories range from levels 1 to 9, with level 1 consisting of the identification of a basic need for the technology, while level 9 constitutes the successful operation of a developed technology in a spaceflight mission. At the start of this project, our technology was categorized as TRL level 3, with the formulation of an initial design and completion of proof-of-concept studies. However, as this work has progressed, and with the completion of a suborbital flight, the technology level has been raised to a TRL level 7. Our device has been operated in a suborbital flight, i.e. relevant environment, and has shown that it is able to sustain cell cultures and continued monitoring of EPHYS activity before and following the flight. Many microfluidic devices intended for use in space environments have validated their device using simulated microgravity15,16, or brief exposures via drop towers17. Though these constitute relevant environments for assessing technology readiness in microgravity, exposure of our device to microgravity via an actual suborbital flight pushes the limits of microgravity research one step further. Our work and its outcomes clearly support NASA’s Space Technology Mission Directorate (STMD) priorities of developing new technologies to enable human and robotic exploration of the Moon, Mars, and beyond, thus enhancing research and development to contribute to U.S. leadership in space technology through partnerships with academia and industry.
To assess the EPHYS functionality of the microfluidic device and Neuropixels system, EPHYS acquisitions were collected from Rat Hippocampal neurons seeded in microfluidic devices and Axion MEA 6-well plates at a density of 200,000 cells/25 µL Matrigel. Neuronal activity was detected in the microfluidic devices and 2D controls on Day 11 (Fig. 3A, B). Confocal imaging of these samples, Fig. 3C, shows intricate neurite outgrowth and network formation, indicating the presence of healthy neurons capable of spontaneous firing. This is confirmed in Fig. 3D, which shows the number of spikes per minute and mean firing rate (Hz) for 2D neuronal cultures. Increasing firing rate and number of spikes were observed with increased time in culture, although the trends were not statistically significant, with a p-value of 0.4305 for the number of spikes and a p-value of 0.5258 for the mean firing rate (Hz) when assessed between day-11 and day-14.
EPHYS Acquisitions were collected from a microfluidic device containing Rat Hippocampal neurons seeded in Matrigel at a density of 200,000 cells/25 µL Matrigel. A shows sample waveforms collected from an acquisition at Day 11. B shows a sample waveform collected from rat hippocampal neurons cultured in 2D at Day 11. EPHYS data was collected using an Axion Maestro Edge. C Shows 2D rat hippocampal neuronal cultures maintained for sustained periods. D shows the firing activity of the 2D rat hippocampal cultures.
Currently, there are 131 distinct models related to hippocampal cells that have been employed by a wide range of researchers to characterize electrophysiological hardware and models18,19. These cell types have yielded valuable experimental data providing insights into their physiological behavior under various conditions. On the contrary, human iPSC-derived Glu neurons often exhibit poor firing rates and lower robustness, requiring several weeks of maturation under specialized culture conditions to produce reliable electrophysiological (EPHYS) responses20. This is why we chose rat hippocampal neurons for this part of our study.
To verify the capacity of the microfluidic device in sustaining the culture of human Glu neuronal cell types, a live/dead assay was performed on human glutamatergic neurons suspended in Matrigel and cultured in the device and a 96-well plate for 7 days. Results show the presence of live neurons, which display intricate network formation throughout the hydrogel at the end of the culture period (Fig. 4). The microfluidic device also shows elevated cell viability (98.22% viability) in comparison to cultures maintained in the 96-well plate (77.69% viability). Statistical significance was calculated for cell viability in the 96-well plate at 7 days, using a two-tailed test, with (p < 0.0001). Similarly, cell viability in the microfluidic device at 7 days also showed a statistically significant result, with (p < 0.0001). This may have been due to medium flow through the device washing away dead cells during exchanges, which is a drawback of our current device, however, it can be controlled or regulated with a filter embedded in the flow circuit to prevent this from happening in the future. Neuronal networks cultured within the microfluidic device are comparable to those observed in the 2D controls. Additional confirmation of adequate network formation in neurons cultured in microfluidic devices after 8 days is shown in Supplementary Figs. 4 and 5. Similarly, immunostaining of these cultures reveals the expression of MAP-2 and highlights the ability of neurons to form networks in a 3D platform (Fig. 5 and Supplementary Fig. 4). Fluorescence intensity within the device is compromised to some extent due to a thicker layer of Matrigel which is confined within the hydrogel chamber in comparison to 2D controls by which the hydrogel can spread. Similarly, imperfections within the Ostemer itself during casting may lead to reduced image clarity.
Immunostaining of human Glu neurons cultured in a 96-well plate and Ostemer device (8000 cells/mL in Matrigel) after 7 days. The cells were stained using antibodies for the neuronal marker MAP-2 (red) and counterstained using the nuclear stain DAPI (blue). Quantification of the MAP-2 expression is depicted alongside.
