Abstract
The main hazards astronauts face in space collectively affect their health, especially increasing the carcinogenesis risk. However, it is still unclear how these hazards, especially microgravity and space radiation, induce the carcinogenic transformation of normal cells. In the simulated microgravity (SMG) environment, although radiation could inhibit SMG-accentuated target cell proliferation, increase genomic instability (GI) and carcinogenic transformation rate dose-dependently, we found that for bystander cells, radiation-induced damage could be reduced, GI and the probability of carcinogenic transformation could also be decreased at lower doses (below 0.1 Gy for X-rays and 0.3 Gy for carbon ions). After filtration and KEGG analysis, five differentially expressed genes (DEGs) relating to carcinogenesis were screened out from the transcriptomic sequencing results. Based on the Cancer Genome Atlas (TCGA) from NCI, we found that AREG was closely related to the occurrence and development of lung cancer. Using AREG knockdown or overexpression cell lines, we further validated the significant correlation between abnormal expression of AREG and GI. Our findings indicate that AREG plays a substantial role in GI and carcinogenic transformation following exposure to SMG and radiation.
Similar content being viewed by others
Introduction
Space flight is a dangerous and arduous task. With more successful execution of short-distance space missions, long-distance and duration space missions have become a new challenge. However, many external or internal factors in the space environment affect human health1. Internal factors include sleep disorders caused by changes in circadian rhythms and mental health disorders (such as depression and anxiety) caused by constraints, isolation and lack of social interaction2,3. Most internal factors cannot be separated from the influence of external factors. External factors mainly include long-term exposure to space radiation and the microgravity environment4. Long-term exposure to the microgravity environment brings many potential health risks, including bone loss or bone formation disorders5,6, lung injury, and immune system damage7,8,9. Space radiation also poses risks to human health through various potential mechanisms, such as changes in cardiovascular function10, the occurrence of neurodegenerative diseases11, and radiation-induced carcinogenic effects12,13. Some research aims to describe the health risks that astronauts may face more accurately by simulating the combined effects of microgravity and space radiation14,15,16. The combined effects of simulated microgravity (SMG) and X-rays exacerbate osteoblast apoptosis and cause bone loss through the small protein MafG17,18, promote lung epithelial cell death by inducing upregulation of RAC219. The combined effects of SMG and γ-rays encourage the accumulation of lymphocyte damage through the Gadd4520,21. More importantly, the combination of SMG and carbon ions may increase the probability of chromosomal abnormalities and genomic instability (GI) in normal cells by passing cells with DNA damage through G1/S and G2 checkpoints22,23, which suggests that microgravity may intensify the radiation-induced cancer risks24.
In addition, in studies using low-dose or low-dose-rate radiation, the importance of radiation-induced non-targeted effects (NTEs) on cell damage has been continuously emphasized. NTEs act mainly through two pathways, that is, irradiated target cells transmit signals to bystander cells through gap junctional communication or by secreting soluble signaling molecules to affect bystander cells25. Some studies have shown that exposure to low-dose (<1 Gy) heavy ions such as neon ions, argon ions, and carbon ions under microbeam irradiation can significantly increase the micronucleus (MN) frequency of bystander cells26,27. Other studies have found that irradiated target cells regulate signaling pathways or transcription factors and cause carcinogenic phenotypes in bystander cells by secreting signaling molecules such as TGF-β, TNF-α or ROS through medium transfer or co-culture methods28,29,30. Long-term space missions inevitably need to face low-dose and low-dose-rate space radiation, so the NTEs can not be ignored. After predicting the space radiation risk of cancer mortality among white Americans and Asian American Islanders in the United States during the 6-month International Space Station, 80-day lunar mission, and Joint International Space Station Moon mission, it was found that NTEs increased cancer risk by ~2.3 times compared to models ignored NTEs31. Data on the occurrence of Harderian gland tumors in mice also showed that compared to radiation-induced targeted effects (TEs), prediction models considering NTEs increased cancer risk by about two times32,33. However, the carcinogenic risk of NTEs caused by the combination of microgravity and radiation is still unknown but attractive at present.
Our research aimed at the TEs and NTEs on carcinogenic transformation in the microgravity environment. We used the 3D-clinostat to simulate microgravity and then irradiated cells to explore the biological mechanisms. By establishing a co-culture system, we also observed the effects of diffusing soluble molecules secreted from target cells on bystander cells to study NTEs. We found that in the SMG state, radiation increases target cells’ GI and transformation frequency (TF), but the low-dose radiation appears to make target cells secrete signaling molecules that protect bystander cells from damage. Our study also primarily explored the biological mechanisms of TEs and NTEs in the SMG environment, aiming to explain the combined effects of the two on the carcinogenic transformation of normal cells.
Results
In the SMG environment, radiation inhibits SMG-accentuated target cell proliferation and promotes GI and carcinogenic transformation
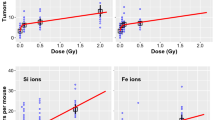
We first used BEAS-2B cells to explore the TEs on normal cells in the SMG state. Compared to the control group, the survival fraction (SF) of cells in the SMG state significantly increased, suggesting that SMG may stimulate normal cells to secrete growth-promoting factors or activate related signaling pathways to promote proliferation (Fig. 1a, d). The slight increase of MN frequency in the SMG state showed as the proliferation rate accelerates, the occurrence of GI also increased (Fig. 1b, e). Meanwhile, radiation caused the SF and MN frequency to change dose-dependently regardless of whether cells were in normal gravity or SMG conditions (Fig. 1a, b, d, e). The SF and MN frequency of cells irradiated with different doses of X-rays or carbon ions after SMG were significantly higher than that of the irradiation-only group (Fig. 1a, b, d, e and Supplementary Fig. 1a, b). Those indicate that SMG may prevent radiation-induced target cell damage or death and cause GI. GI is one of the main markers of tumorigenesis and heterogeneity, which increases the possibility of normal cells developing into tumor cells by increasing the mutagenicity and acquiring genetic diversity34,35. Based on the upregulation of GI and survival rate simultaneously, we speculated that normal cells may undergo carcinogenic transformation effects. By soft agar colony formation experiment, we found that the TF of cells irradiated with X-rays or carbon ions alone showed a dose-dependent increase but after SMG treatment, irradiated target cells’ TF increased more significantly (Fig. 1c, f and Supplementary Fig. 2a, b), further indicating that the SMG may exacerbate carcinogenic transformation of normal cells induced by irradiation. Using the two-factor analysis of variance to analyze the interaction of SMG and radiation, we found the interaction between carbon ions and SMG was more significant than that of X-rays (Table S1), suggesting that heavy ions had greater effects on survival rate and GI in the SMG environment. These results emphasize that the combination of heavy ions and SMG may have very serious consequences on carcinogenic transformation.
