Abstract
Parkinson’s disease (PD) is a devastating neurodegenerative disorder with growing prevalence worldwide and, as yet, no effective treatment. Drug repurposing is invaluable for detecting novel PD therapeutics. Here, we compiled gene expression data from 1231 healthy human brain samples and 357 samples across tissues, ethnicities, brain regions, Braak stages, and disease status. By integrating them with multiple-source genomic data, we found a PD-associated gene co-expression module, and its alignment with the CMAP database successfully identified drug candidates. Among these, meclofenoxate hydrochloride (MH) and sodium phenylbutyrate (SP) are indicated to be able to prevent mitochondrial destruction, reduce lipid peroxidation, and protect dopamine synthesis. MH was validated to prevent neuronal death and synaptic damage, improve motor function, and reduce anhedonic and depressive-like behaviors of PD mice. The interaction of MH with a PD-related protein, sigma1, was confirmed experimentally. Thus, our findings support that MH potentially ameliorates PD by interacting with sigma1.
Similar content being viewed by others
Introduction
Parkinson’s disease (PD) is a chronic neurodegenerative movement disorder characterized by a large number of motor symptoms, including resting tremor, rigidity and postural instability. The non-motor symptoms of PD include autonomic, psychiatric, sensory and cognitive impairments, as well as dementia1. As one of the most common neurodegenerative disorders, PD affects 2–3% of 65-year-olds and is responsible for more than 100,000 deaths worldwide each year2. PD can be broadly categorized into two genetic forms monogenic PD, caused by mutations in single genes (5–10%), and complex forms of PD resulting from the interplay between multiple genetic risk factors and environmental influences (90–95%). The heritability of PD is approximately 27–34%3,4,5. However, our understanding of the etiology of PD remains incomplete. Thus, treatments for PD are still limited in their efficacy, e.g., dopamine replacement therapy, the most commonly used therapeutic strategy for PD, is capable of improving clinical symptoms but is unable to halt disease progression6.
Thanks to decades of research, it is clear that PD exhibits considerable locus heterogeneity7; an increasing number of disease genes/pathogenic mutations are being identified in PD patients by means of whole genome sequencing (WGS) or whole exome sequencing (WES) studies8, and both autosomal recessive and dominant forms have been described among the monogenic forms of PD. Such marked locus heterogeneity underlying PD not only represents a major obstacle in identifying common disease mechanisms, but also restricts our options for appropriate therapeutic intervention. This notwithstanding, many mutations have been repeatedly identified among PD patients9,10,11, thereby linking the familial and sporadic forms of PD mechanistically12,13. Thus, mutations in the α-synuclein (SNCA) gene appear to be involved in both the familial and sporadic forms of PD; SNCA function/homeostasis is modulated by various contributory risk factors for PD, including oxidative stress, mitochondrial dysfunction, post-translational modifications, and concentrations of fatty acids14,15. It has therefore been reasoned that perturbation of the molecular networks involving multiple genes might commonly underlie the pathogenesis or progression of PD, and that the elucidation of these networks could facilitate the future development of therapeutic interventions. So far, the global effort toward this goal has led to the establishment of multiple valuable sources of information that have facilitated the compilation of such gene networks16,17. Instead of focusing on single genes, considerable emphasis has been placed on utilizing data-driven frameworks at the system or network level to generate biologically/clinically meaningful gene modules comprising sets of functionally associated genes whose homeostasis may be altered by specific pathophysiological events15,16,17,18,19. For instance, the weighted gene co-expression network analysis (WGCNA) and Differential Co-expression (DiffCoEx) are designed to identify gene co-expression modules by analyzing gene expression through coefficient test18,19. The application of such an approach has led to the identification of network modules that are implicated in neurodevelopmental processes, metabolism, and the immune system20. An analysis of GEO data of PD (n = 128) identified modules associated with RNA metabolism pathology as a potential cause of PD by sorting differentially active pathways between brain transcriptomics samples from PD patients and controls21,22. However, these studies were generally based on patient gene expression data, and may have been biased due to insufficient numbers of samples and inter-patient heterogeneity.
One important application of the molecular network is in drug repurposing. Drug repurposing represents an attractive avenue in drug discovery due to its relatively low cost and fewer safety concerns. By definition, drug repurposing is designed to redirect new or additional indications for three kinds of therapeutic molecules i.e., drugs approved for a particular indication, drugs that have already been well-characterized during their clinical development and accompanied by thorough post-market surveillance data, and drugs which have undergone some clinical development but were subsequently abandoned23,24. Often, biological networks combined with Genome-Wide Association Studies (GWAS) are the most commonly employed sources of information for drug repurposing, as GWAS studies are intended to impartially link controlled factors to genetic or transcriptomic alterations in human subjects with no specific emphasis on a single gene or fixed set of genes25,26. Thus, developing a method for detecting the gene network perturbations caused by PD-associated variants through combining large-scale human genomic data, including functional interactions between genes from healthy humans, is emerging as a useful approach to drug repurposing.
To test the effects of medicines for PD, an animal model is often used. The most common PD model involves a neurotoxin approach, such as the rotenone-induced PD model27,28,29. From 2000 onwards, researchers used rotenone to create a PD animal model, and it has been proven to be very informative. However, there are limitations of the rotenone-based model, which are low reproducibility and acute toxicity30,31. The chemical-induced models, like rotenone, may not fully recapitulate the genetic diversity observed in PD patients. In contrast, genetic models, such as those involving mutations in SNCA, LRRK2, or PINK1, specifically replicate familial forms of PD but may not capture the environmental factors implicated in sporadic cases. Thus, the chemical models may lack genetic relevance, while genetic models may not reflect the complexity of sporadic PD32. From 2000 onwards, researchers used rotenone to create a PD animal model33, and it has proven to be very informative. It is thought to cause dopaminergic degeneration by inducing oxidative stress, as well as inducing in vivo aggregation of α-synuclein, which is the major component of Lewy bodies34. Recently, Ahn et al. constructed a rotenone-induced PD mouse model in order to explore the role of δ-secretase in cleaving both α-Syn at N103 and Tau at N36835. Moreover, multiple studies have used the rotenone-induced mouse model in the study of PD-targeted medicines. For example, Liu et al. have investigated the protective effects of piperlongumine in rotenone-induced PD cell and mouse models36. Another study that used the rotenone-induced C57Bl/6 J mouse model indicated the potential role of anle138b in the treatment of PD37.
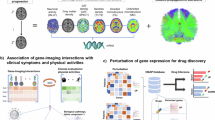
To explore drug repurposing for Parkinson’s Disease (PD), we developed a computational framework called iGOLD, which integrates multi-source genomic data with gene co-expression modules (Fig. 1). This framework (available at https://github.com/fanc232CO/iGOLD_pipline) was used to identify gene co-expression modules affected by PD-related genes and SNPs. We evaluated the gene co-expression modules that were significantly enriched in PD-associated genes and SNPs through conservation analysis using seven gene expression datasets spanning various ethnicities, brain regions, tissues, Braak stages, and PD disease status. The highly conserved modules were then utilized for drug discovery. Subsequent experiments involved rotenone-induced primary neuronal cells and a mouse model to assess the efficacy of the identified drugs in promoting neuronal survival, enhancing hippocampal function, and modifying PD-related behaviors. Finally, we conducted additional experiments focused on mitochondrial functions and metabolic factors to elucidate the specific mechanisms through which these drugs exert their effects.
Step 1: constructed gene co-expression modules using 1231 healthy human samples from ten brain regions. CCM—concurrently co-expressed modules expressed in ten brain regions, SCM--brain region-specific co-expressed modules. Step 2: from the constructed gene co-expression modules, we selected the PD-associated module through enrichment of PD-associated genes, enrichment of PD-associated SNPs (by Chi-square test and stratified LDSC (sLDSC) analysis, respectively), and fraction of PD-associated differential expression genes. DEGs differentially expressed genes. Step 3: testing the conservation of the gene co-expression relationships in the validated modules across ethnicities, tissue, and disease development stages. Step 4: inside the selected PD-associated module, the differentially expressed genes in the PD patients were used for drug discovery with the Connectivity Map (CMAP). The enrichment of up-regulated and down-regulated genes by the drug-induced gene expression profiles was tested, and drugs that reverse the differential gene expression in PD were considered as the lead compound candidates.
Results
Overview of the study
Here, we first developed a computational architecture integrating multiple-source genomic data with gene co-expression modules for drug repurposing (iGOLD) for drug repurposing. The source code of iGOLD is available at (https://github.com/fanc232CO/iGOLD_pipline). As shown in Fig. 1, it comprises by four main steps: (1) using gene expression data of 1231 healthy human brain samples across ten brain regions38,39,40 to construct gene co-expression modules associated with normal brain functions (Table 1); (2) identifying the co-expression modules enriched with disease-associated genes, SNPs and genes expressed significantly different in patients and controls; (3) examining the conservation of the selected co-expression modules in the gene expression data from brain tissues across different brain regions, disease status, and ethnicities, and in the gene expression data from blood and multiple other cell types; (4) aligning the highly conserved modules to the gene expression profiles perturbed by small molecular compounds in CMAP database41,42, and identifying the gene expression profiles enriched in genes from the conserved modules. The small molecular compound was considered a drug candidate. The drug candidate was further validated by primary neurons and mouse models. The binding targets of the candidate drugs were predicted by DStruBTarget43. The interactions between the candidate drug and the drug target were validated experimentally.
The gene co-expression modules in hippocampi and substantia nigra as being associated with PD
Using the gene expression data from the healthy human brain, iGOLD constructed 19 concurrently co-expressed modules expressed in ten brain regions (CCM) (Supplementary Table 1), and 68 modules (brain region-specific co-expressed modules, SCM) specifically expressed in one of the ten brain regions but not in the other nine brain regions (Supplementary Fig. 1). The functional similarity of these modules was then evaluated by determining the number of overlapping genes between each pair of modules expressed in two brain regions. The width of the line in Supplementary Fig. 1 represents the significance of the number of overlapping genes (Chi-square test) between one pair of modules from different brain regions compared to the modules from other brain regions (Supplementary Table 2).
Among these modules, one CCM module, M3, and four SCM modules, BR7M4 (Substantia Nigra), BR9M3 (Thalamus), BR6M3 (Putamen) and BR3M2 (Hippocampus), were suggested as enriched (FDR < 0.05) with both PD-associated genes and PD-associated SNPs (PFisher’s exact test < 0.05 and PsLDSC < 0.05) (Supplementary Fig. 2, Table 1, and Supplementary Tables 1 and 3–5). The principle of selecting SCM modules includes: (1) PD-associated genes/SNPs enriched in the module, (2) the number of PD-associated genes in the module is larger than five, and (3) the module size (number of co-expressed genes inside this module) is less than 3000. A supplementary Excel file (https://github.com/fanc232CO/iGOLD_pipline /tree/main/supplementary_material/Module_enrichment_details.xlsx) is provided to show the significance of the enrichment and the number of PD-associated genes in the module. Accordingly, four SCM modules were selected, which are BR7M4, BR9M3, BR6M3, and BR3M2. We further examined the enrichment of DEGs in these five modules (Fig. 2A). The DEGs were obtained from two GEO gene expression datasets44, GPL96 and GPL97 (Supplementary Table 6). The DEGs in GPL96 and GPL97 were respectively termed GPL96-DEGs and GPL97-DEGs. The proportions of GPL96-DEGs and GPL97-DEGs in BR7M4 are significantly higher than in other modules (Fig. 2A and Supplementary Fig. 4), indicating that the BR7M4 module might best describe the gene expression profile characteristic of PD.
A Enrichment of PD-associated genes and SNPs, and the proportion of differentially expressed genes (DEGs) in co-expression modules that are most likely associated with PD compared to the other modules, specifically expressed in the same brain region. From the outer ring to the inner, the circles sequentially represent the brain regions, the module names, the fraction of DEGs (dark green for GPL96 and light green for GPL97) in the co-expression modules, the enrichment of PD-associated genes by the co-expression modules, the enrichment of PD-associated SNPs by the co-expression modules tested by Chi-square analysis, and the heritability enrichment of PD-associated SNPs tested by sLDSC analysis. TC temporal cortex, TM thalamus, WM white matter, CC cerebellar cortex, FC frontal cortex, HC hippocampus, MD medulla, OC occipital cortex, PM putamen, SN substantia Nigra. B Conservation of the BR7M4 genes in three brain regions of Japanese samples across different Braak stages, including Braak 0, Braak I–II, Braak III–IV, and Braak V–VI. Conservation of BR7M4 genes expressed in brain regions, IPSC-induced dopaminergic neurons, and peripheral blood of PD patients and healthy controls. Gene co-expression conservation of BR7M4 module in the brain regions of hippocampus and substantia nigra, respectively. Red dashed line—high conservation Z summary cutoff of 10. Cyan dashed line—medium conservation Z summary cutoff of 2. EC entorhinal cortex, FC temporal cortex, TC frontal cortex, PM putamen, LC locus coeruleus, iPSC IDN IPSC-induced dopaminergic neurons, PB peripheral blood, SN substantia nigra, HC hippocampus. C Interactions between BR7M4-novel genes and known PD-associated genes. BR7M4-novel genes overlapping with Mouse-DEGs are filled in red. Node size represents the significance of genes in RNA-seq analysis from the ROT group and the NC group. The edge between the two genes represents their expression correlation less than 0.85 (scored by WGCNA), and genes linked by them are highlighted as triangles edged in black. D The gene expression profile of the NC group, ROT group, ROT + MH group, and ROT + SP group. E The gene expression profile was obtained by analyzing GPL96 data.