Human glutamatergic (Glu) neurons suspended in Matrigel were seeded in a microfluidic device at a density of 200,000 cells/25 µL Matrigel and maintained for a period of at least 8 days, prior to EPHYS studies. EPHYS acquisitions of 60-second durations were collected from each device beginning 24 h after culture. Spontaneous action potentials were observed as early as 24 h after seeding in the device and were maintained through the entire culture period (Fig. 6). Captured spikes were sorted using Kilosort3 and quantified using Phy. The number of spikes, n, detected for each acquisition shown in Fig. 6 is n = 113, n = 180, and n = 128. Action potentials are taken from the same electrode, suggesting that the firing is likely from a neuron situated at that position along the probe shank. MEA technology consists of hundreds of closely packed electrodes simultaneously recording EPHYS activity. The proximity of these electrodes to one another enables the recording of action potentials from a single neuron across multiple electrodes, with amplitudes decreasing with increasing distance21. Evidence of the primary action potentials on surrounding electrodes, highlighted with green arrows (Fig. 6), was also observed in all probe banks. Known as spatial decay, this phenomenon further demonstrates the presence of spontaneous firing activity.
Neuronal peaks were detected in a microfluidic device seeded with human Glu neurons in Matrigel over a period of 8 days. Sample waveforms are taken from the same section of the Neuropixels probe. Number of spikes, n, detected for each acquisition are as follows: n = 113 (Day 1), n = 180 (Day 4), and n = 128 (Day 8). White arrows indicate main action potential waveforms, green arrows indicate evidence of spatial decay on surrounding electrodes.
Human Glu neuronal function correlated by studying their gene expression while being cultured within the device was characterized via qPCR focused on the genetic markers, vesicular glutamate transporter 1 (VGLUT1) and vesicular glutamate transporter 2 (VGLUT2) in comparison to 2D cultures normalized to the housekeeping gene GAPDH22. While it was expected that microgravity would affect the expression levels of VGLUT 1 and 2, we did not expect GAPDH levels to be altered based on our prior published works22. The presence of VGLUT 1 and 2 in neuronal cultures signifies the ability of the neurons to transport glutamate within the network, and thus, fire action potentials.
Utilizing the individual efficiency corrected calculation method, an enhancement over the commonly utilized 2−ΔΔCT method, we calculated the average ΔCT of the target genes, VGUT1, VGLUT2, and MAP-2, calculated from at least three data points recorded per 3D sample extract. After that, the average ΔCT of the housekeeping gene reference, GAPDH, was determined using a minimum of three recorded data points. Subsequently, the 2−ΔΔCT method was employed to compute the expression fold change for the target genes. Next, these values were normalized to the control sample values calculated similarly using the steps listed above. It was expected that in the absence of any treatment in the controls, the levels of target and reference gene expression would be relatively stable. As a result, the control sample data shows the expression fold change for the target genes equaling ‘one.’ The experimental sample was shown with the controls, demonstrating the fold change in expression between both groups. Results showed upregulated levels of VGLUT1 and VGLUT2 in neurons maintained in the 3D microfluidic device compared to controls (Fig. 7), suggesting that neuronal network formation is more robust in a 3D biodevice in comparison to 2D neuronal cultures. Statistical significance was calculated based on average ΔCT values using a two-tailed t-test, and results were statistically significant between 3D microfluidic devices and 2D controls (p-value = 0.004).
Post-suborbital flight outcomes
Understanding how spaceflight might affect the physiological functions of neurons is an important first step in the expansion towards long-term space missions. To assess this, as well as validate the use of our device in exposure to extreme environmental conditions, human Glu neurons seeded in microfluidic devices (200,000 cells/25 µL Matrigel) were maintained under normal conditions for 8 days before being subjected to a suborbital spaceflight. EPHYS acquisitions obtained at the ground, just before integration into the flight locker at T-13 h until launch, show spontaneous neuronal activity. Figure 8 shows a sample waveform obtained from this data, which highlights mean waveforms obtained from a single electrode throughout a 60-second acquisition. At T + 7 h following the suborbital flight, spontaneous action potentials can still be observed in the samples (Fig. 8). This significant finding demonstrates that the neuronal cells maintained their electrical activity despite exposure to microgravity and the stresses of space travel. Observing spontaneous action potentials indicates that the neurons remained viable and functionally active, retaining their capacity to generate and propagate electrical signals essential for normal brain function. This suggests that the microfluidic biodevice and the CubeLab environment effectively preserved the physiological state of the neuronal cells throughout the flight. The retention of spontaneous action potentials post-flight confirms the robustness of the experimental setup and the potential for long-term studies of neuronal function in space. This result provides a foundation for further research into how microgravity affects neuronal signaling and network dynamics, crucial for developing countermeasures to protect astronauts’ brain health during extended space missions. It also opens new avenues for studying neuronal resilience and adaptation to extreme environments, with potential implications for treating neurological disorders on Earth.