Low-dose radiation affects the proliferation, GI, and carcinogenic transformation of bystander cells in the SMG environment
We used BEAS-2B cells without any treatment co-cultured with target cells irradiated with X-rays or carbon ions, shown as \(\frac{{\rm{X\; ray}}}{{\rm{Ctrl}}}\) and \(\frac{{\rm{Carbon}}}{{\rm{Ctrl}}}\) and used bystander cells after SMG treatment co-cultured with target cells irradiated with X-rays or carbon ions after SMG, showed as \(\frac{{\rm{SMG}}+{\rm{X\; ray}}}{{\rm{SMG}}}\) and \(\frac{{\rm{SMG}}+{\rm{Carbon}}}{{\rm{SMG}}}\). The results showed that in the SMG state, the SF of bystander cells (\(\frac{{\rm{SMG}}+{\rm{X\; rays}}}{{\rm{SMG}}}\) and \(\frac{{\rm{SMG}}+{\rm{Carbon}}}{{\rm{SMG}}}\)) was higher than those in the normal gravity group, indicating SMG still has a significant promoting effect on the proliferation of bystander cells (Fig. 2a, d and Supplementary Fig. 1a, b). However, the MN frequency after SMG was generally lower than those in the normal gravity group (Fig. 2b, e), showing the protective effect of target cells on bystander cells to maintain their genomic stability. Although all treatment groups did not show obvious dose-dependent changes in the SF results, the SF of those bystander cells (\(\frac{{\rm{SMG}}+{\rm{Carbon}}}{{\rm{SMG}}}\)) after SMG seemed to increase slightly after co-culturing with low-dose irradiated target cells (carbon ions at 0.2 and 0.3 Gy) (Fig. 2a, d and Supplementary Fig. 1a, b). In the normal gravity environment, the MN frequency of bystander cells (\(\frac{{\rm{X\; ray}}}{{\rm{Ctrl}}}\) and \(\frac{{\rm{Carbon}}}{{\rm{Ctrl}}}\)) dose-dependent increased slowly and carbon ions treatment seemed to cause higher MN frequency compared to X-rays (Fig. 2b, e). After SMG, the MN frequency of bystander cells (\(\frac{{\rm{SMG}}+{\rm{X\; ray}}}{{\rm{SMG}}}\) and \(\frac{{\rm{SMG}}+{\rm{Carbon}}}{{\rm{SMG}}}\)) showed a non-linear change and it seemed to exist a inflection point (X-rays at 0.1 Gy, carbon ions at 0.3 Gy). When the dose was less than the inflection point, the MN frequency seemed to decrease, but when the dose exceeded the inflection point, the MN frequency started to increase in dose dependence, and carbon ion treatment also showed a more obvious transition than X-rays (Fig. 2b, e and Table S1). To sum up, in the SMG environment, bystander cells with low-dose radiation show the GI decrease, while the survival rate increases. This means that in the SMG state, target cells irradiated with low-dose radiation may secrete some signaling to protect bystander cells and reduce their damage. The TF results further verified this idea. Below the inflection point, the TF of bystander cells in the SMG state showed a dose-dependent decrease. Once the dose exceeded the inflection point, the TF increased with the dose (Fig. 2c, f and Supplementary Fig. 2a, b). Based on a two-factor analysis of variance, we found that the protective effect of carbon ion-irradiated target cells’ activity on bystander cells in the SMG state seemed to be more obvious compared to X-rays (Table S1), suggesting that in the SMG environment, low-dose heavy ions appear to stimulate target cells secret some signaling to protect normal bystander cells from damage and reduce their probability of GI and carcinogenic transformation but this is overwhelmed at higher doses which induce TF in a dose-dependent fashion.
SF (a, d), MN frequency (b, e) and TF (c, f) of bystander cells co-cultured with irradiated target cells in the normal gravity or SMG environment. \(\frac{{\rm{X\; ray}}}{{\rm{Ctrl}}}\) and \(\frac{{\rm{Carbon}}}{{\rm{Ctrl}}}\) mean bystander cells without any treatment co-cultured with target cells irradiated with different doses of X-rays and carbon ions, respectively. \(\frac{{\rm{SMG}}+{\rm{X\; ray}}}{{\rm{SMG}}}\) and \(\frac{{\rm{SMG}}+{\rm{Carbon}}}{{\rm{SMG}}}\) mean bystander cells after SMG treatment co-cultured with target cells irradiated with different doses of X-rays and carbon ions after SMG. All experiments used BEAS-2B cells. Data are represented as mean ± standard, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
mRNA screening based on the biological trends of carbon ions-induced TEs and NTEs in the SMG environment
High and low doses of radiation have been reported to elicit different gene expression patterns, different DNA damage detection and repair kinetics, etc. that can lead to non-linear responses with inflection points and thresholds36. The above different patterns of TEs and NTEs in the SMG environment prompted us to further explore the mechanisms at genetic levels and we were curious whether there is a gene simultaneously regulating TEs and NTEs in the SMG state. Based on the inflection point in NTEs, we chose target cells irradiated with 0.3 Gy and 1 Gy carbon ions after SMG and bystander cells co-cultured with target cells for transcriptomic sequencing and further study.