The associations between BR7M4 and PD were tested using seven unrelated publicly available brain expression datasets38,45,46,47,48,49 that together cover gene expression information across different ethnic backgrounds, brain regions, tissues, and disease status (Supplementary Table 7). As shown in Fig. 2B, module BR7M4 displays medium conservation in the entorhinal cortex (EC), frontal cortex (FC), and temporal cortex (TC) at four Braak NFT stages (0, I–II, III–IV, and V–VI) in Japanese samples. We examined the conservation of BR7M4 in putamen, locus coeruleus and IPSC-induced dopaminergic neurons using Spanish samples, and found that the BR7M4 module exhibits medium conservation in the putamen, high conservation in the locus coeruleus, and medium conservation in IPSC-induced dopaminergic neurons (Fig. 2B). When the BR7M4 module was tested in both hippocampus38 and substantia nigra samples38 from Europeans, it exhibited high conservation in both brain regions (Fig. 2B). By contrast, BR7M4 showed low conservation in the peripheral blood of American samples (Fig. 2B). Thus, unsurprisingly, the PD-associated functions of the BR7M4 module appear to be expressed through brain regions (e.g., substantia nigra and hippocampus) and dopaminergic neurons rather than through peripheral blood.
BR7M4 enriched with PD-associated SNPs from GCST007780 (P-value = 0.024) by LDSC analysis, and from GCST007780 (P-values = 0.041) and GCST010765 (P-values = 0.020) by Chi-square analysis (Supplementary Fig. 2). Compared to other modules, BR7M4 is significantly (PFDR-adjust < 0.05) enriched with genes and SNPs (P-value < 0.05) from the largest number of resources. Thus, we choose BR7M4 for further validation.
Figure 2B is to shows the conversation scores of the module BR7M4 across tissues from multiple brain regions. The conservation score is to represent the enrichment of genes expressed in specific tissues and is estimated by modulePreservation, a function in the WGCNA R package50. The higher score (Zsummary)means higher conservation, and a Zsummary higher than 10 is suggested as highly conserved. When we performed the conservation analysis for module BR7M4 across the tissues from multiple brain regions of health controls, the module BR7M4 has shown the highest conservation in Hippocampus (HC) (Zsummary = 10.53) and Substantia Nigra (SN) (Zsummary = 33.93). In comparison, it has shown conservation in putamen (Zsummary = 7.98), locus coeruleus (Zsummary = 10.35) and IPSC-induced dopaminergic neurons (Zsummary = 1.85) using Spanish samples(Fig. 2B). The lack of significance in the brain regions could be attributed to their relatively lower expression of genes involved in the core pathological processes of PD.
The module BR7M4 contained 399 genes whose interactions are shown in Supplementary Figure 5. The association of BR7M4 with PD was examined using hippocampal samples from mice since the expression of BR7M4 is highly conserved in the hippocampus (Fig. 2B, Zsummary = 10.53). We performed RNA sequencing (RNA-seq) on the hippocampi of ten mice into two groups of mice: DMSO (NC; n = 5) and ROT (rotenone-induced group; n = 5) (Supplementary Table 8). The ROT-induced C57 L/J mouse model can recapitulate many features of human PD, including anatomical, neurochemical, behavioral, and neuropathological features35,51,52. RNA-seq data analysis identified 2195 genes (Mouse-DEGs) that were expressed significantly [adjusted P < 0.05, absolute value of fold-change (FC) greater than 2] differently between the NC and ROT groups. Among the Mouse-DEGs, 50 genes were present in the BR7M4 module (termed Mouse-DEGs-Mod), which is significantly (single-tailed binomial test P = 0.016) more than the genes that were not expressed significantly differently between the NC and ROT groups (termed Mouse-non-DEGs) (Supplementary Fig. 5). Moreover, 39 of the Mouse-DEGs-Mod genes were not PD-associated genes. Nevertheless, the expression levels of these 39 genes were found to be closely correlated (WGCNA TOM similarity > 0.15) with those of 41 PD-associated genes in BR7M4 (Fig. 2C). Thus, the co-expression module BR7M4 is strongly associated with PD.
In this study, we have used the gene expression data from different ethnicities and tissues to evaluate the module conservation. To evaluate the gender effects on the module conservation, we first divided the samples into male and female groups to perform the module conservation analysis. The result indicated that the module conservation (Zsummary) in the male group (905 samples) is 18.44, and in the female group (326 samples) is 18.91. According to the widely agreed standard that Zsummary > 10 indicates high module conservation, this result suggests that gender does not play a vital role in module conservation in this study. As to the age factor, because most of the samples are from elder PD patients, the module conservation analysis has not been performed to check the influence of age.
Meclofenoxate hydrochloride (MH) and sodium phenylbutyrate (SP) restore the normal expression levels of PD-associated genes via different mechanisms
From BR7M4, we extracted DEGs for the discovery of PD candidate therapeutics (Supplementary Table 9), from which two drugs, SP (connectivity score −0.963 and ranked in top 0.05% of 6100 drugs) and MH (connectivity score of −0.814 and ranked in top 0.2% of 6100 drugs), were considered for further validation since they were not only top-ranking candidates but were also able to pass through the blood brain barrier. We have included the druglike and ADME data of the potential candidates of Supplementary Table 9 in a separate supplementary Excel file (“https://github.com/fanc232CO/iGOLD_pipline/tree/main/supplementary_material/candidates_druglike_ADME.xlsx”). Both the druglike and the ADME were predicted using the model of CMPNN (Communicative Message Passing Neural Network, https://github.com/SY575/CMPNN)53. The CMPNN predicted 52 parameters to evaluate the drugs, as shown in the file candidates_druglike_ADME.xlsx. The drug candidates were ranked by their connectivity scores. From them, we selected those (absolute values of connectivity scores higher than 0.8) ranked in the top 14 are shown in Supplementary Table 9. Among them, Cyanocobalamin, SC-58125, Dexamethasone, and Rofecoxib are well-studied PD drugs, suggesting the reliability of our method in identifying drugs for PD. Out of the remained drug candidates, SP and MH have been reported as having the ability to pass through the blood-brain barrier, while Carteolol has been explicitly with low penetrability to pass the brain-blood barrier54. Thus, SP and MH were selected for further validation. To assess the impact of MH or SP on PD-associated gene expression, we performed RNA-sequencing on the hippocampi of mice from the NC, ROT, ROT + SP, ROT + MH, SP, and MH groups (Supplementary Table 8). We found 91 genes to be expressed significantly (Bonferroni-corrected P < 0.05 and |log(FC)| > 2) differently between the ROT group and the NC group, as well as between the ROT + SP group and the ROT group. These genes were termed the SP-ROT set. Meanwhile, we found 666 genes that were expressed significantly (Bonferroni-corrected P < 0.05 and |log(FC)| > 2) differently between the ROT group and the NC group, as well as between the ROT + MH group and the ROT group. These genes were termed the MH-ROT set. Among them, 28 were in the BR7M4 module and displayed the same direction of regulation as the GPL96 dataset. The expression of these genes in the ROT group, the NC group, the ROT + MH group and the ROT + SP group are shown in Fig. 2D. The expression of these genes in the GPL96 dataset is shown in Fig. 2E. The expression profile of these gene in controls in GPL96 is similar to that of the NC group, whereas the gene expression profile of the PD individuals in GPL96 is similar to that of the ROT group (Fig. 2D, E). Thus, after MH or SP treatment, gene expression in the ROT group was restored such that it approximated the characteristics of the NC group, suggesting a specific effect of MH or SP in remodeling the expression pattern of PD-associated genes.
Of the genes in the MH-ROT set, 129 genes were not in the SP-ROT set, whilst 74 genes from the SP-ROT set were not in the MH-ROT set, which were then termed the Uni-MH-ROT set and Uni-SP-ROT set, respectively. A STRING analysis was performed to detect the networks of protein-protein interactions (PPIs) in the Uni-MH-ROT set and the Uni-SP-ROT set, respectively. The PPIs of the Uni-MH-ROT genes were mainly enriched in synapse-related functions (Supplementary Fig. 6A), whereas the PPIs of the Uni-SP-ROT group were enriched in mitochondrial electron transport and mitochondrial respiratory chain complex I assembly functions (Supplementary Fig. 6B). Thus, the effect of MH on gene transcription is potentially distinguishable from that of SP in terms of its modulatory effect on genes with synapse-related functions in murine hippocampus.
The potential targets of MH and SP were further examined by DStruBTarget55 to predict those proteins that could directly bind to MH or SP (Supplementary Material, Supplementary Table 10, and Supplementary Fig. 7). All these proteins in Supplementary Table 10 are predicted as binding with MH by the DStruBTarget model that has been developed based on the fusion of protein-drug interaction and ligand similarity methods. DStruBTarget indicated that the top ten predicted proteins binding to MH were enriched in neuroactive ligand-receptor interactions and neurotransmitter receptor activity functions (P = 1.3 × 10−7), whereas the top 10 DStruBTarget predicted proteins binding to SP were enriched in inflammation-related functions (Supplementary Table 11). Thus, MH and SP may bind to different targets for restoring the normal expression levels of PD-associated genes in the hippocampi of mice. Among the predicted binding targets, DRD4, 5-HT1A, 5-HT2A, Sigma1 (σ1), PPARG, CNR1 and CNR2 have been reported to be PD associated by previous studies56,57,58,59,60,61,62. The target proteins of MH have not been reported anywhere, and require further experimental validation. The predicted MH-protein interactions, if validated, may at least partially underlie the protective effect of MH in treating PD.
Both SP and MH protect neurons against ROT-induced neurodegeneration
The neuronal nuclear protein (NeuN) is often used as a positive marker for the functional state of postmitotic neurons. Thus, the NeuN-positive rate of neurons is usually used to assess neurodegeneration63,64,65. Here, immunohistochemical (IHC) staining with anti-NeuN was performed on the dentate gyrus (DG), dentate gyrus2 (DG2), and cornu ammonis (CA1) of the hippocampus from six groups of mice (NC, ROT, SP, ROT + SP, MH, and ROT + MH), with four mice in each group. As shown in Fig. 3A, B, the average relative numbers of NeuN-negative cells [quantified by ImageJ66] (29.5%) increased by 27.9% in the ROT group as compared to those of the NC group (1.6%) (P = 2.7 × 10−3). In the SP + ROT and MH + ROT groups, the average relative numbers of NeuN-negative cells (3.1% for SP treatment and 2.4% for MH treatment) were respectively reduced by 26.4% and 27.1% (P = 1.9 × 10−3 and P = 1.3 × 10−2, respectively) compared to the ROT group (Fig. 3A, B). The average relative numbers of NeuN-negative cells in the DG2 structure of the hippocampus in the ROT group (30.6%) increased by 26.7% compared to the NC group (3.9%) (P = 1.5 × 10−2). In the SP + ROT and MH + ROT groups, the average relative number (5.2% and 4.6%, respectively) of NeuN-negative cells in DG2 was reduced by 25.4% (P = 2.3 × 10−2) and 26.0% (P = 3.6 × 10−2), respectively, compared to the ROT group (Fig. 3A, C). In the CA1 substructure of the hippocampus, the average relative number (54.7%) of NeuN-negative cells in the ROT group increased by 50.9% compared to the NC group (3.9%) with P = 1.7 × 10−2. The average relative numbers of NeuN-negative cells of ROT + SP (3.0%) and ROT + MH (3.1%) in the CA1 substructure of the hippocampus were reduced by 51.7% and 51.7%, respectively (P = 1.8 × 10−2 and P = 3.5 × 10−2) compared with the ROT group (Fig. 3A, D). Thus, MH and SP treatments reduce the number of NeuN-negative cells in different parts of the hippocampus.
A IHC representation of NeuN in the DG, DG2, and CA1. Magnification 20×. Scale bar = 10 μm. B The relative number of NeuN-negative cells in the DG structure of the hippocampus in different groups. C The relative number of NeuN-negative cells in the DG2 structure of the hippocampus in different groups. D The relative number of NeuN-negative cells in the CA1 structure of the hippocampus in different groups. E IHC representation of TH positive cells in the substantia nigra striatum region of the experimental mice (N ≥ 3 mice/group). TH-positive cells were reduced in the ROT-induced mice, and increased in the MH-treated mice as compared with the NC. Magnification 10×. Scale bar = 50 μm. F The TH-positive cells in the ROT group were significantly lower than those in the NC group. SP treatment and MH treatment can increase the TH-positive cells in the ROT-induced group. G GFAP in the hippocampus of each group. The number of GFAP-labeled astrocytes was significantly increased in the ROT group as compared to the NC group. The number of GFAP-labeled astrocytes was significantly reduced in the ROT + SP-treated mice and ROT + MH-treated mice as compared to the ROT group. Magnification 10×. Scale bar = 50 μm. H The relative number of GFAP-labeled astrocytes in the hippocampus structure in different groups. I Fluorescence image of glucose metabolism capacity shown by PET of the mouse brain. J The average change of SUVs in each group. K The maximum change of SUVs in each group. SP and MH prevented ROT-induced neurodegeneration. [18F]-FDG 18F-fluorodeoxyglucose, α-SYN α-synuclein group, eGFP enhanced green fluorescent protein, PET positron emission tomography, A anterior, P posterior, L left, R right. *P < 0.05, **P < 0.01, ***P < 0.001.