Spontaneous EPHYS data measured from rat hippocampal neurons at 11 days (Fig. 3) captured 106 spikes recorded in a single unit on the probe over 60 s with mean peak amplitudes comparable to those measured in 2D studies at 15 days, as published by others19. This could indicate that the culture of cells in a 3D culture platform may provide a preferential environment for cell maturation, while also displaying the capacity of our device to support the culture of firing neuronal cell types and recording spontaneous EPHYS activity. In our 2D studies, EPHYS data collected from 3D microfluidic devices also showed elevated spike counts, peak amplitudes, and firing rates in comparison to neuronal cultures in an Axion well plate (Fig. 3B, D). Following this trend, EPHYS data collected from human Glu neurons in 3D microfluidic devices show similar firing rates to neurons in 2D cultures as published by others23, with an increased number of spikes detected across 8 days (Fig. 6). The number of spikes, firing rate, and peak amplitude remained relatively stable across the culture period. Post-flight data (Fig. 8) revealed increased firing activity in the number of spikes, peak amplitude, and firing rate in comparison to pre-integration metrics. This may be due to the progressive maturation of neurons with longer time in culture, as well as aging on a suborbital spaceflight. It is important to note that secondary effects of the suborbital flight, such as vibration and hypergravity experienced by the cells during launch and landing, could not be controlled during this experiment. Still, this work provides a more accurate approach to the different types of environmental stressors undergone by cells during a flight study. Maximum hypergravity conditions were experienced during booster separation, with G-forces ranging from 1.05 G to 2.9168 G over 2.14 min, and upon landing with G-forces ranging from 1.05 G to 5.106 G over the course of 1.28 min as shown in the flight log (supplementary data). The samples were stored perpendicularly to the launch vector and were secured within the CubeLab using metal clamps. The CubeLab was designed to intentionally protect the devices against the maximum dynamic loads the payload would experience. Additionally, the Neuropixels probe is cast within the Ostemer, preventing drift.
Samples exposed to microgravity were analyzed for the expression of VGLUT1, VGLUT2, and MAP-2 in comparison with the GAPDH gene. Sample extraction included two biological replicates, and each biological replicate being used for three technical replicates for the experiment (n = 6). Samples exposed to microgravity (n = 2) showed slightly altered expression of VGLUT1 and VGLUT2 in comparison to ground controls (n = 2) (Fig. 9) while the expression of MAP-2 was seemingly upregulated in samples exposed to microgravity in comparison to ground controls. Statistical significance was calculated using a two-tailed t-test. For VGLUT 1 and 2, the results were not statistically significant (p-value = 0.6791), whereas for MAP-2 the results were significantly different between 3D flight samples and ground controls (p-value = 0.0001). In summary, results suggest that exposure to microgravity and suborbital flight influences the expression of specific genes, with some showing slight alterations and others showing more significant changes compared to ground controls. Reducing gravity levels during Drosophila metamorphosis in space (on the ISS) leads to significant changes in gene expression, with a large number of differentially expressed genes (DEGs) observed compared to controls under normal gravity (1 g)24. However, the spaceflight environment introduces additional stresses (due to preparation procedures and non-ideal conditions on the ISS), which also affect gene expression. Therefore, further ground studies in a simulated microgravity environment should be conducted in the future to fully distinguish the effects of space-induced microgravity from the effects of vibration and launch.
qPCR analysis of human Glu neurons exposed to microgravity in a 3D microfluidic device versus Earth’s Gravity. Analysis was performed to assess the relative expression of neuronal markers MAP-2, VGLUT1, and VGLUT2, normalized to GAPDH for each sample set. Data is represented as expression fold-change in the alteration of gene expression in samples exposed to microgravity in comparison to ground controls.
Discussion
From this recent suborbital flight experiment utilizing this technology, it was revealed that human Glu-neurons that were exposed to a limited duration of microgravity and radiation showed an altered gene expression of the vesicular glutamate transporters (VGLUTs) 1 and 2 while maintaining a sustained and an enhanced expression of the neuronal differentiation marker, microtubule-associated protein 2 (MAP-2). Cell viability was retained, and EPHYS recordings collected prior to and post-flight confirmed the physiological fidelity of the biodevice for the entire study. Since homeostatic control and regulation of glutamate signaling is essential for normal brain functioning, its altered modulation may lead to dysfunction of normal brain functions as well as neuro-inflammation25. Therefore, findings from our recent study can be further expanded to study pathological mechanisms involved in neuronal degeneration and neurological disorders on Earth, such as Alzheimer’s and Parkinson’s disease. Although a limited number of studies are underway, no well-established human neuronal tissue-on-a-chip platform currently exists that can be used as a basis for such studies. We hypothesize that such an engineered ‘neuronal’ biology platform can be extremely useful in probing the effects of long-term space-induced neurological changes. Furthermore, it can also offer a window into the neuro-pathophysiology of brain degenerative processes in humans, which could potentially benefit life on Earth. This interdisciplinary study merging biomedical engineering and neuroscience aims to advance both fields, benefiting terrestrial applications. Findings could aid in developing technologies to mitigate the effects of prolonged exposure to microgravity and space radiation on CNS neurons. Additionally, the research may lead to the creation of organ-on-a-chip platforms or biosensor devices for screening potential therapies against neurodegenerative disorders on Earth and studying disease progression in space stations. Enhancing methods for the early detection of neurodegenerative diseases can aid in prompt diagnosis and intervention for these conditions. Studies show that space missions accelerate the aging process of the human brain, similar to their effects on the heart and skeletal muscles. However, the precise mechanisms through which space exploration may influence the development of diseases such as Alzheimer’s and Parkinson’s remain poorly understood.