We first paid attention to DEGs related to TEs and identified all DEGs after comparing the SMG-only group and those exposed to both SMG and 0.3 Gy and 1 Gy carbon ions with the control group (Supplementary Fig. 3a–c). Among those groups, there are 189 intersecting DEGs (Fig. 3a), of which 63 genes showed both significantly upregulated or downregulated expression in the SMG group and upregulated or downregulated dose-dependently in those exposed to 0.3 Gy and 1 Gy carbon ions after SMG groups (Fig. 3d, e). Wondering whether one or more of these 63 genes that regulate TEs after SMG might be involved in NTEs, since the MN frequency and TF of bystander cells co-cultured with target cells irradiated with 0.3 Gy carbon ions after SMG were decreased, we defined the screening criterion as that the gene expression level of this group needed to be simultaneously lower than that of the SMG group and the group co-cultured with target cells irradiated with 1 Gy carbon ions after SMG. There are 12 DEGs that were screened out (Fig. 3b). Through KEGG pathway analysis and qRT-PCR, we further focused on 5 genes (AREG, CDKN1A, ETV4, ETV5, FAS) related to tumor occurrence and development (Fig. 3c, f–j). AREG belongs to the epidermal growth factor (EGF) family, fully expressed in epithelial and mesenchymal cells and could stimulate cell proliferation, promoting invasion, and metastasis in tumor cells37,38. CDKN1a is a checkpoint gene and target of p53, which is central in DNA repair and would contribute to the survival and MN endpoints39. ETV4 and ETV5 are highly conserved polyomavirus enhancer activator 3 subfamily members in ETS family transcription factors and are involved in mechanotransduction and cell fate determination. ETV4 is more prominent in density perception during embryonic development, tumor resistance and inflammatory pathway activation40 and it detects cell density (in the sense of confluence) due to mechanical deformation, which can also result from altered gravity41. The FAS gene is the core molecule that maintains immune homeostasis and tissue health and regulates cell apoptosis through its encoded death receptor. The abnormal expression of FAS may accelerate tumor progression by influencing the cellular inflammatory response42.
a Venn map of DEGs between target cells treated only with SMG, irradiated with 0.3 Gy and 1 Gy carbon ions after SMG and the control group. b Heatmaps of DEGs after screening TEs and NTEs groups. c KEGG pathway analysis of DEGs screened in both TEs and NTEs groups. d, e Heatmaps of dose-dependent upregulation (d) and downregulation (e) of DEGs in TEs groups. f–j The relative expression level of mRNA about AREG, CDKN1A, ETV4, ETV5, FAS in TEs and NTEs. Data are represented as mean ± standard, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
All of the above genes may regulate TEs and NTEs in the SMG environment. Since the cell lines we used belong to the lung, we found AREG had the most significant relationship with the survival rate of patients with lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) using the TCGA database (Fig. 4a, b and Supplementary Fig. 4a–h). The expression levels of AREG in LUAD cells A549 and HCC827 also showed significantly higher than those in normal lung epithelial cells BEAS-2B (Fig. 4c). To further validate the role of AREG in space environment-induced carcinogenesis, we first knocked down (shAREG) or overexpressed (oeAREG) in BEAS-2B cells (Fig. 4d). In TEs, the expression level of AREG and MN frequency in normal cells (NC) and shAREG cells both increased in a dose-dependent manner after carbon ion treatment (Fig. 4e, g). Compared with the normal gravity group, after the same-dose irradiation, the expression level of AREG and MN frequency further increased in the SMG environment, indicating that in the SMG state, carbon ions further promote AREG expression which may increase normal cells’ GI. In addition, the MN frequency of shAREG cells was almost always lower than that of NC under the same treatment, indicating that the decrease in AREG expression level is beneficial for maintaining genomic stability (Fig. 4g). However, the MN frequency of shAREG cells was still higher than that of the NC group without any treatment, indicating that extremely low expression levels of AREG may induce GI. In the SMG state, after co-culturing with target cells irradiated with 0.3 Gy carbon ions, the expression level of AREG in shAREG bystander cells increased towards normal expression levels but in oeAREG bystander cells it decreased (Fig. 4f), while the MN frequency was both reduced (Fig. 4h). And compared to shAREG cells, the MN frequency of oeAREG cells was higher in the same treatment group, which means high expression of AREG has a more negative impact on genomic stability. The above results all demonstrate that AREG plays a key regulatory role in the carcinogenic effects of TEs and NTEs in the SMG environment. High expression of AREG has shown a significant correlation with tumorigenesis. In the SMG environment, target cells treated with low-dose carbon ions may restore the AREG expression in bystander cells to a steady state through signaling molecules, thereby maintaining genomic stability.
a, b The overall survival rate of LUAD (a) and LUSC (b) patients with different expression levels of AREG in the TCGA database. c Comparison of relative mRNA expression levels between normal lung epithelial cells BEAS-2B and LUAD cells A549 and HCC827. d The expression levels of AREG in BEAS-2B cells after overexpressing (oeAREG) and knocking down (shAREG) AREG. e, g AREG expression changes (e) and MN frequency (g) of normal (NC) and shAREG target cells after SMG alone, irradiation alone, or SMG combined with carbon ion treatment. f, h AREG expression levels (f) and MN frequency (h) of shAREG (\(\frac{{\rm{NC}}}{{\rm{shAREG}}}\)) and oeAREG (\(\frac{{\rm{NC}}}{{\rm{oeAREG}}}\)) bystander cells after SMG co-cultured with NC irradiated with 0, 0.3 and 1 Gy carbon ions after SMG. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
The upstream transcription factors and interacting proteins that may interact with AREG
To more easily explore specific mechanisms of AREG, we used transcription factor prediction websites PROMO (ALGGEN-PROMO (upc.es)) and hTFDB (HumanTFDB (hust.edu.cn)) which integrate multiple databases to predict transcription factor binding sites and have been used extensively to predict AREG’s upstream transcription factors43,44. These two websites provided predicted binding sites of transcription factors and the scores of their binding sites. The undirected weighted graphs were plotted by adjusting the size of nodes based on the scores of binding sites and the width of edges based on the number of transcription factor binding sites (Supplementary Fig. 5a, b). There are 11 intersected predicted transcription factors between the two website results (Supplementary Fig. 5c) and 7 of them (STAT4, VDR, MAZ, ATF3, YY1, STAT3, USF2) had varying expression levels in different groups in our transcriptomic sequencing results (Supplementary Fig. 5d). Among them, STAT3 and STAT4 are core members of STAT family and are involved in tumorigenesis by regulating cell proliferation, immune response and metabolic pathways, respectively. Aberrant activation of STAT3 is a driver of multiple cancers, while STAT4 polymorphism significantly affects HCC susceptibility45,46. As a nuclear receptor, VDR mainly mediates the biological effects of 1,25-dihydroxy vitamin D3. The activated VDR can regulate the homeostasis of calcium and phosphorus metabolism and the proliferation and differentiation of normal cells47, but its abnormal regulation may also be related to tumorigenesis48. Myc-associated zinc-finger protein (MAZ) is a DNA-binding transcription factor that activates the transcription of target genes by recognizing specific DNA sequences (such as G-4 structure) and regulates the expression of cell cycle and proliferation-related genes (such as CCND1), which plays a role in promoting metastasis in a variety of cancers49. YY1 is also from the zinc-finger protein family, which has dual functions of activating or repressing gene expression. It dynamically regulates chromatin states by recruiting histone modification complexes (such as histone deacetylase or acetyltransferases), thereby affecting the transcriptional activity of target genes50. USF2 affects the stability of signaling pathways such as TGF-β/BMP by inhibiting the transcription and protein expression of ubiquitin ligase SMURF1251, or by regulating the expression of PKM2 (a key enzyme in glycolysis), and participates in the reprogramming of tumor cell energy metabolism52.