IHC analyses were performed to examine the presence of tyrosine hydroxylase (TH) positive cells in the substantia nigra tissues of the mice, as TH is generally considered as an indicator of dopamine production in neurons. As shown in Fig. 3E, F, the number of TH-positive cells in the ROT group (19.50) was significantly lower than in the NC group (52.80) (P = 3.1 \(\times \,10\)−3). Similarly, in the ROT + MH group, the number of TH positive cells was 56.83, nearly 3 times higher than in the ROT group (P = 4.0 \(\times \,10\)−4) (Fig. 3F). In the MH group, the number of TH positive cells was 64.00, which was nearly 2-fold higher than in the ROT + SP group (36.83) (P = 3.6 \(\times \,10\)−2). The number of TH-positive cells in the MH group was comparable to the NC group, significantly higher than in the ROT group (P = 1.0 \(\times \,10\)−4). Although SP treatment increased the number of TH positive cells, it did not significantly improve the damage to the substantia nigra. In contrast, MH treatment improved the dopamine-producing capacity of neurons.
Subsequently, IHC staining with anti-GFAP (glial fibrillary acidic protein) was also performed to visualize the intermediate filament (IF) protein expressed in numerous cell types of the central nervous system (CNS) including astrocytes and ependymal cells, with the number of GFAP-positive (GFAP+) cells serving as an indicator for the activation of the neuroinflammatory pathway in the murine hippocampus (Fig. 3G). As shown in Fig. 3G, the proportion of GFAP+ cells was markedly increased in the hippocampus of the ROT group (50.17 ± 9.88) compared to that in the NC group (25.50 ± 6.80) (P = 3.83 \(\times \,10\)−7). By contrast, the number of GFAP+ cells in the ROT + SP and ROT + MH groups was significantly lower than that of the ROT group [SP: 29.08 ± 4.81 (P = 1.16 \(\times \,10\)−6) and MH: 26.89 ± 8.87 (P = 2.23 \(\times \,10\)−5)]. However, little or no difference was observed in terms of the number of GFAP+ cells between the ROT + SP and ROT + MH groups (Fig. 3H). Similar results were obtained for the interleukin 1 complex (IL-1), another proinflammatory cytokine, and GFAP, in the murine striatum (Supplementary Fig. 8). Taken together, it is clear that both MH and SP repress ROT-induced neuroinflammation in the hippocampus, suggesting an anti-inflammatory effect of these drugs.
MH and SP both upregulate glucose metabolism in the brains of ROT-induced PD mice
To measure glucose metabolism in mouse brains, mice from both the NC and ROT groups (Supplementary Table 8) were subjected to neuro-imaging through [18F]-fluorodeoxyglucose positron emission tomography (18F-FDG PET) (Supplementary Materials). The cross-sectional small animal PET images of mice from the six groups are presented in Fig. 3I and are quantified in Fig. 3J. In Fig. 3J, the average and maximum standardized uptake values (SUV) of the ROT group are 1.19 and 1.30 which were decreased by 0.18 (P = 2.3 \(\times 10\)−2) and 0.18 (P = 3.2 \(\times \,10\)−2), respectively compared to the NC group (average SUV = 1.37 and maximum SUV = 1.48) (Fig. 3K), indicating higher intensity of [18F]-FDG uptake in the ROT-induced group than in the control group (NC). These data clearly indicate that ROT treatment markedly down-regulated glucose metabolism of the murine neurons. For the mice in the SP + ROT group, the average SUV was 1.48 (Fig. 3J), whilst the maximum SUV value was 1.62 (Fig. 3K), which were decreased by 0.28 (24.4%, P = 2.67 \(\times \,10\)−6) and 0.32 (24.6%, P = 1.67 \(\times \,10\)−6), compared with the ROT group. For the mice in the ROT + MH group, the average SUV value was 1.48, with a maximum SUV value of 1.54, 0.32 (24.6%) and 0.24 (18.5%) higher than for the ROT group (P = 5.04 \(\times \,10\)−5 and (P = 5.29 \(\times \,10\)−4), respectively (Fig. 3J, K). Thus, treatment with either SP or MH appears to significantly promote glucose metabolism in neuronal cells in ROT-induced PD mice.
Validating the action of SP and MH in preventing ROT-induced cell damage and preserving neuronal cell morphology
We then tested the potential effects of SP and MH on the survival and morphology of cells in primary neuron culture. The proportions of viable cells, as well as the relative volume of cell bodies, were calculated by ImageJ66. The volumes of the neuronal cell bodies appeared to shrink to 25.9% of the NC in the presence of ROT (Fig. 4Aa, b). After SP and MH treatment, the volumes of the neuronal cells increased by 4.78 (P = 7.9 \(\times \,10\)−3) and 6.59 (P = 5.1 \(\times \,10\)−3), respectively, compared to the ROT group (Fig. 4Ab). The average number of surviving cells in the ROT-induced group (9.14) was significantly lower than in the NC group (154.43) (P = 6.2 \(\times \,10\)−4) (Fig. 4B). Remarkably, the treatment of the ROT-induced cells with either SP or MH increased cell survival from 9.14 to 63.71 or 47.43, respectively (Fig. 4B), strongly indicating that SP and MH has the potential to protect neurons from ROT-induced damage.
A a: Representative scans from immunocytochemical preparations acquired with 4× and 100× objective lenses. SP protects NeuN+ and mouse neurons against the deleterious effects of ROT with an ensuing increase in NeuN and tubulin expression in PD-containing neurons. Insets correspond to high magnification images. Data are representative of 20–40 neurons per group obtained from seven independent cultures. b: The relative volume of cells in the ROT group was significantly smaller than that in the other three groups. B The number of tubulin+ cells with cell bodies and synapses in more than three visual fields of each group. SP and MH effectively prevent cell death caused by ROT-induced cytotoxicity and increase the number of viable cells in each field. C The relative expression of fluorescence of the ROT group was significantly lower than that of the NC group. The relative expression of fluorescence increased after SP and MH treatments (ROT + SP and ROT + MH groups). D The length of the nerve synapse of the ROT group was significantly lower than that of the NC group, and the length of the nerve synapses increased after SP and MH treatments (ROT + SP and ROT + MH groups). Scale bar: 20 μm in 4× and 500 μm in 100×. On average, 25–30 neurons per condition were tested from three independent cultures. ***P < 0.001, **P < 0.01 and *P < 0.05.
Furthermore, the relative expression of NeuN fluorescence (fluorescence/area) in the ROT-induced group was 0.007, significantly lower than that in the NC group (P = 2.0 \(\times \,10\)−2), which increased to 0.013 and 0.018 after SP and MH treatment, respectively, significantly higher than in the ROT-induced group (P = 1.8 \(\times \,10\)−2 and 1.2 \(\times \,10\)−2, respectively) (Fig. 4C).
Moreover, we found that the average lengths of the nerve synapses in the ROT-induced group were approximately 12.75 μm, significantly shorter than that in the NC group (~44.98 μm) (P = 1.3 \(\times \,10\)−2). After SP and MH treatment, the lengths of the nerve synapses increased to 20.19μm (P = 2.6 \(\times \,10\)−2) and 37.06 μm (P = 2.3 \(\times \,10\)−2), respectively, significantly higher than for the ROT-induced groups. Interestingly, the average length of the synapses in neurons treated with MH was longer than that treated with SP (P = 2.1 \(\times \,10\)−2) (Fig. 4D).
MH and SP improved motor function, anhedonia, and the depression-like behaviors of PD mice
Both MH and SP significantly improved motor function, reduced anhedonia, and alleviated depression-like behaviors in PD mice. To further evaluate the potential of MH and SP as drug candidates for PD treatment, we also examined their effects on PD-like behavior traits in a mouse model induced by ROT, a known cause of motor function abnormalities. The mice from each of the six groups mentioned above underwent a motor function behavior test (for a detailed description of the test, see Supplementary Material). As shown in Fig. 5A, the footprints of the NC group (red) were straight, whereas the footprints of the ROT group (orange) were irregular, indicating an unstable motor function. The motor function behavior of the SP- (yellow) or MH-treated (cyan) groups was also compared to that of the NC or ROT group. In the ROT + SP group (green), the footprints were nearly straight, although the walking directions of the front and rear feet on the same side were not exactly parallel to each other. By comparison, for the mice in the ROT + MH group (blue), the front and rear feet on the same side were precisely in parallel, with the lines of the footprints being straighter than those of the mice in the ROT + SP group (Fig. 5A).
A The influence of SP and MH on the behavior of the PD animal model constructed by ROT (footprinting test). PD behavioral changes were mainly disclosed as motor function changes, whilst the repetition rate of footprints decreased. B Stride length of mice. The stride length of PD mice treated with ROT + SP and ROT + MH was significantly less than that of PD mice. C Stride width of mice. The stride width of mice treated with ROT + SP and ROT + MH was significantly less than that of PD mice, respectively. D Angular change per step in each group. E Effects of SP and MH pretreatment on ROT-induced anhedonic behavior evaluated by means of the sucrose preference test (SPT). F Effects of SP and MH pretreatment on ROT-induced depressive-like behavior of mice evaluated by the forced swim test (FST). G Effects of SP and MH pretreatment on ROT-induced depressive-like behavior of mice evaluated by the tail suspension experiment (TSE). H Results of the open field test for each group. I Statistics on the number of center crossings. J Activity levels of mice in each group. K Total distance traveled in the open field test for each group. *P < 0.05, **P < 0.01, ***P < 0.001.
To quantitatively assess the motor function behavior of the mice, we measured the stride lengths. As shown in Fig. 5B, the stride lengths of the NC, SP, and MH groups were 5.00 mm, 5.18 mm, and 6.46 mm, respectively, with no statistically significant difference evident between them (P å 0.05). However, the average stride length of the ROT group was 12.67 mm, 2.5 times longer than that of the mice in the NC group (P = 2.0 \(\times \,10\)−3). SP treatment served to completely reverse the ROT-induced increase in stride length, as the average stride length of the ROT + SP group was 5.22 mm, almost 2.5 times shorter than that of the ROT group (P = 1.0 \(\times \,10\)−4). Similarly, in the ROT + MH group, the average stride length was 7.73 mm, significantly smaller than that of the ROT group (P = 1.0 \(\times \,10\)−4) (Fig. 5B). Turning to the stride width, those of the NC, SP and MH groups were 3.49 mm, 2.55 mm and 4.06 mm, again not statistically different from each other (P å 0.05) (Fig. 5C). Whilst the average stride width of the ROT group was 6.35 mm, significantly greater than that of the NC group (P = 1.0 \(\times \,10\)−3), the average stride widths measured for the SP + ROT and MH + ROT groups were 4.59 mm (P = 1.4 × 10−3) and 3.97 mm (P = 1.7 \(\times \,10\)−2), respectively, suggesting a narrowing effect of SP and MH on the stride width of the animals (Fig. 5C). The ROT group exhibited an angular change of approximately 3.24° per step, while the NC group showed an angular change of 0.88° per step (P < 0.1 \(\times \,10\)−4). The SP group demonstrated an angular change of 1.03° per step (P < 0.1 \(\times \,10\)−4), and the ROT + SP group exhibited an angular change of 1.56° per step (P < 0.1 \(\times \,10\)-4). The MH group displayed an angular change of 0.97° per step (P < 0.1 \(\times \,10\)−4), while the ROT + MH group exhibited an angular change of 1.16° per step (P < 0.1 \(\times \,10\)−4) (Fig. 5D). Taken together, these data strongly indicate that the administration of SP or MH can efficiently restore the ROT-induced changes in mouse motor function behavior, suggesting that these drugs hold promise for alleviating the motor function abnormality typically observed in PD patients.
Clinically, anhedonia (lack of interest) and depression are commonly noted in PD patients. Whilst a reduction in the sucrose preference ratio in an experimental group compared to controls is held to be indicative of anhedonia in animals67, the forced swimming test (FST) and the tail suspension test (TST) have been devised to assay depression-like behavior in the preclinical mouse model for PD68,69. The sucrose preference test was performed to measure the percentage of sugar water intake in 24 h for the mouse groups (Supplementary Material). The sucrose preference of the ROT group (65.4%) was approximately 25% lower than that of the NC group (81.4%) (P = 2.8 \(\times \,10\)−2). Interestingly, the sucrose preference of the ROT + MH group (77.5%) increased by approximately 20% compared to the ROT group (P = 2.8 \(\times \,10\)−2), whilst the sucrose preference of the ROT + SP group increased to 72.4%, but this increase was subtler and did not attain statistical significance (P = 1.2 \(\times \,10\)−1) (Fig. 5D).
In the forced swimming test (FST), the “immobility” time indicates the state the experimental animals eventually adopt to avoid the stressor (water, in this case), which could be quantified to indicate depression-like behavior of mice68. As shown in Fig. 5E, the immobility time for the ROT group (13.86 s) was longer than for the NC group (7.38 s) (P = 8.0 \(\times \,10\)−3). The immobility time of the ROT + MH group was significantly lower (9.07 s) than for the ROT group (P = 1.5 \(\times \,10\)−2). However, the average immobility time (11.42 s) of the ROT + SP group was comparable to that of the mice that received only ROT (Fig. 5E).