By establishing a unified infrastructure for automated EPHYS recordings and conducting differential gene expression analysis, the research also seeks to correlate physiological data with molecular mechanisms associated with neurodegeneration, paving the way for future prevention and treatment strategies. A reductionist approach was employed to study neuronal network activity, focusing on responses to microgravity and radiation. By correlating parameters such as electrical activity measurement and gene expression alteration, mechanistic signaling changes in individual neurons or selective neuronal networks can be studied in the future26. Through this focused investigation, fundamental cellular processes and mechanisms underlying neural circuits, behavior, and diseases can be elucidated. This strategy offers a more detailed understanding of cellular function, providing valuable insights into the broader complexities of biological systems. Specifically, it will shed light on the variable effects of microgravity and radiation on different sets of CNS neurons, determining whether these effects are intrinsic to neuronal cell properties or associated with complex neuronal circuit behavior observed in vivo.
Through this focused investigation, the study offers versatility in aiming to elucidate fundamental cellular processes and mechanisms underlying neural circuits, behavior, and diseases. Specifically, it sheds light on the variable effects of microgravity and radiation on different sets of CNS neurons, enabling the determination of whether these effects are intrinsic to neuronal cell properties or associated with complex neuronal circuit behavior observed in vivo. Instead of human Glu neurons, the cell type can be replaced entirely with other types of CNS neurons or mixed populations used for future studies. Given the crucial role of Glu-neurons in modulating glutamate signaling for normal brain function, any disruptions in their regulation might result in brain dysfunction and neuroinflammation. Based on such findings, we hypothesize that studying the functional roles of glutamate or glutamic acid transporters, including VGLUTs in CNS neurons and their alterations, might generate new insights into the treatment of PD for which no effective drugs have been developed. VGLUT1 and VGLUT2 are specifically expressed in Glu-neurons and dopaminergic (DA) neurons27. Most studies on PD have mainly focused on DA neurons. However, a comprehensive understanding of the role and function of these glutamate transporters in the pathophysiology of PD based on a diverse array of neuronal cell types, such as Glutamatergic neurons, GABAergic and cholinergic neurons, as therapeutic targets will help in the development of new methods for the treatment of PD. So, our overarching goal is to highlight the relevance of microgravity in the development of PD via dysregulation of the VGLUTs, as shown by our recent study. Furthermore, EPHYS data collected prior, on spaceflight and the ISS-NL, as well as post-flight may reveal new information on the physiological transport mechanisms of VGLUT and how that might be altered during spaceflight studies leading to exacerbation of microgravity-induced neurodegeneration. Neurodegenerative diseases and space medicine may have a stronger connection than previously understood.
Since neurodegenerative disorders such as AD, PD, and ALS pose significant societal and individual burdens, early diagnosis, potentially facilitated by additional biomarkers, offers benefits including improved quality of life and research advancement. This project aims to use cutting-edge tools and technologies to study neuronal degeneration and aging in a novel engineered tissue platform, shedding light on physiological processes, such as cytoskeletal changes observed during long-duration space flights. The data collected could offer unique insights into disease mechanisms for neurodegeneration, particularly in early-onset diseases like PD. Understanding alterations in excitatory neurotransmission dynamics may lead to improved disease diagnosis and treatment strategies, ultimately benefiting societal and individual well-being.
This study reports the development of a novel microfluidic device capable of capturing spontaneous action potentials from human neuronal cultures in a 3D microfluidic device for the first time. This will become a valuable tool for studying the effects of microgravity and extreme environmental conditions on neuronal function in space studies. The optimal design features a central hydrogel chamber surrounded by a closed-loop medium channel with inlets for grounding, effectively capturing electrophysiological recordings while maintaining a favorable environment for tissue viability and function. This is supported by data showing sustained cell viability and neuronal network formation over a one-week culture period. Gene expression analysis confirms the presence of VGLUT 1, 2, and MAP-2, indicating electrically active neuronal networks in both ground-maintained and microgravity-exposed samples. The device successfully captures spontaneous action potentials from neuronal networks for extended periods, validating its use in extreme environmental conditions.
Since several neurodevelopmental and neurodegenerative diseases, such as AD, PD, Huntington’s, and amyotrophic lateral sclerosis (ALS), are known to emanate from deficiencies in appropriate cytoskeletal assembly, this study will lead to new knowledge on how the effective regulation of cytoskeletal and axonal architecture is critical for normal EPHYS function in neurons. It is also known that several neurodegenerative diseases share various molecular and cellular pathologies, including protein aggregation, mitochondrial dysfunction, and neuro-inflammation, presenting challenges for treatment strategies that often target only one aspect of the pathology while neglecting others. Thus, understanding the influence of altered gravity at the cellular and network level is of high importance. This study will offer new insights into studying neurodegenerative processes on Earth, potentially benefiting humanity’s understanding and treatment of such conditions by utilizing an innovative engineered tissue platform.