In addition, using the STRING website (STRING: functional protein association networks (string-db.org)), we also constructed the protein network diagram to find proteins that might interact with AREG (Supplementary Fig. 5e). Every node means a protein which has its color, the greater the number of lines between nodes, the greater the interaction of the protein. And we also found five genes, namely EPGN, EREG, ICAM1, ERBB2, ERBB3 have varying degrees of gene expression in different groups; the expression of EPGN is almost over tenfold among them, indicating the high probability of interaction with AREG (Supplementary Fig. 5f). EPGN, EREG, ERBB2 (HER2), and ERBB3 (HER3) are all members of the EGFR family, and their high expression levels have been reported to be significantly correlated with the occurrence and development of tumors53,54. ERBB2 and ERBB3 also play important roles in regulating cell proliferation, differentiation and survival by forming heterodimers and activating downstream signaling pathways (e.g., PI3K-AKT, MAPK)55. As a key adhesion molecule, ICAM2 is also very important in cancer metastasis suppression by epigenetic regulation, such as enhancing the ubiquitination and degradation of RDX56. Overall, the prediction results based on databases combined with our sequencing data give us more clarified exploration directions at the gene and protein levels and facilitate further exploring the carcinogenic formation mechanisms of TEs and NTEs in microgravity environments. However, we still need more experiments to identify the specific transcription factors and proteins that are critical to AREG.
Discussion
Space is known for lacking the gravity vector, which affects the body at the organ, tissue, and cellular levels. Microgravity, a condition of apparent weightlessness, is a significant space stressor known to significantly impact human health, such as bone loss and cardiac deconditioning6,8. Although there are more complex intercellular interactions in the response of cells to long-term microgravity in real space environments, current research indicates that the effects of microgravity environments on cytoskeleton remodeling, metabolic activity and differentiation, and tissue engineering exist under both simulated and real conditions57. The three-dimensional (3D) clinostat that we used in our study has been commonly used for SMG. By controlling the rotation of two axes, a 3D-clinostat cancels the cumulative gravity vector at the center of the device and produces an environment with an average of 10−3 g over time58. This device has been used as a tool to simulate the microgravity environment in vitro and study the changes in lung epithelial cells under SMG19, the influence of SMG on mammary epithelial cells’ proliferation58, as well as the impact of SMG on antiviral immunity59. Through simulated microgravity and real microgravity experiments, the impact of microgravity on cancer cells has also been a growing focal point of interest60. Microgravity has been shown to suppress the activity of immune cells and disrupt multibody systems, which may increase the risk of developing cancer61,62. The large implication that SMG has on cellular progression, proliferation, and apoptosis in myriad tumor cell lines, including lung cancer, has also been well documented and reviewed63.
During deep space missions, the main radiation sources of space radiation are GCR and SPEs. Space radiation, especially the HZE particle component, has the characteristics of high energy, high LET and high relative biological effectiveness (RBE), which can induce more complex clustered DNA damage and a higher probability of tumorigenesis12. Radiation-induced activation of proto-oncogenes, inactivation of tumor suppressor genes, and imbalance of expression of important genes involved in cell cycle regulation play roles in radiation-induced malignant transformation of target cells and bystander cells64. Space radiation also has the characteristics of low dose (defined as doses <100 mGy) and low-dose rate. The effective dose for a 180-day lunar mission is about 0.17 Sv65. On traveling to Mars, the effective radiation dose during flight is about 1.84 mSv/day, and about 0.64 mSv/day on the surface of Mars, amounting to a total effective dose of ∼1.01 Sv for a round trip to Mars with a 180-day one-way flight and 500-day stay at the surface of Mars66. At the same time, NTEs are often induced by low-dose or low-dose-rate radiation and cause the GI and carcinogenic transformation potential in the constant environment30,67,68. So, exploring the role of NTEs in the space environment is also a research focus.