The tail suspension experiment (TSE) was quantified using a ratio of static time to moving time. As shown in Fig. 5F, the ratio of static time to moving time in the ROT group (84.5%) was approximately 20% greater than in the NC group (63.0%) (P = 3.3 \(\times \,10\)−2). Remarkably, the ratio of static time to moving time in the ROT + MH group (70.0%) was significantly lower than in the ROT group (P = 3.2 \(\times \,10\)−2), whereas the average ratio of static time to moving time (80.5%) in the ROT + SP group scarcely changed from that of the ROT group (Fig. 5F).
We further employed the open field test to validate the preventive effects of MH and SP on PD (Fig. 5H). The ROT group exhibited a significantly reduced number of center crossings compared to the NC group (51.00 vs 84.80, P = 2.7 \(\times \,10\)−2). The number of center crossings in the SP group was similar to that of the NC group (100.00 vs 84.80, P = 7.0 \(\times \,10\)−1), while the ROT + SP group showed a marked increase in center crossings compared to the ROT group (91.25 vs 51.00, P = 1.1 \(\times \,10\)−2). Similarly, the number of center crossings in the MH group was comparable to that of the NC group (107.60 vs 84.80, P = 2.4 \(\times \,10\)−1), and the ROT + MH group showed a significant increase compared to the ROT group (90.00 vs 51.00, P = 8.4 \(\times \,10\)−3) (Fig. 5I).
The activity level in the ROT group was significantly lower than that of the NC group (9.77 vs 12.75, P = 3.3 \(\times \,10\)−2), whereas the activity level in the SP group was similar to the NC group (15.36 vs 12.75, P = 1.1 \(\times \,10\)−1). The ROT + SP group demonstrated a significant increase in activity compared to the ROT group (14.10 vs 9.77, P = 2.0 \(\times \,10\)−3). The MH group displayed higher activity than the NC group (16.94 vs 12.75, P = 1.5 \(\times \,10\)−3), and the ROT + MH group showed a significant increase compared to the ROT group (12.93 vs 9.77, P = 2.1 × 10−2) (Fig. 5J).
In terms of movement distance of mice, those in the ROT group showed a significant decrease compared to the NC group (2333 mm vs 3472 mm, P = 4.7 × 10−2). The total movement distance of mice in the SP group was similar to that of the NC group (4291 mm vs 3472 mm, P = 3.1 \(\times \,10\)−1), while the ROT + SP group exhibited a significant increase compared to the ROT group (3671 mm vs 2333 mm, P = 2.3 × 10−2). The movement distance of mice in MH group was similar to that of the NC group (4294 mm vs 3472 mm, P = 2.5 \(\times \,10\)−1), and the ROT + MH group showed a significant improvement compared to the ROT group (3675 mm vs 2333 mm, P = 1.4 \(\times \,10\)−2) (Fig. 5K), suggesting effectiveness of the MH in alleviating the movement symptoms of PD mice.
Taken together, we conclude that MH can significantly alleviate both anhedonia and depression-like behavior in the ROT-induced mouse model for PD, with considerably greater efficiency than SP.
Exploring the mechanisms by which MH protects mitochondrial function, postsynaptic density, and synaptic functions in neuronal cultures and mouse brain
Having found that the impact of MH on the behavior of PD mice was significantly greater than that of SP (Fig. 5A–F), we next directed our efforts toward elucidating the underlying mechanisms by which MH treatment might be efficacious. We therefore sought to ascertain the impact of MH on mitochondrial homeostasis and function in the hippocampus of the ROT-induced PD mouse model. The mitochondria presented themselves as continuous filaments in the hippocampal neurons of the NC group, whilst they showed more obvious fragmentation after treatment with ROT (Fig. 6Ac). Additionally, ROT treatment appeared to lead to a concentration of clumped mitochondria in the cell bodies of the neurons that were broken and lumpy in shape (Fig. 6Aa–c). Remarkably, compared to the disrupted mitochondria in the neurons from the ROT group, the ROT + MH group appeared to have significantly restored the mitochondrial morphology in the neurons, with most of them being maintained in the soma, and continuous in shape (Fig. 6Ae, f). Interestingly, when the neuronal cells were treated with MH alone, mitochondria were observed with many more synapses in the neurites than in the NC group (Fig. 6Ad).
Consistent with a protective effect of MH on the morphology of neuronal mitochondria, MH treatment also appeared to improve mitochondrial function of the hippocampi in the mice of the ROT + MH group compared to those in the ROT group (Fig. 6B–D). The most commonly used markers for mitochondrial functions include malondialdehyde (MDA) and glutathione (GSH), and ATP. As shown in Fig. 6B, ROT increased the MDA level from 1.00 to 1.22 in the hippocampi (P = 3.0 \(\times \,10\)−3) of mice, but such a trend was efficiently reversed by MH treatment, as the MDA level of the ROT + MH group decreased to 0.91 (25.4%) (P = 6.1 \(\times \,10\)−3) (Fig. 6B). Meanwhile, ROT administration significantly decreased the GSH level in the hippocampus (P = 2.3 \(\times \,10\)−3) compared to NC, whereas in the MH + ROT group, the GSH level (P = 3.8 \(\times \,10\)−2) was elevated to 152.27, rather higher than the 100.77 in the ROT groups (Fig. 6C). Moreover, in the ROT-induced mice, the ATP level was found to have decreased from 124.02 to 75.61 in the hippocampi (P = 5.8 \(\times \,10\)−3) compared to the NC group. In the ROT + MH group, we observed a significant elevation of the ATP level from 75.61 to 103.26 (P = 2.2 \(\times \,10\)−2) compared to the ROT group (Fig. 6D).
A Protective effects of MH on mitochondrial morphology in a ROT-induced PD cell model. a: Mitochondrial morphology of control neurons. b: Mitochondrial morphology in a ROT-induced PD cell model with 200 μM ROT. c: Mitochondrial morphology in a ROT-induced PD cell model with 400 μM ROT. d: Mitochondrial morphology of neuronal cells treated with MH alone. e: Mitochondrial morphology of the PD neuron model induced by 200 μM ROT and treated with MH. f: Mitochondrial morphology of the PD neuron model induced by 400 μM ROT and treated with MH. B–E MH protects mitochondrial function in neurons. Effects of MH pretreatment on ROT-induced oxido-nitrosative stress assessed by B malondialdehyde (MDA) level, C reduced glutathione (GSH) level, and D ATP levels of the hippocampus of the mice in each group. E–H According to the results of mitochondrial dysfunction, we performed western blotting for mitochondrial-related proteins. The western blot (E) was quantified for α-syn (F) and NeuN (G) in the murine hippocampi. The bands were quantified using Sigma Gel software, and the differences are represented by a histogram. GAPDH was used as a loading control. The results showed that both MH and SP could restore the abnormal expression of α-syn (F), NeuN (G), and VDAC (H). The western blot (I) was quantified for TH (J) and HSP60 (K) in the substantia nigra. The bands were quantified using Sigma Gel software, and the differences are represented by a histogram. GAPDH was used as a loading control. The results showed that both MH and SP could restore the abnormal expression of TH (J) and HSP60 (K). All values are expressed as mean ± SEM. All experiments were repeated more than three times individually. Scale bar = 500 μm in 100×. *P < 0.05, **P < 0.01, and ***P < 0.001.
To better characterize the protective effect of MH in the brain, we examined the impact of MH on the homeostasis of mitochondrial marker proteins (Fig. 6E). As shown in Fig. 6F, the level of α-synuclein exhibited by mice in the ROT group increased from 1.06 to 1.35 (P = 3.5 \(\times \,10\)−3) as compared to the NC group, whilst expression of α-synuclein decreased from 1.35 to 1.00 in the ROT + MH group (P = 4.0 \(\times \,10\)4) compared to the ROT group. The NeuN level of mice in the ROT group decreased from 0.99 to 0.50 (P = 2.5 \(\times \,10\)−2) as compared to the NC group, whereas the ROT + MH groups displayed significant increases (from 0.50 to 0.82) in NeuN expression (P = 2.5 \(\times \,10\)−2) as compared to the ROT group (Fig. 6G). The VDAC1 level in the ROT group decreased from 1.49 to 1.06 (P = 1.4 \(\times \,10\)−2) as compared to the NC group, whereas the ROT + MH groups displayed significant increases (from 1.06 to 1.26) in VDAC expression (P = 4.1 \(\times \,10\)−2) as compared to the ROT group (Fig. 6H).
We further utilized primary cells from the substantia nigra (SN) to verify the effects of SP and MH on PD. We found that ROT significantly decreased the expression of TH (P = 1.0 × 10−4) and HSP60 (P = 1.8 \(\times \,10\)−3) in primary neurons from the SN (Fig. 6I), indicating that ROT impairs the ability of SN neurons to synthesize TH and reduces mitochondrial content within the cells. SP effectively prevented the ROT-induced reduction in SN neuronal TH synthesis (P = 2.9 \(\times \,10\)−2), and MH similarly prevented this reduction (P = 2.0 \(\times \,10\)−4) (Fig. 6J). Furthermore, SP effectively prevented the ROT-induced decrease in HSP60 in SN neurons (P = 4.1 \(\times \,10\)−3), and MH also successfully prevented this reduction (P = 9.2 \(\times \,10\)−3) (Fig. 6K).
To improve our understanding of the mechanism(s) whereby MH improves mitochondrial function in murine hippocampi and neurons, we used qRT-PCR to test a set of genes (Ndufa12, Cox6b1, Atp5k, Src, Ndufb10, and Dlgap3) in the Uni-MH-ROT set that are known to be involved in the pathways pertaining to mitochondrial function and postsynaptic density (Supplementary Fig. 7A). As shown in Fig. 7A–D, the levels of Ndufa12, Cox6b1, Atp5k, and Src expression were restored to normal after treatment with MH compared to the ROT group.
MH can prevent the dysregulation of lipid oxidation-related genes in the brains of mice treated by ROT. Effects of MH on regulating mitochondria-related genes were detected by qRT-PCR in the hippocampus of mice, including A Ndufa12, B Cox6b1, C Atp5k, and D Src. The expression values are given as mean ± SEM. The experimental data were taken from more than three independent experiments. MH can prevent the dysregulation of synapse-related genes in the brains of mice treated by ROT. Effects of MH on regulating synapse-related genes were detected by qRT-PCR in mouse hippocampus, including E Chrna4, F Syt11, G Cdh8, and H Sncg. Expression values are expressed as mean ± SEM. The experimental data were taken from more than three independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001. The effect of MH on mitochondrial metabolites of ROT-induced PD primary neuron model. I OPLS VIP (variable influence on projection for the orthogonal projections to latent structures model) and Student’s t-test P-values of 22 metabolites whose VIP was higher than 1 and t-test P-values lower than 0.1. *P < 0.05, **P < 0.01; ***P < 0.001. J KEGG pathway enrichment analysis on 22 metabolites (OPLS VIP > 1 and t-test P < 0.1) combined with 10 genes (Chrna4, Syt11, Cdh8, Sncg, Ndufa12, Cox6b1, Atp5k, Src, Ndufb10, and Dlgap3) that are indicated in this study as expressed significantly differently in hippocampal regions of the ROT and MH + ROT groups. Compound 1: 2-{(3S)-1-[4-(trifluoromethyl)benzyl]-3-pyrrolidinyl}-1,3-benzoxazole. Compound 2: 2-heptyl-4-hydroxyquinoline-N-oxide. Compound 3: 5,5-dimethyl-2-{[(2-phenylacetyl)amino]methyl}-1,3-thiazolane-4-carboxylic acid. Compound 4: trans-cinnamoyl beta-D-glucoside. Compound 5: 6,9-dioxo-11R,15S-dihydroxy-13E-prostenoic acid. Compound 6: 17beta-hydroxy-5beta-androstan-3-one. NAFLD non-alcoholic fatty liver disease, RE signaling retrograde endocannabinoid signaling, IMR of TRP channels inflammatory mediator regulation of TRP channels, SH steroid hormone, P.R.T biosynthesis phenylalanine, tyrosine and tryptophan biosynthesis.
When analyzed the RNA-seq data from the brain tissue of mice treated with ROT and the mice treated with both MH and ROT by GSEA analysis, we found that genes down-regulated in brain tissue of ROT + MH mice are significantly enriched with genes annotated as “regulation of postsynaptic neurotransmitter receptor activity” in Gene Ontology database (GO0098962), with P-value as 0.0016** and adjusted P-value as 0.0145* comparing to the brain tissue of mice treated with ROT (Supplementary Fig. 9). Among the 21 genes annotated with GO0098962, the differential expression of Src had been validated by RT-PCR assay in mouse brain sample, as shown in Fig. 7D. The genes related to neurotransmitter regulation are collected from the Gene Ontology database (Supplementary Table 12). The detailed methods for the GSEA analysis are shown in the Supplementary Material.
To better understand how MH might impact synaptic function in the PD mouse model, qRT-PCR was performed to assess the drug’s effect on the expression of several genes (Chrna4, Syt11, Cdh8, and Sncg) that are known to play a role in synaptic pathways70,71,72,73,74 in the NC, ROT, MH or MH + ROT groups. As shown in Fig. 7E–H, the expression levels of Chrna4, Syt11, and Cdh8 were significantly reversed in the ROT + MH group as compared to the ROT group (more detailed results are given in Supplementary Material and Supplementary Fig. 10).