Methods
Microfluidic device design and fabrication
3D printed molds (Clear Resin) of the device were designed and printed using Formlabs Form3B (Somerville, MA, USA). These were used to cast silicone molds from SylgardTM 184 (Dow Chemical Company, Midland, MI, USA). From this step, final devices are cast from Ostemer® 322 (Mercene Labs, Stockholm, Sweden) following the vendor’s recommendations. Uncured Ostemer was poured into the Sylgard 184 molds and cured using Ultraviolet A (UVA) with a wavelength ranging from 320 to 400 nm. The UVA curing was done for 8 s at a 40% intensity using an IntelliRay 600 UV flood system (Uvitron International, Springfield, MA, USA). After the completion of the UV curing step, the product was placed onto a microscope slide with a Neuropixels 1.0 probe (imec, Leuven, Belgium) set in place, then further oven-cured for 1 h at 90 °C. Fresh Ostemer was carefully pipetted into gaps around the Neuropixels probe and cured using the previously described protocol to prevent hydrogel and medium leakage through this channel. After all curing steps were completed, 1X PBS was passed through the channels to ensure proper bonding between Ostemer layers, as well as to assess flow through each channel. Fully cured devices were rinsed three times with 70% Ethanol, three times with 1X PBS, and UV sterilized for a minimum of 30 min in a biosafety cabinet before use in any applications involving cell culture.
Cell culture and sample preparation
Rat Brain Hippocampus neurons (R-HI-501) and their culture medium were purchased from Lonza (Basel, Switzerland). Cells were thawed and 4.75 mL pre-warmed maintenance medium (PNBM Basal Medium, 2mM L-Glutamine, 50 µg/mL Gentamycin, 37 ng/mL Amphotericin, and 2% NSF-1) containing 5% FBS was added to the initial 250 µL suspension. 750 µL of this suspension was collected in a 1.5 mL Eppendorf tube, for six samples, and centrifuged at 100 G for 5 min. The supernatant was removed, and cells were resuspended in 5 µL complete maintenance medium containing 5% FBS. 25 µL Matrigel (Corning Inc., Corning, New York, USA) was added to the suspension and gently mixed by pipetting to achieve a density of 150,000 cells/25 µL Matrigel. The gel was pipetted into the microfluidic device gel chamber and incubated for 30 min at 37 °C, 5% CO2. Control samples were pipetted onto an Axion Microelectrode array (MEA) 6-well plate (Axion Biosystems, Atlanta, GA USA). A complete maintenance medium containing 5% FBS was added to the device. After 4 h, the medium was replaced with a complete maintenance medium not containing FBS. A 50% medium exchange was performed every 3–4 days thereafter. The cells were maintained in culture for at least 2 weeks to stabilize their EPHYS behavior, before being subjected to any further experiments.
Human glutamatergic neurons derived from iPSCs (iCell® GlutaNeurons) and their culture medium were purchased from Cellular Dynamics International (Madison, WI, USA). Cells were thawed and resuspended in 9 mL complete BrainPhys maintenance medium (BrainPhys Neuronal Medium, iCell DopaNeurons Supplement, iCell Nervous System Supplement, N-2 Supplement, Laminin, and Penicillin–Streptomycin). Cells were centrifuged at 400 rpm for 5 min and resuspended in 500 μL complete maintenance medium. 100 μL of this suspension was then taken and placed into a 1.5 mL Eppendorf tube and centrifuged again. Cells were then resuspended in 5 μL complete maintenance medium. 25 μL Matrigel was added to the suspension and gently mixed by pipetting to achieve a density of 200,000 cells/25 μL Matrigel. The gel was pipetted into the microfluidic device gel chamber and incubated for 30 min at 37 °C, 5% CO2. Control samples were pipetted onto an Axion Microelectrode Array (MEA) 6-well plate (Axion Biosystems, Atlanta, GA, USA). A complete maintenance medium was then added to the device and exchanged every 1–2 days. The cells were maintained in culture for at least 2 weeks to stabilize their EPHYS behavior before being subjected to any further experiments.
The use of MEA plates to culture and study cells, particularly neurons, was utilized to analyze their electrical activity. MEA-culture plates with Rat Hippocampal neurons were used to record extracellular electrical signals from the 2D cultured neuronal networks, allowing us to validate their electrophysiological properties and functional connectivity of these networks, before 3D EPHYS probe integration (d.).
Microfluidic tissue-on-a-chip sample preparation for the suborbital flight study
Four devices were made as previously described in preparation for the suborbital flight, two samples were integrated into an enclosed experimental platform named CubeLab developed by SpaceTango, and two remained on the ground to be used as ground controls. During culture, luer connector components (McMaster Carr #51525K211) served as hydrostatic reservoirs. Upon integration into the CubeLab, these components were replaced with alternate connectors (McMaster Carr #5463K36) fitted with tubing to accommodate pumps, which will facilitate automated media exchange during the flight. The CubeLab layout could only accommodate four devices, including two for biology validation and two for hardware validation only.
Electrophysiology (EPHYS) measurements using MEA-based techniques
EPHYS data in 2D was collected using an Axion Maestro Edge from Rat Hippocampal neurons seeded on an Axion MEA 6-well plate following the protocols described earlier. After 24 h in culture, the well plates were placed into the Maestro Edge and data was collected using the Axis Navigator software. Spontaneous neural recordings were captured using the default program settings. The resulting data was then exported and processed using the Neural Metrics software developed by Axion. The final data was then plotted using the Axion Plotting Tool and utilized for analysis.