To further understand health risks during long-term space missions, our studies explored the carcinogenic transformation of low-dose radiation from 0–2 Gy in SMG environments. For TEs, the results that target cell survival, GI and TF were all increased after SMG (Fig. 1a–f) are consistent with some published research. In vitro, exposure simultaneously to high-LET radiation and SMG has been demonstrated to increase chromosome aberrations rate23 and a higher incidence of LUAD was also found in a mouse lung cancer susceptibility model that received high-LET mixed-beam irradiation64. However, there seems to be no study to explore the NTEs in the microgravity environment at present. Our results showed that in the SMG state, co-cultured with low-dose (0.1 Gy for X-rays and 0.3 Gy for carbon ions) irradiated target cells could reduce bystander cell damage, GI and TF (Fig. 2a–f). As the dose continued to increase, GI and TF still increased in a dose-dependent manner. X-rays, as low LET radiation, can not be used to infer biological effects relevant to space radiation, but we could more clearly understand that carbon ions cause a more serious influence in TEs and NTEs by comparing X-rays with carbon ions, emphasizing that in the SMG environment, low-dose carbon ions appear to promote the probability of irradiated normal cells transforming into tumor cells and enhance protective effects in the low-dose range but not high to reduce bystander cells from damage and reduce their probability of GI and carcinogenic transformation. However, this study only used carbon ions and one cell line, our in vitro results can not be directly applied to the health assessment of astronauts without considering the systemic physiological effects. It is important to investigate the carcinogenic transformation effects of other high-LET radiation on more normal cell types and animal models under microgravity conditions.
Based on transcriptomic analysis and experimental verification, we propose that the AREG gene may play an important regulatory role in the carcinogenic transformation induced by carbon ions in the SMG environment (Figs. 3 and 4). As a member of the EGF family, AREG was originally identified as a key factor involved in several physiological processes such as bone formation, keratinocyte proliferation, mammary gland development, fibrosis, and so forth, it is also identified as a key pro-oncogenic factor playing an important role in tumor progression and metastasis69,70. Low expression of AREG has also been reported to diminish α-smooth muscle actin, fibronectin and intercellular adhesion molecule 1, which always make significant changes in the microgravity environment71. Our results indicate that in the SMG state, target cells’ AREG expression increases in a dose-dependent manner and is significantly correlated with GI (Fig. 4e, f). In the SMG state, target cells irradiated with low-dose carbon ions may help regulate bystander cells’ AREG expression level toward a steady state through signal molecule transduction, thereby maintaining genomic stability (Fig. 4g, h). These results demonstrate the important role of AREG in the carcinogenic effects of space radiation, and AREG may be a potential biomarker for effectively monitoring the physical health status of astronauts or a potential indicator for predicting the carcinogenic risk of space missions. It is necessary to further explore the mechanisms by which AREG plays a role in both TEs and NTEs.
Overall, our study conducted a preliminary exploration of the biological mechanisms underlying the carcinogenic effects of the space environment and explored the NTEs in the SMG environment for the first time. We also find a target gene AREG which may play a key role in regulating the TEs and NTEs in the SMG environment. These results may give a reference for exploring the risk assessment of astronauts carrying out space missions.
Methods
Cell culture
The human bronchial epithelial cell line BEAS-2B and human non-small cell lung cancer cell lines A549 and HCC827 were all purchased from the American Type Culture Collection (CRL-9609) and maintained at a concentration of 10% fetal bovine serum (Gibco, Grand Island, NY, USA), penicillin (100 U/mL) and streptomycin (100 μg/mL) in DMEM medium (Gibco, Grand Island, NY, USA) at 37 °C (Thermo Fisher Scientific, Waltham, MA, USA) in a fully humidified incubator with 5% carbon dioxide.
SMG
We used a 3D-clinostat (SM-31, Center for Space Science and Applied Research, Chinese Academy of Sciences) to simulate microgravity for 48 hours at 37 °C with a random rotating speed of 0–10 r.p.m72. The cells were seeded in φ35-mm dishes (NEST Biotechnology, Hangzhou, China) or T25 culture flasks (Corning, New York, NY, USA). Before SMG treatment, it was filled with culture medium and sealed with sealing film. The clinostat with cell culture vessels was placed in a humidified incubator (5% CO2, 37 °C) and was connected to the control console outside of the incubator.
Radiation
X-ray irradiation was performed at room temperature in an X-RAD SmART Biological Irradiator (Precision X-Ray Inc., North Branford, CT, USA) at a dose rate of 1.0 Gy/min. The irradiator’s tube voltage and tube current are 225 kV and 2.99 mA, respectively. Carbon ion irradiation was performed at the Shanghai Proton and Heavy Ion Center (SPHIC) with a dose-averaged LET of 50 keV/μm within the spread-out Bragg peak (SOBP) (156 MeV/u).
For the combined treatment of SMG and irradiation, cells were first subjected to SMG for 48 h and then exposed to X-rays or carbon ions. For NTEs, the processed cells were digested and resuspended. Unirradiated cells were seeded into a 24-well plate (Thermo Fisher Scientific, USA) at 2 × 104 cells/well, and irradiated cells were seeded into a Transwell chamber (NEST Biotechnology, Hangzhou, China) at 1 × 104 cells/well. The Transwell chamber was inserted into the 24-well plate and co-cultured with the unirradiated cells for about 48 h until further analysis.
Colony formation assay
The processed BEAS-2B cells were immediately digested with trypsin and counted. Then they were seeded into a six-well plate (Thermo Fisher Scientific, USA), added with ~5 mL of culture medium, and returned to the incubator for growth for about 14 days. After 14 days, the cells were fixed with 75% ethanol for 5 min and then stained with 0.5% crystal violet for about 10 min. After washing away the dye, colonies containing more than 50 cells were counted.
MN assay
Measure MN using the cytokinesis blockade technique. First, cells were treated with cytochalasin B at 4 μg/ml for 24–28 h and then fixed with methanol/acetic acid (9:1 v/v) for about 5 min. After drying, the cells were stained with Giemsa and observed under a fluorescence microscope (Lecia, Wetzlar, Germany). Micronuclei present in at least 1000 binuclear cells were counted (Eq. (1)).
Soft agar colony formation assay
The processed cells were passaged for 8–10 days and then digested and counted. Mix 1.2% agarose gel thoroughly with 2×DMEM medium (containing 20% FBS and 2% penicillin/streptomycin) and add to a six-well plate, with 2 mL per well. After it solidifies, add 2 mL of well-mixed 0.6% agarose gel and 2×DMEM medium which contains 10,000 cells to each well. It was placed in the incubator for about 21 days and supplemented with medium every 5 days. Colonies containing more than 50 cells formed in different treatment groups were counted (Eq. (2)). The TF of the other different treatment groups was normalized based on the TF of 0 Gy treatment in the only irradiation group.