Identification of a metabolic signature in mitochondria linked to aminoacyl-tRNA and phenylalanine, tyrosine, and tryptophan biosynthesis in an MH-treated PD neuronal cell model
To study the metabolic alterations in mitochondria after MH treatment of ROT-induced PD neuronal cells, a global metabolomic analysis was performed in the mitochondria of ROT-induced PD neurons, as well as in ROT + MH-treated neuronal cells. Interestingly, eight metabolites measured in whole neuronal cells were significantly (P < 0.05) altered in both ROT-induced PD neuronal cells and ROT + MH-treated neuronal cells, and with variable influence on projection (VIP) more than 1.0 calculated by the Orthogonal Projections to Latent Structures (OPLS) method (Fig. 7I and Supplementary Table 13). Among them, the compound of 2-{(3S)-1-[4-(Trifluoromethyl)benzyl]-3-pyrrolidinyl}-1,3-benzoxazole was newly collected in the PubChem database in May 2021, whilst the other seven metabolites have been reported as being related to PD therapeutic or neuroprotective effects75,76,77,78,79,80,81,82,83,84,85,86,87.
We performed KEGG analysis using MetaboAnalytic88,89 by combining the 22 metabolites (OPLS VIP > 1 and P < 0.1) and 10 genes (Chrna4, Syt11, Cdh8, Sncg, Ndufa12, Cox6b1, Atp5k, Src, Ndufb10, and Dlgap3) involved in mitochondrial function or synaptic function, which were expressed significantly differently between the ROT and ROT + MH groups in murine hippocampus. As shown in Fig. 7J, these metabolites and genes are significantly (P < 0.05) enriched in seven typical KEGG pathways including mitochondrial function pathways (thermogenesis and oxidative phosphorylation), brain disease pathways (PD, Alzheimer’s disease and Huntington disease), pathways closely related to PD mechanisms such as NAFLD90, and the prolactin signaling pathway91. Therefore, mechanistically, MH may protect neurons from ROT-induced damage by modulating the function of neuronal mitochondria with an impact on mitochondria-linked metabolic pathways. KEGG pathways with 8 out of 22 metabolites significantly different between ROT+/− MH groups, removed genes of Nfugb10 and Dlgap3 that were not significantly different between groups, were shown in Supplementary Fig. 11.
The pharmacological mechanism of MH in the treatment of PD
As shown in Supplementary Table 10, ten proteins are predicted as the most possible drug targets binding with MH by DStruBTarget55, among which DRD4, 5-HT1A, 5-HT2A, σ1, PPARG, CNR1, and CNR2 are known PD-related proteins, suggesting the potential molecular mechanisms of MH in influencing PD. We additionally identified the proteins that are known to bind with drugs in a similar structure (Tanimoto score > 0.4) as MH. These proteins are THRα, Beta3, and Alpha1D. The interactions of these proteins and MH were examined by four assays. Among the assays used in this study, TR-FRET Thyroid Receptor alpha Coactivator Assay (Invitrogen, Cat PV4587) is used for detecting the interactions between MH and THRα92, TR-FRET PPARγ Competitive Binding Kit (Invitrogen, Cat PV4894) is used to evaluate the interactions between MH and PPARγ93, and cAMP Detection Kit (Cisbio Cat 62AM4PEJ)94,95 is used for identifying the interactions between MH and Beta3 (Supplementary Table 15). The interactions of MH with σ1, DRD4, CNR2, 5-HT1A, and Alpha1D are evaluated by Filtration Binding Assay (Supplementary Table 14). In Supplementary Fig. 12, we provided the details on the inhibition rates of the reference drugs on these predicted target proteins. All the results and experimental assays are shown in Supplementary Table 14. As shown in Supplementary Table 14, the experiments indicated that the MH (1 μM) significantly reduced the activity of σ1 with an inhibition rate of 75.4%, suggesting potential interaction between σ1 and MH. Here, the inhibition rate = (1 − (Sample Well − LC)/(HC − LC)) × 100% (sample well: the interaction between MH and the target protein experimentally measured by the sample in one well; LC: positive control; HC: DMSO). In comparison, the inhibition rates of MH on other protein targets are close to 0 or negative. Thus, MH has not shown obvious effects on inhibiting the activity of other drug targets.
SN neurons were stained using Sigma1-Receptor (S1R) immunofluorescence. The S1R levels were significantly increased in the MH group, while ROT reduced S1R levels compared to the NC, SP, and MH groups. Thus, MH is able to protect the S1R in the SN from ROT-induced damage (Fig. 8C).
To further validate interaction between MH and σ1, we measured half maximal inhibitory concentration (IC50) of MH in inhibiting σ1 by filtration binding assay. Figure 8A is the IC50 of positive control, Haloperidol, in inhibiting σ1. Figure 8B describes the potential of MH in binding with σ1, indicating MH (with an IC50 value of 1.6 μM) binds to σ1, reducing the interaction between σ1 and radiolabeled ligand. No existing study has been found to report the interactions between MH and σ1. DMSO functioned as the negative control, while haloperidol served as the positive control. Detailed results can be found in Supplementary Table 14 and Supplementary Fig. 12.
Discussion
We established a method, integrating gene co-expression modules in normal human brain with disease-associated genes or SNPs, to identify disease-associated gene co-expression modules that were further used for drug repurposing. iGOLD is able to interpret the impact of individual genes on disease, especially those genes expressed differentially in patients and controls (DEGs) obtained using insufficient samples, which are unable to provide information on the signaling circuitry of disease-associated pathways96,97,98. It can also explain the role of SNPs in regulating gene co-expression modules in the normal human brain. These SNPs were discovered as part of large population-based GWAS studies, and their functional roles in regulating gene co-expression modules from normal human brain have remained unclear99,100. When these genes are used for drug discovery, iGOLD determines the drug efficiency not in terms of its target proteins but rather in terms of its ability to restore the normal gene expression profile. This makes iGOLD a powerful tool in drug discovery by dint of its considering multiple genes in one network. This approach can be generally applied to repurposing drugs for other brain disorders simply by connecting any disease-associated genes or SNPs with the gene co-expression modules associated with normal human brain samples.
When we used the conservation score to represent the enrichment of genes expressed in specific tissues, we found that the conservation score of the BR7M4 module in SN is 33.93 and in HC is 10.53. The conservation score larger than 10 is defined as highly conserved by a previous study50. The conservation analysis of the gene co-expression modules across tissues is to find the modules showing high expression in specific tissues, especially brain tissues. These modules have high potential to be related to PD. Because we assume PD is a disease directly related to the brain, the gene co-expression modules that have shown high expression in other tissues are not further investigated, although they may represent another mechanism of PD. In addition to BR3M2, it is highly conserved in the HC brain area. However, the BR3M2 is not highly enriched with PD-associated genes compared to the BR7M4. The result of the enrichment analysis of PD-associated genes in these two modules is shown in Supplementary Table 5. Thus, we selected BR7M4 for further study. This result had been exhibited in Fig. 2A and Supplementary Table 5.
To test sensitivity of the PD associated gene co-expression modules identified by this study, we have performed RNA-seq analysis for the brain tissues from hippocampi and substantia nigra brain regions of PD mouse model. The results were compared to the mice in the controls. The result indicated that the module, BR7M4, highly conserved in hippocampi, is enriched with genes expressed differently (DEGs) in the PD mice and the controls, indicating the reliability of our method in identifying PD-associated brain regions. Moreover, gene expressions of 39 DEGs in BR7M4 are significantly correlated with the PD-associated genes. The roles of these genes in PD require further clarification in the future.
Although both hippocampus and the substantia nigra have shown high potential to be associated with PD in the module conservation analysis, the RNA-seq analysis is performed on the hippocampus of mice.
Indeed, BR7M4 shows the highest conservation in two brain regions, Substantia Nigra and hippocampus that are also among the most affected brain regions in PD. The commonly accepted mechanism of PD is the degeneration of dopaminergic neurons in the Substantia Nigra101. McGregor and Nelson101 highlight the crucial role of SN in motor control and its profound involvement in PD pathogenesis. The loss of these neurons in substantia Nigra in PD is well-documented in both clinical and preclinical studies by Dauer and Przedborski29. In 2017, Surmeier et aldiscuss the selective vulnerability of substantia Nigra dopaminergic neurons in PD, emphasizing the molecular and cellular mechanisms that make this region particularly susceptible to neurodegeneration102. Furthermore, Poewe et al. provide a comprehensive review of PD pathophysiology, including the progressive loss of dopaminergic neurons in substantia nigra and its impact on motor dysfunction103. These studies collectively reinforce the pivotal role of the Substantia Nigra in PD progression and pathology.
Hippocampus is also critically implicated in PD, especially concerning non-motor symptoms such as cognitive decline, depression, and anxiety. Hijaz and Volpicelli-Daley discuss how α-synuclein aggregation, particularly in regions like the hippocampus, contributes to cognitive deficits in PD104. A wide range of cognitive impairments in PD patients is considered associated with the hippocampal dysfunctions105. Regarding the molecular mechanisms of PD, the change of NOX4 in the hippocampus is known to be involved in PD by investing human PD patients106. Our findings of significant enrichment in hippocampal modules are in line with these studies, suggesting a deeper connection between PD-related cognitive dysfunction and hippocampal gene expression.
Our study also found that BR7M4 has shown marginal conservation in the other two regions, the Thalamus and the Putamen. The Thalamus and Putamen, both regions, are integral parts of the motor control circuit, often implicated in PD due to their involvement in motor and sensory processing. Dorsey et al. provide evidence for the important role of these regions in PD progression, particularly the putamen, which is affected by dopaminergic dysfunction107. The Thalamus, as part of the basal ganglia-thalamocortical circuit, also shows evidence of alterations in PD patients, further supporting our findings.
The commonly used drug for PD is levodopa. However, the shortcoming of levodopa is the drug resistance. In this study, we found that MH treatment was associated with improving PD-related behaviors. It has the potential to be an alternative choice for PD patients showing resistance to levodopa. MH is known as a psychostimulant in the nootropic agent group, and is an accepted treatment for traumatic cataphora, alcohol poisoning, anoxia neonatorum, and children’s enuresis108. Oral administration of MH to rats in chronic hypoperfusion improved behavioral dysfunction, suggesting an ability of MH to attenuate neuronal damage after ischemia109. Previous studies have examined the potential effect of MH on the states of α-syn in yeast, and similar effects were also observed in dopaminergic neurons of worms expressing110. Additionally, MH was also found to improve muscle tone and brain lipid peroxidation in a rat model111. However, it remained entirely unclear whether MH would have any effect on PD-related behaviors or symptoms, as neither yeast nor worm was an ideal model for PD, and the etiology and progression of PD were far more complicated than deregulated muscle tone or brain lipid peroxidation. Here, we constructed a rotenone-induced mouse model to validate the biological effects of MH. Many studies have employed rotenone to generate an experimental animal model of PD to mimic the PD-like symptoms, such as motor deficit, cognitive decline, and depression34,35,112,113,114. Most of the previous studies focus on the functions of MH in improving memory115,116,117,118. Our findings demonstrate that MH effectively improves PD-related behaviors of mice, as measured by changes in motor function, sucrose preference, forced swim test, and the tail suspension experiment. Recent research has provided evidence supporting a correlation between motor dysfunction observed in PD and the hippocampal region, as it has been confirmed that the hippocampal region can project to the midcingulate motor area and the supplementary motor area119. Furthermore, there is a strong association between the hippocampus and non-motor symptoms of PD. A study120 demonstrated that increased iron levels in the early stages of the hippocampus can trigger the occurrence of non-motor symptoms. Another review121 discussed the link between the hippocampus and non-motor symptoms, including depression and fatigue. Therefore, our research on the hippocampus aims not only to verify the motor symptoms of PD but also to demonstrate the role of MH in protecting the hippocampus and improving PD-related non-motor symptoms. Moreover, we have provided extensive evidence to support the mechanisms underlying the beneficial effects of MH. Our findings demonstrate that MH can prevent neuronal death, synaptic damage, and mitochondrial destruction, reduce lipid peroxidation, protect dopamine synthesis, and reverse abnormal mitochondrial metabolism. These results highlight the ability of MH to improve both mitochondrial metabolism and brain function, thus ameliorating the most overt symptoms of PD. Moreover, no study has reported the binding target of MH. This is the first study providing evidence that MH plays a role in PD through binding with σ1.
Although many psychostimulant and cholinergic drugs were reported to promote the REDOX metabolism of brain cells, and MH is prescribed in China and elsewhere to treat a variety of CNS conditions, the actual effect of MH on mitochondrial function has never been tested before. We then set out to test whether and how MH may act on mitochondria-associated metabolic pathways in primary neurons. First, as shown in Fig. 6A, MH was found to restore mitochondrial morphology that was altered upon ROT treatment, indicating its positive impact on overall mitochondrial homeostasis in primary neurons. Consistently, MH seemed to also protect mitochondrial function from ROT-induced oxido-nitrosative stress in neurons. Remarkably, when we examined the metabolites of mitochondria in primary neurons, the MH treatment significantly (Student t-test P < 0.05) decreased the homeostatic levels of 17a-Ethynylestradiol, L-Indospicine, 2-{(3S)-1-[4-(Trifluoromethyl)benzyl]-3-pyrrolidinyl}-1,3-benzoxazole, cotinine, cypridinaluciferin, 5-HETE, D-(+)-maltose and oxprenolol, comparing to the primary neurons only treated with ROT (Supplementary Table 13). As these metabolites have been shown as involved in the pathways associated with PD in previous studies75,77,79,81,82,122, our data thus collectively supported the notion that MH did significantly protect mitochondrial function, which may at least partially underlie its effects on PD.