Electrophysiology measurements were obtained using a Neuropixels 1.0 probe with a metal cap (imec, Leuven, Belgium) integrated into the device. Each probe features 384 recording channels capable of addressing 960 low-impedance TiN6 sites arranged along a 10-mm long, 70 × 20-μm cross-section shank, all fabricated on a single chip within the 6 × 9-mm probe base. Voltage signals undergo filtration, amplification, multiplexing, and digitization on the basis, facilitating the direct transmission of noise-free digital data. The dense recording sites and high channel count enable the isolation of spiking activity from hundreds of neurons per probe when implanted in rodent brains, with over 700 well-isolated single neurons recorded simultaneously using two probes2. Data acquisition was performed using a OneBox (imec, Leuven, Belgium) operated by a Python-based code designed specifically for this project. A headstage and cable were connected to each probe at the time of recording. A wire connected to a grounded outlet was also inserted into the grounding ports of each device. Acquisitions were collected from banks 0 and 1, using a gain of 500 and ean xternal reference.
Data acquisitions began 24 h after the seeding of cell-laden hydrogels into the microfluidic devices. Acquisitions were facilitated via a headstage, cable, and data collection system designed and developed by imec. These components are connected to microfluidic biodevices and provide the Neuropixels probe with a power supply, bi-directional communication capabilities, and data transmission to a PC.
Microfluidic device biocompatibility testing
A live/dead assay was performed to assess cell viability and overall device biocompatibility for the ostemer materials used. iCell human glutamatergic neurons suspended in Matrigel (200,000 cells/ 25 µL Matrigel) were cultured for 7 days in the ostemer based device. Medium exchanges were performed every 1–2 days. iCell human glutamatergic neurons suspended in Matrigel (60,000 cells/7 µL Matrigel) were seeded in a 96-well plate and maintained for 7 days as a control. Live/Dead assay was performed using a kit (Invitrogen, Gibco, CA, USA) and used according to the manufacturer’s protocol. Images were obtained using an LSM 700 microscope (Zeiss AXIO Observer 5; Zeiss, Oberkochen, Germany) at 20X magnification and were processed using ImageJ to quantify the percent viability of the cells studied.
Bright-field imaging
Bright-field images were used to visualize the cells and their density in gels within the biodevice and were obtained using an EVOS XL Core Imaging System (ThermoFisher Scientific, Waltham, MA USA) at 20X magnification using 4/10 contrast.
Immunofluorescence
The Rat Hippocampal neurons (R-HI-501) or the human Glu (iCell® Gluta)-Neurons in Matrigel were seeded and maintained in an Ostemer device at a density of 200,000 cells/25 μL for 7 days. Cells were fixed using 4% paraformaldehyde (ThermoFisher Scientific, Waltham, MA) for 30 min. Samples were washed and blocked using 1X PBS, 5% BSA, and 0.3% Triton-X solution for 1 h. Immunocytochemistry was performed by using MAP2 Monoclonal Antibody (M13) as a primary antibody (ThermoFisher Scientific, Waltham, MA) diluted in 1X PBS, 1% BSA, and 0.3% Triton-X at a ratio of 1:400. Samples were incubated overnight at 4 °C. Alexa Fluor Plus 647 (Invitrogen, USA) was used as a secondary antibody diluted to 1:400. Secondary antibodies were incubated for 1 h at 25 °C, protected from light. Samples were washed, and DAPI Fluoromount G (Southern Biotech, Birmingham, AL, USA) was applied. Images were acquired using an LSM 700 confocal microscope.
Quantitative PCR (qPCR) analysis
Human Glu neurons were used to study the gene expression of the neuronal markers, Microtubule-Associated Protein 2 (MAP-2) and Vesicular Glutamate Transporter 1 & 2 (VGLUT 1 and VGLUT 2) and their relative variation across 2D and 3D cultures as well as comparison between 3D cultures maintained at Earth’s Gravity (Ground controls) versus microgravity (suborbital flight). As mentioned in section 2.3 earlier, four 3D EPHYS devices were made out of which two samples were adopted for the spaceflight, and the remaining two were used to serve as ground controls. In addition, cells were also studied in 2D MEA well plates as described above. For this procedure, 20 ng of total RNA was converted to cDNA using the First Strand cDNA Synthesis Kit (OriGene Technologies Inc., Rockville, Maryland, USA) in a 20 μL reaction following the manufacturer’s instructions. The resulting cDNA was quantified using a NanoDrop OneC spectrophotometer (ThermoFisher Scientific, Waltham, MA, USA), and absorbance ratios at 260/280 nm and 260/230 nm were recorded. RT-qPCR reactions were carried out on a Quantstudio 3 system from Applied Biosystems, Invitrogen. All samples, including controls (human Glu-neurons in 2D wells) and experimental (human Glu-neurons in 3D microfluidic devices) samples, were run in triplicate using qPCR Tubes, 8 strips, 0.2 mL, with optical strip caps from Pure AMP PCR plastics, MTC™ Bio Incorporated, in a 20 μL reaction volume. Each reaction contained 4000 ng of cDNA, 10 μL of Go Taq qPCR Master Mix SYBR, 0.2 μL of supplemental CXR Ref. Dye from Promega, 1 μL of 10 mM primer mix (Fw, Rv) from Origene Tech Inc., and nuclease-free water. The primer sequences corresponding to the genes for evaluation are shown in Table 1 below.