RNA collection and sequencing
After SMG, cells were immediately irradiated with 0.3 Gy and 1 Gy carbon ions. Continue to co-culturing irradiated cells with non-irradiated cells for 48 h. After 48 h, all target cells and bystander cells were collected, and the total RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The total RNA samples were sent to Azenta (Suzhou, China) for transcriptomic sequencing. Cutadapt (version 1.9.1) software was used to filter the raw sequencing data, and the adapter sequences or sequences with low-quality 3’ end bases of reads were removed due to excessively long sequencing lengths. Gene expression was calculated by using HTseq software (V0.6.1) and the FPKM (Fragments per kilobases per million reads) method73. Gene differential analysis was performed using the Bioconductor software package DESeq2 (V1.26.0), and the results were screened according to the significance criteria of DEG changes of more than two times and Q value ≤ 0.05.
qRT-PCR
Total RNA was collected from cells using the Trizol reagent. RNA was reverse transcribed into cDNA using HisyGo RT Red SuperMix reagents (Vazyme Biotech, Nanjing, China). Quantitative PCR was performed by using Taq Pro Universal SYBR qPCR Master Mix (Vazyme Biotech, Nanjing, China). PCR amplification was performed by Life Technologies System (Vii7A, Grand Island, NY, USA). Amplified PCR products were quantified and normalized using GAPDH. The data were analyzed using the C(t) value comparison method and normalized by expression levels of the control group in each sample. The primer sequence for gene amplification is displayed as follows:
AREG: 5′-GCACCTGGAAGCAGTAACATGC-3′ (forward),
5′-GGCAGCTATGGCTGCTAATGCA-3′ (reverse);
CDKN1A: 5′-GTGGGGTTATCTCTGTGTTAGGG-3′ (forward),
5′-CCCTGTCCATAGCCTCTACTGC-3′ (reverse);
ETV4: 5′-CCGCTCGCTGCGATACTATT-3′ (forward),
5′-CGGTCAAACTCAGCCTTCAGA-3′ (reverse);
ETV5: 5′-CAGCACACGGGTTCCAGTCAC-3′ (forward),
5′-TGGCAGTTAGGCACTTCTGAATCG-3′ (reverse);
FAS: 5′-AGATTGTGTGATGAAGGACATGG-3′ (forward),
5′-TGTTGCTGGTGAGTGTGCATT-3′ (reverse);
GAPDH: 5′-AGCCACATCGCTCAGACAC-3′ (forward),
5′-GCCCAATACGACCAAATCC-3′ (reverse).
Statistical analysis
Data obtained from at least three independent experiments with at least three parallel samples per experiment were statistically analyzed. Data were expressed as mean ± standard deviation. Statistical analysis was conducted using GraphPad Prism 9.0. Non-paired t test was used to compare the significant differences between two groups, one-way analysis of variance was used to calculate the significant differences between three or more groups, and two-way analysis of variance was used to calculate the interaction between two treatment factors. Python 2017 was used to analyze and predict transcriptomic sequencing and other database results. All analyses with a P value < 0.05 are considered statistically significant.
Data availability
Data are provided within the manuscript or supplementary information files.
Abbreviations
- DEG :
-
differentially expressed genes;
- EGF :
-
epidermal growth factor;
- GI :
-
genomic instability;
- LUAD :
-
lung adenocarcinoma;
- LUSC :
-
lung squamous cell carcinoma;
- MN :
-
micronucleus;
- NTE :
-
non-targeted effects;
- SF :
-
survival fraction;
- SMG :
-
simulated microgravity;
- TE :
-
targeted effects;
- TF :
-
transformation frequency
References
Krittanawong, C. et al. Human health during space travel: state-of-the-art review. Cells 12, 1–32 (2023).
Marazziti, D., Arone, A., Ivaldi, T., Kuts, K. & Loganovsky, K. Space missions: psychological and psychopathological issues. CNS Spectr. 27, 536–540 (2022).
Arone, A. et al. The Burden of space exploration on the mental health of astronauts: a narrative review. Clin. Neuropsychiatry 18, 237–246 (2021).
Patel, Z. S. et al. Red risks for a journey to the red planet: the highest priority human health risks for a mission to Mars. NPJ Microgravity 6, 1–13 (2020).
Rosen, C. J. et al. The mechanosensitive Piezo1 channel is required for bone formation. eLife 8, e47454 (2019).
Ha, P. et al. Bisphosphonate conjugation enhances the bone-specificity of NELL-1-based systemic therapy for spaceflight-induced bone loss in mice. NPJ Microgravity 9, 75 (2023).
Chen, Y., Xu, C., Wang, P., Cai, Y. & Ma, H. Effect of long-term simulated microgravity on immune system and lung tissues in Rhesus macaque. Inflammation 40, 589–600 (2017).
Baevsky, R., Funtova, I. I. & Luchitskaya, S. Role of the right and left parts of the heart in mechanisms of body adaptation to the conditions of long term space flight according to longitudinal ballistocardiography. Acta Astronaut. 178, 894–899 (2021).
Karlsson, L. L., Van Muylem, A. & Linnarsson, D. Lung diffusing capacity for nitric oxide in space: microgravity gas density interactions. Front. Physiol. 14, 1161062 (2023).
Brojakowska, A. et al. Lifetime evaluation of left ventricular structure and function in male ApoE null mice after gamma and space-type radiation exposure. Front. Physiol. 14, 1292033 (2023).
Miller, K. B. et al. Ionizing radiation, cerebrovascular disease, and consequent dementia: a review and proposed framework relevant to space radiation exposure. Front. Physiol. https://doi.org/10.3389/fphys.2022.1008640 (2022).
Guo, Z., Zhou, G. & Hu, W. Carcinogenesis induced by space radiation: a systematic review. Neoplasia32, 100828 (2022).
Suman, S. et al. Relative biological effectiveness of energetic heavy ions for intestinal tumorigenesis shows male preponderance and radiation type and energy dependence in APC1638N/+ mice. Int. J. Radiat. Oncol. Biol. Phys. 95, 131–138 (2016).