Additionally, we found that MH may prevent the further deterioration of Parkinsonian symptoms by improving mitochondrial function, such as impacting the expression of markers for lipid peroxidation and mitochondrial proteins. Moreover, it is widely believed that mitochondrial-associated neurodegenerative diseases involve the perturbation of calcium flux or energy generation123,124. Thus, we measured the ATP levels in the hippocampi of the mice in each group, and noted that MH significantly restores the ATP level in the ROT + MH group to a level comparable to that of the NC group (Fig. 6D). The improved mitochondrial function consequent to MH treatment might also be due, at least in part, to the restoration of normal expression of Ndufa12, Cox6b1, Atp5k, and Src genes in the ROT-induced PD mouse model. The NDUFA12 gene has been shown to encode a key member of the mitochondrial respiratory chain125,126. Low expression of the Cox6b1 gene has been associated with Alzheimer’s disease127. In a similar vein, ATP5K is known to be involved in mitochondrial ATP synthesis-coupled proton transport128.
We also investigated the impact of MH on mitochondria by means of mitochondrial metabolomics, and disclosed several specific metabolites that were regulated by MH, suggesting that MH may influence mitochondrial function by reprogramming metabolic pathways. Understanding drug-metabolite associations is crucial for research into pharmacoepidemiology and for improving drug efficiency129. One recent study has demonstrated that metabolic abnormalities can alter neuronal excitability in the brain130. We found that MH treatment can restore normal levels of several metabolites associated with PD pathogenesis, including 5-HETE and L-indospicine. For example, 5-HETE (OPLS VIP = 1.250 and t-test P = 3.9 × 10−2) has been reported as a biomarker of oxidative damage in PD; 5-HETE interacts with SRC, regulates the TRPV1 gene, which has been reported to be associated with PD development131,132,133,134,135,136. Another compound, L-indospicine (OPLS VIP = 1.107, P = 5.0 × 10−4), has been reported to be a potent inhibitor of arginase that can cause a shift in L-arginine metabolism to the NOS pathway64 closely related to PD development137,138,139.
Damage to synaptic plasticity is also known to be related to the onset and progression of both the motor and cognitive symptoms of PD140. Previous studies have employed immunohistochemistry to investigate the protective potential of MH in relation to synapses141,142. However, these studies could not determine the true length and number of synapses. In order to confirm the protective action of MH on synapses, we performed in vitro and in vivo experiments, as well as cluster analysis to demonstrate that MH can protect synapses in terms of synaptic length. Additional q-PCR experiments indicated that MH treatment of ROT-induced PD primary neurons restores normal expression of the Chrna4, Syt11, and Cdh8 genes. These genes have been previously shown to encode proteins with functions pertaining to synaptic function71,72,73,143,144,145,146. Thus, our study supports the view that MH may protect synapses by impacting the pathways in which both mitochondria-related genes, and metabolic factors such as maltose and cotinine, are involved.
To further reveal the molecular mechanism of MH in affecting PD, we used our previously developed method, DStruBTarget, to predict MH and protein interactions. DStruBTarget has provided the top 10 most MH-protein interactions. Among them, the MH-σ1 interaction is validated by the Filtration Binding Assay. Usually, σ1 is considered a crucial target for preventing and treating PD56,147,148. σ receptors have been recognized as unique receptors, initially thought to be a subtype of opioid receptors149,150. σ1 receptor is implicated in aging and various diseases, including schizophrenia, depression, Alzheimer’s disease, and ischemia148,151. Other studies suggest that the σ1 receptor is involved in regulating dopamine synthesis and release152,153,154. Our research predicts, on one hand, that MH can interact with the σ1. The interaction was further validated through Filtration Binding Assay, demonstrating robust interaction of MH to σ 1 with IC50 = 1636 nM (Fig. 8B). This is the first study confirming MH plays roles in PD through σ 1.
The Filtration Binding Assay used in this study is the radiolabeled binding assay that is often applied to evaluate protein-drug interactions. This method has been used in many previous studies155,156,157. All these studies suggested that the filtration binding assay is reliable in evaluating the interactions between drugs and proteins by filtering receptor samples using a vacuum processing system, assessing their ability to interfere with the specific binding of a radiolabeled ligand to the receptor. This method can be employed for accurate and universal high-throughput screening.
Our studies do, however, have several limitations. Firstly, the tissue chip can only interrogate part of the synapses, and is unable to fully observe the protective effect of MH on the murine synapse. New technology for observing whole synapses will be required to confirm the protective effect of MH. Secondly, the CMAP database only includes a limited number of drugs, which may hinder the identification of more effective drugs for repurposing. Thirdly, the effectiveness of the drugs themselves still requires further supporting evidence from clinical studies. One of the limitations of the study is that the iGOLD is dependent on the gene co-expression module analysis. The inaccurate modules influence the determination of the effective drugs.
Another limitation of this study is that we have not performed a dopamine transporter molecular imaging to clarify the most responsive region of MH, although the molecular experiments have indicated the roles of MH in the hippocampus. Finally, no clinical validation has yet been performed in this study.
In conclusion, this study revealed MH as a potential drug candidate for PD. Subsequent experiments indicated that MH is able to improve PD-related behavior and protect neurons by regulating mitochondrial-related genes, synaptic pathways, and metabolite pathways. Thus, it would appear that MH may help to arrest the progressive deterioration of Parkinsonian symptoms.
Methods
Study design
Here, we designed a computational architecture, iGOLD, for drug repurposing. This approach involved the construction of gene co-expression modules of normal human brain by applying weighted gene co-expression network analysis (WGCNA)18 and DiffCoEx19, and analyzing gene expression data of 1231 brain samples from ten brain regions of healthy humans. The sample size for each brain region is shown in Table 1. Then, iGOLD was used to identify the modules enriched in PD-associated genes and PD-associated SNPs by employing 11 datasets encompassing PD-associated genes, SNPs, and DEGs between PD and controls. The identified modules were evaluated in relation to their expression conservation in brain samples across ethnicities, brain regions, and disease stages of PD by ModulePreservation, a function in the WGCNA R package. This analysis was based upon seven datasets with sample sizes ranging from 4 to 57. The highly conserved modules were used for drug repurposing by CMAP41,42. The drug candidates were ranked by their connectivity scores. From them, we selected those ranked in the top 15 (Supplementary Table 9) and having the ability to pass through blood blood-brain barrier for further validation. The source code of iGOLD and related data used in this study are available at https://github.com/fanc232CO/iGOLD_pipline.
The experimental validations of the drug effects were conducted in primary neurons and a mouse model. The primary neurons were obtained from the hippocampi of mice on postnatal days 0–3. We used Rotenone (ROT) to treat the primary neurons as described previously in ref. 158 since ROT has been shown to induce PD-like symptoms in human27and animal models28,29. The effectiveness of drugs in protecting neuronal damage was evaluated by immunofluorescence marking the tubulin of synapses and primary neurons, and a mitochondrial fluorescent probe for mitochondrial morphology. The protective effects of the drug on mitochondrial functions were tested by determining the malondialdehyde (MDA) level, the reduction in the glutathione (GSH) level, ATP levels, and mitochondrial proteins. The regulatory effects of drugs on mitochondrial metabolites were assessed in murine primary neurons.
The mouse model was constructed using ROT-induced C57BL/6J male mice (n = 86, 8-weeks-old) (Supplementary Material). We used PET/CT imaging to examine glucose metabolism in the brains of mice (Supplementary Material). The protective ability of the drugs on cranial nerve damage was evaluated by immunohistochemistry of NeuN (in the DG, DG2, and CA1), NeuN-negative cells in the DG structure of the hippocampus, NeuN-negative cells in the DG2 structure of the hippocampus, NeuN-negative cells in the CA1 structure of the hippocampus, and TH in the hippocampus. Expression of mitochondrial-related proteins was measured by western blotting for α-syn(F) and NeuN(G) in the murine hippocampi. RNA-seq and qRT-PCR were used to measure the expression of mitochondrial-related genes in the hippocampi of mice. The effectiveness of the drugs was further examined in terms of their influence on the PD behaviors of mice, including the footprint test, sucrose preference test, and forced swim test.
Gene expression data used for identifying modules associated with PD
Gene expression data were obtained from the publicly available GEO dataset159. From GEO, we downloaded eight gene expression datasets [one for co-expression module building, five for module conservation analysis, and two for enrichment analysis of Parkinson’s disease (PD)-associated DEGs].
The co-expression modules were constructed by WGCNA analysis on the GSE6086238,39,40, a gene expression dataset obtained from the platform of Affymetrix Human Exon 1.0 ST Array. The samples covered ten brain regions, including cerebellar cortex, frontal cortex, occipital cortex, temporal cortex, hippocampus, putamen, thalamus, medulla, white matter and substantia nigra, from 1231 individuals of European descent collected by the UK Brain Expression Consortium (UKBEC).
The module conservation analysis was performed on six GEO datasets, GSE13161745,160, GSE2329046, GSE3451647, GSE5192248, GSE1883849, and GSE34865 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi), which were obtained from the platform of the Affymetrix Human Exon 1.0 ST Array. These datasets covered multiple ethnicities, brain regions, tissues, Braak stages, and PD disease status. In detail, GSE131617 includes transcriptome data from 213 post-mortem brain tissue specimens (=71 subjects × 3 BRs), which covered three brain regions (entorhinal, temporal and frontal cortices) of 71 Japanese brain-donor subjects in four Braak stages161,162,163 (0, I–II, III–IV, and V–VI). GSE23290 included putamen tissues from the 8 idiopathic PD (IPD) patients, 3 LRRK2-associated PD (G2019S mutation) patients, 5 neurologically healthy controls, and one asymptomatic LRRK2 mutation carrier46, the asymptomatic carrier (GSM745539) was removed from analysis. GSE34516 included the locus coeruleus post-mortem tissues from idiopathic PD (IPD) and LRRK2-associated 6 European PD patients47. GSE51922 is built on the RNA profile of IPSC-derived dopaminergic neurons from idiopathic and genetic forms (LRRK2) of PD43. GSE18838 included peripheral blood collected from 18 PD patients and 12 healthy controls49. GSE34865 included gene expression data of substantia nigra samples from 57 healthy adults. Details of these datasets can be found in Supplementary Table 7.
GSE839744,164 were downloaded for the generation of PD-associated differentially expressed genes (DEGs). GSE8397 is built on the gene expression of substantia nigra split into medial and lateral portions, and frontal cortex from 24 PD patients and 15 controls. Gene expression was accessed through two platforms, GPL96 and GPL9744,164. Details of these datasets can be found in Supplementary Table 6.
Conservation analysis of co-expression gene module
Conservation of modules was estimated by module preservation through the calculation of Zsummary. The Zsummary is determined by estimating the density and connectivity of the test modules and the reference module. Briefly, the calculation of Zsummary is based on permutation tests to assess the mean and variance of Z statistics under the null hypothesis of no relationship between the module assignment in reference and test modules. The reference gene expression data in this study were derived from seven GEO databases (Supplementary Table 7), while the test modules are the PD-associated modules suggested by iGOLD. Modules with Zsummary scores above 10 were interpreted as being highly conserved, Zsummary scores between 2 and 10 were deemed to be moderately conserved, whilst Zsummary scores below 2 were regarded as incompletely conserved.
Stratified LD score regression (sLDSC) analyzing PD-associated SNPs
sLDSC analysis165 was conducted using the parameters and pipelines provided by tutorials in LDSC (https://github.com/bulik/ldsc/wiki). First, we mapped all SNPs to the co-expressed modules if they were within 10 kb of the locations of exon probes. The LD score and heritability were calculated for each co-expressed module. The enrichment of the SNPs in the co-expression modules was defined as the summation of SNP heritability divided by the number of SNPs in that module. Standard errors of the SNP enrichment in the co-expression modules were estimated by a block jackknife166 and were further used to evaluate Z-scores, P-values, and false discovery rates (FDRs) of the SNP enrichment in the co-expression modules87.
Construction of co-expression networks for the normal human brain
The GSE6086238,39,40 dataset was used to construct the gene co-expression networks. The genes expressed in ten brain regions were grouped into co-expression modules by two different approaches, consensus weighted gene co-expression network analysis (WGCNA)18 and DiffCoEx19. The WGCNA was applied to detect co-expression modules common to all ten brain regions (consensus co-expressed modules, CCM). DiffCoEx was used to identify gene modules specifically expressed in each of the ten brain regions compared to the other nine brain regions (specific co-expressed modules, SCM).
The function blockwiseModules in WGCNA, was utilized to construct the co-expression modules as previously described in refs. 167,168. The parameters were set as follows, β = 7 (chosen based on the scale-free topology criterion r2 > 0.8), minModuleSize = 30, mergeCutHeight = 0.25, maxBlockSize = 6000, and corType = ‘pearson’. For each pair of genes, the topological overlap matrix (TOM) was calculated and scaled based on the adjacency matrix. The component-wise minimum of the TOMs in each brain region was then extracted to generate a consensus TOM. This TOM was clustered by using the average hierarchical clustering method to obtain a consensus TOM, defining it as a 1-consensus TOM according to the difference of genetic connectivity. A consensus co-expression module was defined as a branch of a cluster tree generated by a dynamic tree cut.
The differential co-expression network analysis was carried out by the DiffCoEx method in R software as previously described19. To identify gene co-expression differences between transcripts from the substantia nigra brain region and transcripts from the other nine brain regions, we used the function of DiffCoex based on the WGCNA framework by calculating a TOM generated from a matrix of adjacency differences between these brain regions.