To prevent contamination and primer-dimer formation, a no-template control was included. The reaction protocol consisted of an initial denaturation step at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 15 s as per the Origene protocol. The Quantstudio 3 software automatically calculated quantification cycle (Ct) values. Gene expression was evaluated using the comparative Ct method (2−∆∆CT), with average Ct values of GAPDH serving as the endogenous control to assess the stability of target gene expression. We adopted the individual efficiency corrected calculation method, which represents an enhancement over the commonly utilized 2−ΔΔCT method. This method computes an individual efficiency for each sample, thereby averting potential issues associated with estimating background fluorescence. Consequently, this novel approach ensures greater accuracy in relative gene expression quantification28. RNA extraction was performed from human Gluta-neurons for genes mentioned in Table 1.
Statistical analysis
In this study, statistical analysis was conducted to evaluate the significance of various experimental results. Plots were presented as mean ± SD, and statistical significance is depicted as ∗p < 0.05 and was considered to indicate a statistically significant difference. A one-way ANOVA was employed to assess the statistical significance of differences in the number of spikes and the mean firing rate (Hz). For cell viability and gene expression measurements, two-tailed tests were utilized. Sample size was n = 2 for biological replicates and n = 3 for technical replicates per biological sample. This implies that for each biological sample, there were two independent biological replicates, and for each biological replicate, three technical replicates were performed. This experimental design ensures that both biological variability and technical reproducibility are taken into account in the analysis.
Data availability
Source data for this study are all included in the manuscript and the online supporting information.
Code availability
Datasets for this work were acquired and processed using MatLab and Python-based codes. These algorithms are publicly available and can be accessed on the following pages. Data Acquisition: EPHYS Flight Code (Spyder) [Code sharing is pending approval from IMEC]. Data Processing: Kilosort3 (MatLab) [https://github.com/MouseLand/Kilosort]. Phy (Python): [https://github.com/cortex-lab/phy].
Abbreviations
- EPHYS:
-
Electrophysiology
- iPSC:
-
Induced pluripotent stem cells
- 2D:
-
Two-dimensional
- 3D:
-
Three-dimensional
- MEA:
-
Microelectrode arrays
References
Hupfeld, K. E., McGregor, H. R., Reuter-Lorenz, P. A. & Seidler, R. D. Microgravity effects on the human brain and behavior: dysfunction and adaptive plasticity. Neurosci. Biobehav. Rev. 122, 176–189, https://doi.org/10.1016/j.neubiorev.2020.11.017 (2021).
Jun, J. J. et al. Fully integrated silicon probes for high-density recording of neural activity. Nature 551, 232–236, https://doi.org/10.1038/nature24636 (2017).
Pachitariu, M., Sridhar, S., Pennington, J. & Stringer, C. Spike sorting with Kilosort4. Nat. Methods 21, 914–921, https://doi.org/10.1038/s41592-024-02232-7 (2024).
Roy-O’Reilly, M., Mulavara, A. & Williams, T. A review of alterations to the brain during spaceflight and the potential relevance to crew in long-duration space exploration. NPJ Microgravity 7, 5. https://doi.org/10.1038/s41526-021-00133-z (2021).
Sprugnoli, G., Cagle, Y. D. & Santarnecchi, E. Microgravity and cosmic radiations during space exploration as a window into neurodegeneration on Earth. JAMA Neurol. 77, 157–158, https://doi.org/10.1001/jamaneurol.2019.4003 (2020).
Wevers, N. R. et al. High-throughput compound evaluation on 3D networks of neurons and glia in a microfluidic platform. Sci. Rep. 6, 38856. https://doi.org/10.1038/srep38856 (2016).
Mhatre, S. D. et al. Neuro-consequences of the spaceflight environment. Neurosci. Biobehav. Rev. 132, 908–935, https://doi.org/10.1016/j.neubiorev.2021.09.055 (2022).
Clément, G. R. et al. Challenges to the central nervous system during human spaceflight missions to Mars. J. Neurophysiol. 123, 2037–2063, https://doi.org/10.1152/jn.00476.2019 (2020).
Padilla, A. E., Hovell, C., Mares, J., Reumers, V. & Joddar, B. Electrophysiological recording of human neuronal networks during suborbital spaceflight. bioRxiv https://doi.org/10.1101/2022.10.25.512608 (2022).
Pletser, V., Migeotte, P. F., Legros, J. C., Deneyer, B. & Caron, R. The suborbital research association: using suborbital platforms for scientific and student experiments. Microgravity Sci. Technol. 28, 529–544, https://doi.org/10.1007/s12217-016-9502-0 (2016).
Selimovic, A., Erkal, J. L., Spence, D. M. & Martin, R. S. Microfluidic device with tunable post arrays and integrated electrodes for studying cellular release. Analyst 139, 5686–5694, https://doi.org/10.1039/C4AN01062K (2014).
Wu, X.-T. et al. Cells respond to space microgravity through cytoskeleton reorganization. FASEB J. 36, e22114. https://doi.org/10.1096/fj.202101140R (2022).
Buccino, A. P. et al. SpikeInterface, a unified framework for spike sorting. Elife 9, e61834, https://doi.org/10.7554/eLife.61834 (2020).