Yatagai, F., Honma, M., Dohmae, N. & Ishioka, N. Biological effects of space environmental factors: a possible interaction between space radiation and microgravity. Life Sci. Space Res. 20, 113–123 (2019).
Beheshti, A. et al. Genomic changes driven by radiation-induced DNA damage and microgravity in human cells. Int. J. Mol. Sci. https://doi.org/10.3390/ijms221910507 (2021).
Basirun, C. et al. Microgravity × radiation: a space mechanobiology approach toward cardiovascular function and disease. Front. Cell Dev. Biol. https://doi.org/10.3389/fcell.2021.750775 (2021).
Wang, H. et al. The small protein MafG plays a critical role in MC3T3-E1 cell apoptosis induced by simulated microgravity and radiation. Biochem Biophys. Res. Commun. 555, 175–181 (2021).
Dong, J. et al. The combined effects of simulated microgravity and X-ray radiation on MC3T3-E1 cells and rat femurs. NPJ Microgravity 7, 1–8 (2021).
Tan, S., Pei, W., Huang, H., Zhou, G. & Hu, W. Additive effects of simulated microgravity and ionizing radiation in cell death, induction of ROS and expression of RAC2 in human bronchial epithelial cells. NPJ Microgravity 6, 34 (2020).
Wei, L. et al. Synergistic effects of incubation in rotating bioreactors and cumulative low dose 60Co γ-ray irradiation on human immortal lymphoblastoid cells. Microgravity Sci. Technol. 24, 335–344 (2012).
Canova, S. et al. Modeled microgravity’ affects cell response to ionizing radiation and increases genomic damage. Radiat. Res. 163, 191–199 (2005).
Ikeda, H. et al. Expression profile of cell cycle-related genes in human fibroblasts exposed simultaneously to radiation and simulated microgravity. Int. J. Mol. Sci. 20, 4791 (2019).
Hada, M. et al. Increased chromosome aberrations in cells exposed simultaneously to simulated microgravity and radiation. Int. J. Mol. Sci. 20, 43 (2019).
Xu, Y., Pei, W. & Hu, W. A current overview of the biological effects of combined space environmental factors in mammals. Front. Cell Dev. Biol. 10, 1–10 (2022).
Campa, A., Balduzzi, M., Dini, V., Esposito, G. & Tabocchini, M. A. The complex interactions between radiation induced non-targeted effects and cancer. Cancer Lett. 356, 126–136 (2015).
Suzuki, M., Funayama, T., Suzuki, M. & Kobayashi, Y. Radiation-quality-dependent bystander cellular effects induced by heavy-ion microbeams through different pathways. J. Radiat. Res. 64, 824–832 (2023).
Matsumoto, Y. et al. Dependence of the bystander effect for micronucleus formation on dose of heavy-ion radiation in normal human fibroblasts. Radiat. Prot. Dosim. 166, 152–156 (2015).
Qin, F. et al. Golgi phosphoprotein 3 mediates radiation-induced bystander effect via ERK/EGR1/TNF-α signal axis. Antioxidants 11, 1–13 (2022).
Shao, C., Folkard, M. & Prise, K. M. Role of TGF-Β1 and nitric oxide in the bystander response of irradiated glioma cells. Oncogene 27, 434–440 (2008).
Yang, H., Anzenberg, V. & Held, K. D. The time dependence of bystander responses induced by iron-ion radiation in normal human skin fibroblasts. Radiat. Res. 168, 292–298 (2007).
Cucinotta, F. A. Non-targeted effects and space radiation risks for astronauts on multiple International Space Station and lunar missions. Life Sci. Space Res. 40, 166–175 (2024).
Cucinotta, F. A. & Cacao, E. Non-targeted effects models predict significantly higher Mars mission cancer risk than targeted effects models. Sci. Rep. 7, 1–11 (2017).
Chang, P. Y. et al. Harderian gland tumorigenesis: low-dose and LET response. Radiat. Res. 460, 449–460 (2016).
Bach, D. H., Zhang, W. & Sood, A. K. Chromosomal instability in tumor initiation and development. Cancer Res. 79, 3995–4002 (2019).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Grudzenski, S., Raths, A., Conrad, S., Rübe, C. E. & Löbrich, M. Inducible response required for repair of low-dose radiation damage in human fibroblasts. Proc. Natl. Acad. Sci. USA 107, 14205–14210 (2010).
Simbulan-Rosenthal, C. M. et al. CD133 stimulates cell proliferation via the upregulation of amphiregulin in melanoma. Cells 13, 777 (2024).
Lin, K. et al. Collagen I-induced VCAN/ERK signaling and PARP1/ZEB1-mediated metastasis facilitate OSBPL2 defect to promote colorectal cancer progression. Cell Death Dis. 15, 85 (2024).
Fukazawa, T. et al. Simulated microgravity enhances CDDP-induced apoptosis signal via p53-independent mechanisms in cancer cells. PLoS ONE 14, e0219363 (2019).
Yang, S. et al. ETV4 is a mechanical transducer linking cell crowding dynamics to lineage specification. Nat. Cell Biol. 26, 903–916 (2024).
Ingber, D. E. Tensegrity I. Cell structure and hierarchical systems biology. J. Cell Sci. 116, 1157–1173 (2003).
Yang, F. et al. Fas signaling in adipocytes promotes low-grade inflammation and lung metastasis of colorectal cancer through interaction with Bmx. Cancer Lett. 522, 93–104 (2021).
Messeguer, X. et al. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics18, 333–334 (2002).
Hu, H. et al. AnimalTFDB 3.0: a comprehensive resource for annotation and prediction of animal transcription factors. Nucleic Acids Res. 47, D33–D38 (2019).
Chao, X. et al. A new discovery of STAT4 single nucleotide polymorphisms associated with hepatocellular carcinoma risk in Chinese Han population: a case–control study. Biosci. Rep. 41, BSR20210124 (2021).
Huang, Y. W. et al. Amphiregulin promotes vascular endothelial growth factor-C expression and lymphangiogenesis through STAT3 activation in human chondrosarcoma cells. Cell. Physiol. Biochem. 52, 1–15 (2019).