Module conservation analysis
The conservation of the association between the gene co-expression modules and PD was evaluated according to the enrichment of the modules in DEGs of PD patients and controls from five different sources45,46,47,48,49,160. These datasets were all obtained from the GPL5175 platforms, and the samples in the datasets were divided into healthy control and PD groups. The samples in this dataset are accompanied by information on Braak stages, indicating the disease severity161,162,163. These samples were partitioned in terms of two ethnicities, three brain tissues, five brain regions, and six PD disease states. Further information is provided in Supplementary Table 7. The degree of module conservation was estimated by modulePreservation, a function in the WGCNA R package.
Enrichment analysis of PD-associated genes
We validated the association between PD and CCM and SCM modules by the enrichment analysis of PD-associated genes and PD-associated SNPs. The PD-associated genes were obtained from DisGeNet169,170,171 (https://www.disgenet.org/home/). DisGeNET covers the full spectrum of human genetic diseases, as well as normal and abnormal traits. The currently released version of DisGeNET includes more than 24,000 different genetic diseases and traits, 17,000 genes, and 117,000 genomic variants171. As shown in Supplementary Table 3, searching under the term “Parkinson disease” (UMLS CUI: C0030567) allowed the collation of six types of PD-associated genes by the DisGeNet database, of which “CausalMutation” was filtered out before enrichment analysis because it contained only one gene. Gene enrichment analysis in the remaining five gene sets was evaluated by means of the single-tailed Fisher’s exact test, and further adjusted by the FDR. The background genes for the enrichment analysis are those that are not PD-associated genes by considering a total number of human genes as 20,814.
Enrichment analysis of genes expressed significantly differently in PD patients and controls
Differentially expressed genes (DEGs) between PD patients and controls were obtained by analyzing gene expression data from GEO with access ID GSE839744,164 (Supplementary Table 6). In GSE8397, samples from the whole substantia nigra (combination of samples from lateral and medial substantia nigra regions) were used for DEG analysis. The DEG analysis was performed by means of the GEO2R tool172,173 in the GEO website (https://www.ncbi.nlm.nih.gov/geo/). DEGs were selected with a fold change (FC) threshold of 1.2 and an adjusted P-value threshold of 0.05. Enrichment was evaluated by a single-tailed Fisher’s exact test adjusted by the FDR.
RNA-seq data processing
In the RNA-seq analysis of the hippocampi of mice, we first used FASTP (https://github.com/OpenGene/fastp) to carry out data preprocessing. All parameters of FASTP were kept as default to preprocess the raw data, including stripping adapters, filtering out low-quality reads, correcting mismatched base pairs, and trimming poly G ends. After the preprocessing, the RNAseq data were delivered to the Salmon tool to generate a gene expression matrix by aligning the reads to the GRCm9 (http://asia.ensembl.org/Mus_musculus/Info/Index) gene annotation file downloaded from Ensembl. The genes that exhibited raw counts of 0 in each sample were excluded. Finally, we obtained the counts of 29,324 gene symbols of 28 mouse samples.
Drug discovery by CMAP
First, the DEGs were generated by analyzing gene expression data (GSE8397) of substantia nigra samples obtained from the platform of GPL96 (Affymetrix) (15 healthy controls and 24 PD). The overlapping genes between these DEGs and the genes in PD-related modules were extracted and mapped to the GPL96 probe, and then delivered to the Connectivity Map (CMAP)41,42 (https://portals.broadinstitute.org/cmap/) to predict potential PD drug candidates. We ranked the output drug candidates by their connectivity scores. When the connectivity scores were close to −1, the drugs were deemed to have strong potential to restore the normal gene expression profile of the PD-associated genes. In this study, drug candidates were selected for further analysis if their connectivity scores were lower than −0.8.
Neuronal cell culture
The hippocampal primary neurons were obtained from mice on postnatal days 0–3. Hippocampal neurons were plated on poly-D-lysine (Sigma)-coated chamber slides and Six-hole plates for 2 h to allow neurons to adhere. The cultured neurons were maintained with complete culture medium composed of B27 supplement (Gibco, USA), L-glutamine (Life Technologies), and Neurobasal-A medium (DMEM/F12) (Gibco, USA) at 37 °C in a 7% CO2 incubator for 7 days to ensure the growth of nerve synapses.
Construction of neuron models
The primary neurons were randomly divided into six groups: the normal control (NC) group, the ROT-induced group, the SP-treated group, the MH-treated group, the ROT + SP-treated group, and the ROT + MH-treated group.
To construct the ROT-induced group, ROT with a concentration of 400 nM was applied directly to the culture medium for 24 h. To create ROT + SP(2 μM) and ROT + MH(10 μM) groups, we pretreated murine primary neurons with SP or MH for 2 h, and then used 400 nM rotenone (ROT) to treat the neuronal cells for 24 h. The SP and MH groups of the murine primary neurons were treated with SP or MH for 2 h. We then used immunofluorescence, marking the tubulin of synapses and primary neurons to evaluate the cell damage from ROT.
Construction of a mouse model
C57BL/6 J male mice (n = 86, 8-weeks-old, 30 g) were purchased from the Guangdong Medical Laboratory Animal Center (Foshan, China). The mice were randomly assigned to six groups. Unless otherwise specified, they were provided with ad libitum access to food and water and were housed four to five per cage in a temperature-controlled (23 ± 1 °C) and humidity-controlled room (40–60%) with a 14-h light and 10-h dark cycle. All animal care and experimental procedures were conducted in accordance with protocols approved by the Institutional Animal Care and Use Committee of Sun Yat-sen University. The mice were anesthetized with an intraperitoneal injection of 1% sodium pentobarbital (60 mg/kg) and were euthanized by cervical dislocation.
C57BL/6 J male mice were randomly assigned into six groups, namely the control group (NC), the Sodium phenylbutyrate (SP) group, the Rotenone (ROT) group, the ROT + SP group, the Meclofenoxate-hydrochloride (MH) group, and the ROT + MH group. The mice in the NC group received dimethylsulfoxide (DMSO) (olive oil only); the mice in the SP group were treated with SP (300 mg/kg bw/d; intraperitoneal [i.p.]) for 4 consecutive weeks; the mice in the ROT group were given rotenone (1 μg/g bw/d; i.p.) for 3 consecutive weeks; the mice in the ROT + SP group received SP prophylaxis (300 μg/g bw/d; i.p.) for 1 week followed by ROT (1 μg/g bw/d, i.p.) from 1 week onwards for the next three consecutive weeks; the mice in the MH group were administered with MH (50 μg/g bw/d; i.p.) for four consecutive weeks; the mice in the ROT + MH group received MH prophylaxis (50 μg/g bw/d; i.p.) for 1 week followed by ROT challenge (1 μg/g bw/d, i.p.) from 1 week onwards for the next 3 weeks.
Body weight and food intake of the ROT-induced PD mouse model
In this study, 36 C57BL/6 J mice were randomly divided into six groups that were treated with DMSO (NC), ROT, SP, MH, ROT + SP, and ROT + MH, each group containing six mice. Administration of SP and MH did not elicit any behavioral alterations during the experimental period, nor were any significant changes in food intake evident. Mice treated with ROT showed no decrease in body weight during the treatment period. Mice in other groups did not exhibit any significant decrease in body weight during the treatment period nor any significant changes in food intake, except for the ROT group (Supplementary Table 8).
Hippocampus sample collection
From each group of mice, more than three hippocampi were collected. The mice were sacrificed by anesthetization, and their brains were extracted within 24 h after the last injection of ROT. The hippocampi were then isolated under a microscope.
Biospecimen collection
The striatum brain regions of mice were separated and processed to obtain both cytosolic and mitochondrial fractions, after they were sacrificed by anesthetization, within 2 h. The biochemical investigations were conducted, and 4–5 murine striata (from each group) were processed for histopathological examination.
RT-PCR
Total cell RNA extraction was performed using TRIzol (Invitrogen) and reverse transcribed according to the manufacturer’s protocol (Takara). qRT-PCR was carried out using SYBR Premix qRT-PCR ExTaq™ II (Takara) and analyzed on a Bio-Rad CFX96 real-time PCR cycler (Bio-Rad, Netherlands). The primer sequences are given in Supplementary Table 15. Differences in mRNA expression were calculated by means of the formula N = (2)−ΔΔCT 174.
Immunofluorescence
Samples were washed three times with 0.01 M PBS and fixed with 3.7% paraformaldehyde for 15 min at room temperature. The samples were then permeabilized in 0.5% Triton X-100 for 3 min and blocked with 3% goat serum albumin for 30 min prior to incubation with a primary antibody, namely, anti-α-synuclein, mouse anti-TH, SIGMAR1 Ab (Affinity Biosciences, Cat.#: DF7363) and anti-NeuN, at dilutions of 1:100 overnight at 4 °C. Secondary antibodies, anti-rabbit or anti-mouse (Tianjin Sungene Biotech Co., China) at 1:200 dilution, for 1 h at room temperature. Nuclei were stained with DAPI.
Immunohistochemistry
The slides were washed twice for 15 min in 0.01 M PBS, and proteinase K was added to the tissue and incubated at 37 °C for 5 min. This step was followed by quenching for 10 min in a solution of methanol containing 30% hydrogen peroxidase and further incubating for 1 h in blocking solution containing 5% normal goat serum and 1% Triton X-100 in 0.01 M PBS. After blocking, the slides were incubated overnight in rabbit anti-caspase-3 antibody (Catalog No.: 10842-1-AP, Proteintech), anti-α-synuclein antibody (Catalog No.: 10842-1-AP, Proteintech, China), anti-TH antibody (Catalog No.: 25859-1-AP, Proteintech, China), and anti-NeuN antibody (Catalog No.: A19086, ABclonal, China) diluted 1:100 in blocking solution. Following incubation with primary antibody, the sections were incubated for 2 h in biotinylated goat antirabbit secondary antibody diluted 1:500 in 0.01 M PBS and subsequently incubated with ABC reagents (Standard Vectastain ABC Elite Kit; Vector Laboratories, Burlingame, CA, USA) for 20 min in the dark at room temperature. The sections were washed twice with 0.01 M PBS and incubated in 3,3‘-diaminobenzidine tetrahydrochloride (DAB); sections were washed with distilled water, dehydrated in graded ethanol (70%, 85%, 95%, and 100%), placed in xylen, and cover-slipped using mounting medium. We then analyzed and counted the active caspase-3 positive cells in the DG, DG2, and CA1 regions of the hippocampus using the ImageJ program analysis.
Mitochondrial fluorescent probe staining analysis
Mitochondrial staining was performed with the mitochondrial probe MitoTracker Red CM-H2XRos (Invitrogen, USA) according to protocols provided by the manufacturer. After being washed with 0.01 M PBS, the cells were counterstained with DAPI for 10 min and imaged with an Olympus BX63 microscope (Olympus, Japan).
Neurons from differentiated groups were stained with MitoTracker Deep Red (200 ng/ml) (Yeasen, Shanghai, China) for mitochondria for 60 min, then fixed with 4% paraformaldehyde for 15 min and permeabilised with Triton X-100 at 0.04% as previously described in ref. 175. All cells (nuclei) were stained with DAPI (4‘,6-diamidino-2-phenylindole, 1 µg/ml). Images were obtained using an Olympus BX63 microscope (Olympus, Japan). Quantification and analysis of the neuronal network were performed using Image J software.
Footprint test
The motor function patterns of mice were assessed by the footprint test as described previously in ref. 176. The apparatus comprises an open field (60 × 60 × 40 cm), in which a runway (4.5 × 40 × 12 cm) was arranged to lead out into a dark wooden box. The motor function parameters were measured by wetting forepaws and hindpaws with commercially available non-toxic colored inks and allowing the mice to trot onto a strip of paper on the runway. Pawprints made at the beginning and the end of the run were excluded. Various motor function parameters, such as stride length (Differences in the forward distance between each fore paw and hind paw footprint with each step), stride width (lateral distance between opposite left and right fore paw and opposite left and right hind paw), and foot direction, were measured.
Sucrose preference test
The mice were housed individually and were first trained to adapt to sugary drinking water in a quiet room by putting two water bottles in each cage. Both bottles were filled with 1% sucrose water. The mice were tested with respect to sucrose preference using the following process: (1) the mice were prevented from drinking for 24 h before administration of the Sucrose Preference Test; (2) each mouse was given a pre-quantified bottle of 1% sucrose water and a bottle of distilled water; (3) the position of the two bottles of water was changed every 12 h; (4) the two bottles of water were taken and weighed after 24 h to calculate the consumption of sucrose water, distilled water and total liquid consumption for each mouse. Sucrose water preference (%) = (sucrose water consumption/total liquid consumption) × 100%.
Forced swim test
The mice were placed individually into an open cylindrical container (10 cm diameter, 30 cm height), containing water (25 ± 2 °C) to a depth of 20 cm. Each mouse was forced to swim for 6 min, and the total duration of immobility in seconds was measured during the last 4 min. The water was changed after each animal experiment was finished. After the experiment, the mice were wiped with a towel until their fur was dry. The immobility time was defined in terms of the absence of escape-oriented behavior.