Rossant, C. et al. Spike sorting for large, dense electrode arrays. Nat. Neurosci. 19, 634–641 (2016).
Strods. A. et al. Development of organ-on-a-chip system with continuous flow in simulated microgravity. Micromachines 15, https://doi.org/10.3390/mi15030370 (2024).
Estlack, Z., Golozar, M., Butterworth, A. L., Mathies, R. A. & Kim, J. Operation of a programmable microfluidic organic analyzer under microgravity conditions simulating space flight environments. NPJ Microgravity 9, 41. https://doi.org/10.1038/s41526-023-00290-3 (2023).
Striebel, J. et al. Human neural network activity reacts to gravity changes in vitro. Front. Neurosci. 17, 2023, [Online]. Available: https://www.frontiersin.org/journals/neuroscience/articles/10.3389/fnins.2023.1085282.
Braeken, D. & Prodanov, D. New trends and challenges in the development of microfabricated probes for recording and stimulating of excitable cells. in New Developments in Biomedical Engineering, InTech. https://doi.org/10.5772/7613 (2010).
Miccoli, B. et al. High-density electrical recording and impedance imaging with a multi-modal CMOS multi-electrode array chip. Front. Neurosci. 13, https://doi.org/10.3389/fnins.2019.00641 (2019).
Amin, H. et al. Electrical responses and spontaneous activity of human iPS-derived neuronal networks characterized for 3-month culture with 4096-electrode arrays. Front. Neurosci. 10, https://doi.org/10.3389/fnins.2016.00121 (2016).
Obien, M. E. J., Deligkaris, K., Bullmann, T., Bakkum, D. J. & Frey, U. Revealing neuronal function through microelectrode array recordings, Front. Neurosci. 8, 423 (2015).
Joddar, B., Loyola, C. D., Ramirez, S. P., Muruganandham, A. & Singh, I. Inhibition of ERK 1/2 pathway downregulates YAP1/TAZ signaling in human cardiomyocytes exposed to hyperglycemic conditions. Biochem. Biophys. Res. Commun. 648, 72–80, https://doi.org/10.1016/j.bbrc.2023.01.014 (2023).
Tukker, A. M., Wijnolts, F. M. J., de Groot, A. & Westerink, R. H. S. Human iPSC-derived neuronal models for in vitro neurotoxicity assessment. Neurotoxicology 67, 215–225, https://doi.org/10.1016/j.neuro.2018.06.007 (2018).
Herranz, R. et al. Spaceflight-related suboptimal conditions can accentuate the altered gravity response of Drosophila transcriptome. Mol. Ecol. 19, 4255–4264, https://doi.org/10.1111/j.1365-294X.2010.04795.x (2010).
Zhou, Y. & Danbolt, N. C. Glutamate as a neurotransmitter in the healthy brain. J. Neural Transm. 121, 799–817, https://doi.org/10.1007/s00702-014-1180-8 (2014).
Tripathy, S. J. et al. Transcriptomic correlates of neuron electrophysiological diversity. PLoS Comput. Biol. 13, e1005814, https://doi.org/10.1371/journal.pcbi.1005814 (2017).
Eskenazi, D. et al. Dopamine neurons that cotransmit glutamate, from synapses to circuits to behavior. Front. Neural Circuits, 15, 665386 (2021).
Rao, X., Huang, X., Zhou, Z. & Lin, X. An improvement of the 2^(-DELTA DELTA CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinforma. Biomath. 3, 71–85 (2013).
Acknowledgements
The authors gratefully acknowledge the support provided by the NIH-NIMHD-RCMI Grant No. 5G12MD007592, which facilitated the utilization of the confocal microscopy facility in the BBRC at UTEP. This study was funded via a NASA REDDI grant (#80NSSC21K0336) to BJ via IMEC, USA. The authors would also like to thank the 3DMPSL, headed by Dr. Natividad Diaz, for access to the FormLabs 3B SLA printer. We thank research technician Ms. Carla D. Loyola in IMSTEL for her help with qPCR and analysis. We also gratefully acknowledge help from Ms. Ivana Hernandez for the normalization processing of fluorescent images for Fig. 5 using ImageJ. Jason Rextroat, Twyman Clements, Paul Gamble, and Ben Lumpp are included as co-authors for the development of the CubeLab system, which maintained our samples during the suborbital flight. Without the CubeLab, the study would not have been feasible.
Author information
Authors and Affiliations
Contributions
A.E.P.: data curation, formal analysis, investigation, methodology, project administration, software, visualization, validation, writing—original draft, writing—review & editing; G.C.: formal analysis, writing—original draft, writing—review & editing; C.H. and J.M.: conceptualization, investigation, methodology, writing—review & editing; V.R. and B.J.: conceptualization, data curation, funding acquisition, investigation, methodology, project administration, visualization, resources, supervision, validation, writing—original draft, writing—review & editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests related to the research, authorship, and/or publication of this study. There are no financial or personal relationships that could influence the work presented in this article.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Padilla, A.E., C, G., Hovell, C. et al. Adoption of microfluidic MEA technology for electrophysiology of 3D neuronal networks exposed to suborbital conditions. npj Microgravity 11, 20 (2025). https://doi.org/10.1038/s41526-025-00476-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41526-025-00476-x