Carlberg, C. Vitamin D and its target genes. Nutrients 14, 1354 (2022).
Carlberg, C. & Muñoz, A. An update on vitamin D signaling and cancer. Semin Cancer Biol. 79, 217–230 (2022).
Wang, W. et al. G-quadruplexes promote the motility in MAZ phase-separated condensates to activate CCND1 expression and contribute to hepatocarcinogenesis. Nat. Commun. 15, 1045 (2024).
Wang, X. et al. YY1 restrained cell senescence through repressing the transcription of p16. Biochim Biophys. Acta Mol. Cell Res. 1783, 1876–1883 (2008).
Tan, Y., Chen, Y., Du, M., Peng, Z. & Xie, P. USF2 inhibits the transcriptional activity of Smurf1 and Smurf2 to promote breast cancer tumorigenesis. Cell Signal 53, 49–58 (2019).
Chen, M., Li, X., Du, B., Chen, S. & Li, Y. Upstream stimulatory factor 2 inhibits erastin-induced ferroptosis in pancreatic cancer through transcriptional regulation of pyruvate kinase M2. Biochem. Pharmacol. 205, 115255 (2022).
Aceto, N. et al. Co-expression of HER2 and HER3 receptor tyrosine kinases enhances invasion of breast cells via stimulation of interleukin-8 autocrine secretion. Breast Cancer Res. 14, 1–11 (2012).
Freed, D. M. et al. EGFR ligands differentially stabilize receptor dimers to specify signaling kinetics. Cell 171, 683–695.e18 (2017).
Yun, C. et al. Essential role of Her3 in two signaling transduction patterns: Her2/Her3 and MET/Her3 in proliferation of human gastric cancer. Mol. Carcinog. 54, 1700–1709 (2015).
Tang, X. et al. Intercellular adhesion molecule 2 as a novel prospective tumor suppressor induced by ERG promotes ubiquitination-mediated radixin degradation to inhibit gastric cancer tumorigenicity and metastasis. J. Transl. Med. 21, 670 (2023).
Ren, Z. et al. Biomanufacturing of 3D tissue constructs in microgravity and their applications in human pathophysiological studies. Adv. Healthc. Mater. 12, 2300157 (2023).
Winkelmaier, G. et al. Influence of simulated microgravity on mammary epithelial cells grown as 2D and 3D cultures. Int. J. Mol. Sci. 24, 7615 (2023).
Zhu, L. et al. Attenuation of antiviral immune response caused by perturbation of TRIM25-mediated RIG-I activation under simulated microgravity. Cell Rep. 34, 108600 (2021).
Baghoum, H. et al. Simulated microgravity influences immunity-related biomarkers in lung cancer. Int. J. Mol. Sci. 24, 155 (2023).
Crucian, B. E. et al. Immune system dysregulation during spaceflight: potential countermeasures for deep space exploration missions. Front. Immunol. 9, 1437 (2018).
Yuan, M. et al. Integrative analysis of regulatory module reveals associations of microgravity with dysfunctions of multi-body systems and tumorigenesis. Int. J. Mol. Sci. 21, 1–17 (2020).
Tan, X. et al. Simulated microgravity inhibits cell focal adhesions leading to reduced melanoma cell proliferation and metastasis via FAK/RhoA-regulated mTORC1 and AMPK pathways. Sci. Rep. 8, 3769 (2018).
Luitel, K., Siteni, S., Barron, S. & Shay, J. W. Simulated galactic cosmic radiation-induced cancer progression in mice. Life Sci. Space Res.41, 43–51 (2024).
Cucinotta, F. A. & Durante, M. Essay cancer risk from exposure to galactic cosmic rays: implications for space exploration by human beings. Lancet Oncol. 7, 431–435 (2006).
Zeitlin, C. et al. Measurements of energetic particle radiation in transit to Mars on the Mars Science Laboratory. Science 340, 1080–1080 (2013).
Heeran, A. B., Berrigan, H. P. & O’Sullivan, J. The radiation-induced bystander effect (RIBE) and its connections with the hallmarks of cancer. Radiat. Res. 192, 668–679 (2019).
Autsavapromporn, N. et al. Genetic changes in progeny of bystander human fibroblasts after microbeam irradiation with X-rays, protons or carbon ions: the relevance to cancer risk. Int. J. Radiat. Biol. 91, 62–70 (2015).
Singh, S. S. et al. Amphiregulin in cellular physiology, health, and disease: potential use as a biomarker and therapeutic target. J. Cell Physiol. 237, 1143–1156 (2022).
Busser, B., Sancey, L., Brambilla, E., Coll, J. L. & Hurbin, A. The multiple roles of amphiregulin in human cancer. Biochim Biophys. Acta Rev. Cancer 1816, 119–131 (2011).
Son, S. S. et al. In vivo silencing of amphiregulin by a novel effective self-assembled-micelle inhibitory RNA ameliorates renal fibrosis via inhibition of EGFR signals. Sci. Rep. 11, 1–14 (2021).
Wang, D. et al. A method for the evaluation of microgravity biaxial clinostats [P]. CN 101726426 A, 2012-12-12.
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L. & Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 5, 621–628 (2008).
Acknowledgements
The authors thank the Shanghai Proton and Heavy Ion Center (SPHIC) for help with carbon ion radiation. This work was funded by the National Natural Science Foundation of China under Grant Nos. 32071243, 82192882, 82192883 and a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). The funder played no role in the study design, data collection, analysis and interpretation of data, or the writing of this manuscript.
Author information
Authors and Affiliations
Contributions
Y.D. wrote the main manuscript text and completed the data acquisition and analysis. Y.X., X.W., M.Z., and Q.Z. conducted experiments to help the investigation. C.Y. substantively revised the text. G.Z. provided funding support. WH provided conceptualization, funding support and substantively revised the text. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ding, Y., Xu, Y., Wang, X. et al. Microgravity alleviates low-dose radiation-induced non-targeted carcinogenic effects. npj Microgravity 11, 26 (2025). https://doi.org/10.1038/s41526-025-00484-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41526-025-00484-x