Open field test
Mice were first acclimated in a quiet laboratory with constant room temperature for 1 h. After the acclimation period, each mouse was carefully placed in the center of the open field box, and its spontaneous activity was observed for 5 min. After each test, the box was thoroughly cleaned to remove any debris, and 75% ethanol was used to eliminate residual scents from the previous mouse to avoid influencing the behavior of subsequent mice. The number of center crossings, activity level, and total movement distance were recorded.
PET/CT imaging and data analysis
The PET/CT imaging was performed on 36 C57BL/6 J mice that were divided into NC, SP, ROT, ROT + SP, MH, and ROT + MH groups. Each group contains six mice. All PET imaging studies were performed on a Biograph TrueV (Siemens Healthcare) scanner. This PET/CT device is equipped with a 64-slice spiral CT component. After a 3-h fasting period, mice were injected in the tail vein with a solution of 18F-FDG (16–32 MBq) or 18F-FLT (32–37 MBq). The volume of the syringes was always kept below 0.2 ml in order to meet the requirements of our ethics committee. To minimize muscle and brown fat uptake in the case of 18F-FDG imaging, animals were kept anesthetized under warming lights for a 20-min period after injection. Animals were imaged simultaneously in groups of three with the PET/CT scanner; one was placed at the center of the field of view (FOV) whilst the two others were placed on each side of the central animal, at a 5-cm and at a −5-cm radial offset.
The PET/CT was initiated with a CT scan acquired with the following parameters: 80 mA, 130 kV, pitch 0.8, and 64 × 0.6 mm collimation. Then, an emission scan was obtained in 3D mode. PET images were reconstructed in a reduced FOV (35 cm), applying a scaling factor of 2. Images were reconstructed with an algorithm that models the point spread function of the scanner and leads to a 2.2 mm spatial resolution at the center of the FOV. The following parameters were used: six iterations, 16 subsets, no filtering, and a matrix size of 3362, resulting in a 1.02 × 1.02 × 1 mm voxel size. Scatter and attenuation corrections were applied.
Brain activity was obtained from a volume of interest (VOI) encompassing the entire brain. The VOI was determined by means of an isocontour, which was set so that the VOI matched the apparent brain volume on PET and CT images. When discordance was encountered between the PET metabolic volume and the CT volume, the VOI was drawn according to CT images, so that PET/CT images could be compared to ex vivo counting of the entire hippocampus, for which the entire brain was harvested, irrespective of the presence of non-viable areas. Data were analyzed using Statistical Parametric Mapping software (SPM 12, The Wellcome Trust Center for Neuroimaging, London, UK).
Relative [18F] FDG uptake images were analyzed by using microQ (Siemens/Concorde Microsystems, Knoxville, TN, USA). Subsequently, we utilized a voxel-by-voxel approach to obtain maximal use of information without a priori knowledge, using IRW (Siemens/Concorde Microsystems, Knoxville, TN, USA). In short, we used a flexible factorial design depending on time point (after all treatment) and group (NC, SP, ROT, SP + ROT, MH, and MH + ROT), as previously described (1, 2). T-maps were interrogated at a Pheight ≤ 0.005 (uncorrected) peak level and extended threshold of kE > 200 voxels (1.6 mm3)177. Only significant clusters with Pheight < 0.05 (corrected for multiple comparisons) were retained.
ELISA analysis
α-synuclein was quantified in the striatum region of the mouse brain utilizing the Enzyme-linked Immunosorbent Assay (ELISA) Kit from Invitrogen (Cat No.: KHB0061) following the manufacturer’s instructions.
Western blot (WB) analysis
For WBs, cells were extracted in RIPA buffer (Sigma-Aldrich, USA), separated by 10% SDS-PAGE, and transferred onto a polyvinylidene difluoride membrane (Millipore, USA). After blocking with 5% skimmed milk, the membrane was incubated with specific primary antibodies directed against α-synuclein, TH, and GAPDH (Abcam plc, USA) at a dilution of 1:1000 overnight at 4 °C. The protein expression levels were normalized with GAPDH. The membrane was incubated with horseradish peroxidase-conjugated anti-rabbit secondary antibody for 1 h at 37 °C.
Measurement of oxidative stress markers
The markers of oxidative impairment were studied in the cytosol. MDA (malondialdehyde) contents were quantitatively detected using the Lipid Peroxidation MDA-Assay Kit (Beyotime). The unit weight of MDA was calculated by a MDA standard curve measured at 532 nm. MDA was considered a biomarker of lipid peroxidation in tissues and organs178.
Adenosine triphosphate (ATP) measurement
ATP was measured using ATP assay kits (Beyotime, Shanghai, China). After being diluted with dilution buffer, the ATP detection reagent was added to a 96-well plate. After homogenization followed by centrifugation at 12,000 g at 4 °C for 5 min, the samples were added into the wells and mixed with the detection solution. Then, the levels of ATP were measured with a SpectraMax M5 microplate reader (Molecular Devices, San Jose, CA, USA). The ATP content was normalized to the ATP protein content on the basis of the standard curve.
Quantitative analysis of GSH levels
Estimation of GSH: glutathione (GSH) was measured in the supernatant of the hippocampus tissue in the mouse brain. GSH level was measured with the enzyme-linked immunoassay (ELISA) kit from Elabscience (Bethesda, MD) according to the manufacturer’s instructions.
Sample preparation and library preparation for transcriptome sequencing
Hippocampal RNA was extracted by TRIzol. RNA quantification and qualification were evaluated as follows: (1) RNA degradation and contamination were monitored on 1% agarose gels; (2) RNA purity was checked using the NanoPhotometer® spectrophotometer (IMPLEN, CA, USA); and (3) RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA).
A total of 1 µg RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, USA) following the manufacturer’s recommendations; index codes were added to attribute sequences to each sample. Briefly, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. Fragmentation was carried out using divalent cations under elevated temperature in NEBNext First Strand Synthesis Reaction Buffer (5×). First-strand cDNA was synthesized using a random hexamer primer and M-MuLV Reverse Transcriptase (RNase H-). Second-strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. After adenylation of the 3’ ends of the DNA fragments, NEBNext Adapters with hairpin loop structure were ligated in preparation for hybridization. In order to select cDNA fragments of preferentially ~250–300 bp in length, the library fragments were purified with the AMPure XP system (Beckman Coulter, Beverly, USA). Then, 3 µl USER Enzyme (NEB, USA) was used with size-selected, adapter-ligated cDNA at 37 °C for 15 min followed by 5 min at 95 °C before PCR. Then, PCR was performed with Phusion High-Fidelity DNA polymerase, Universal PCR primers, and Index (X) Primer. At last, PCR products were purified (AMPure XP system) and library quality was assessed on the Agilent Bioanalyzer 2100 system.
Clustering and transcriptome sequencing
The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumina) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina Novaseq platform, and 150 bp paired-end reads were generated.
Metabolite extraction and UPLC-MS/MS analysis
The mitochondrial metabolite tests were performed on murine primary neurons. Global metabolic profiles were obtained from the cells using the Metabolon Platform. The principle of the Metabolon Platform has been previously described138,139,140. Approximately 5 × 106 primary neuronal cells were involved in the test. These cells were extracted from mice at postnatal days 0-3 as described above. The primary neuronal cells were extracted and cultured for 7 days, treated with MH or DMSO for 2 h, and then with ROT or DMSO for 24 h, to form a PD cell model, which was used for the experiments described below. To lyse the cells, 1120 μl lysis system (800 μl pre-cooled methanol + 320 μl ice water) was added to the wells of the six-well plate; the cellular metabolites were placed in a 2 ml EP tube, and 800 μl pre-cooled chloroform was injected into the tube prior to vortexing for 15 min. The mixture was then centrifuged at 12,000 rpm for 15 min before being separated into supernatant and precipitate, which were transferred to other microtubes. The supernatant was then dried by a continuous flow of nitrogen gas to render it solid; the solid was re-dissolved with 100 µl acetonitrile-water (1:1), which was centrifuged at 14,000 rpm for 5 min; a mixture of supernatants of the sample (10 µl) was transferred to a quality control (QC) vial. The samples were kept on ice throughout the procedure unless centrifuged. Ratios above were all according to volume.
For UPLC-MS/MS analysis, each sample was reconstituted with a methanol solution with a density of 80% (80% methanol: 20% water) (by an 80-μl volume of methanol with 20-μl H2O). The methanol solution was centrifuged at 12,000 g for 10 min. The samples were then prepared for liquid chromatography-mass spectrometry. Briefly, the samples were injected into a Waters ACQUITY UPLC BEH Amide Column (2.1 × 100 mm, 1.7 µm) at column temperature, 40 °C, with a flow rate of 0.35 ml/min. The mobile phase includes phase A and B (Phase A: 95:5 (acetonitrile: water) containing 10 mM ammonium formate, 0.1% formic acid; Phase B: 50:50 (acetonitrile: water) containing 10 mM ammonium formate, 0.1% formic acid). The gradient elution ratio is shown in Supplementary Table 16. Subsequent analyses were performed using ThermoScientific Ultimate 3000 UPLC coupled with Orbitrap Exploris 480 MS from Sun Yat-Sen Memorial Hospital. Identification of known chemical entities was based on comparison with metabolomic library entries of purified standards. Each biochemical was rescaled to set the median equal to 1. Values for each sample were normalized by Bradford protein concentration.
Metabolomic instrumentation and analytical conditions
LC-MS/MS was used for detection, with at least three replicates for each experimental condition.
Binding target prediction for MH and SP
Binding targets of MH and SP were predicted using the tool DStruBTarget55. The inputs of DStruBTarget are structures of MH and SP, which were obtained from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) in the format of 3D sdf (compound CID: 4039 for MH and 5258 for SP). The parameters of DStruBTarget were employed as the defaults in the prediction. The top ten binding targets with the highest scores were analyzed in this study.
Experimental validation of interactions between MH and PD-related proteins
The interactions of MH with σ1, DRD4, CNR2, 5-HT1A, and Alpha1D targets were determined by the Filtration Binding Assay. The radiolabeled ligands used by this study include 3H-DTG (Perkin Elmer, Cat. no. PE-NET986250UC) for σ1validation, 3H-methylspiperone (Perkin Elmer, Cat. no. PE-NET856250UC) for DRD4 validation, 3H CP 55940 (Perkin Elmer, Cat. no. PE-NET1051250UC) for CNR2 validation, 3H 8-OH-DPAT (Perkin Elmer, Cat. no. PE-NET929250UC) for 5-HT1A validation, and 3H-prazosin (Perkin Elmer, Cat. no. PE-NET823250UC) for Alpha1D validation. The experimental process includes the following steps: (1) transfer 1 μl of compounds to the assay plate according to the plate layout, which leads to the final concentration of the testing compound in the assay plate being 10 μM in duplicate. (2) Transfer 1 μl of non-specific binding compounds to the plate according to the plate map. Summarize the total binding by transferring 1 μl of DMSO onto the assay plate. (3) Allocating specified volumes of membrane material to the plate. Soaking the Unifilter-96 GF/C filter plates in 0.3% PEI (Polyethyleneimine) at room temperature with 50 μl per well for 30 min. (4) Using the Perkin Elmer Filtermate Harvester to filter the reaction mixture through the GF/C plate, then wash each plate four times with cold wash buffer. (5) Dry the filtration plate at 50° for 1 h. After drying, seal the bottom of the filter plate holes with Perkin Elmer Unifilter-96 backing seal tape. Adding 50 μl of Perkin Elmer Microscint 20 cocktail. Seal the top of filter plates with Perkin Elmer TopSeal-A sealing film. Count 3H trapped on the filter using the Perkin Elmer MicroBeta2 Reader. The key reagents utilized in the Filtration Binding Assay include: Ascorbic acid (TCI-A0537), DTT (Sigma, Cat: 43815, Lot: BCBD7009V), Ultima Gold cocktail (PerkinElmer, Cat: 6013329, Lot: 77-16371), and Microscint 20 cocktail (PerkinElmer- 6013329).
The interaction between MH and THRα was validated by using the LanthaScreen™ TR-FRET co-activator assay (Invitrogen, Cat# PV4587, Lot# 2322026). The LanthaScreen™ TR-FRET has been previously used for detecting the interactions of hERα, and the Tb3 + -labeled anti-GST antibody y (fluorescence donor) with the fluorescein-labeled ligand Fluormone ES2™ (fluorescence acceptor 1)92.
The interaction between MH and PPARγ (Peroxisome Proliferator-Activated Receptor γ) was validated by the LanthaScreen™ TR-FRET competitive binding assay kit (Invitrogen-PV4894). The experimental methods used by this study are the same as those in a previous study for detecting the interactions between PPAR-γ and pioglitazone93.
We used the Cisbio cAMP detection kit to validate the interactions of MH with Beta3. The Cisbio cAMP detection kit (Cat # 62 AM4PEJ) was previously used to identify the interactions between PAGly and β2AR95. Here, we used a similar experimental procedure.
Data availability
The source code of iGOLD and related data are available at https://github.com/fanc232CO/iGOLD_pipline. The RNA-seq data generated by this study have been uploaded to NCBI GEO (GEO ID: GSE295746). The metabolomics data generated by this study are available at https://www.ebi.ac.uk/metabolights/editor/study/REQ20250506210329/files.
Code availability
The source code of iGOLD of this study is available at https://github.com/fanc232CO/iGOLD_pipline.
References
Volta, M., Milnerwood, A. J. & Farrer, M. J. Insights from late-onset familial parkinsonism on the pathogenesis of idiopathic Parkinson’s disease. Lancet Neurol. 14, 1054–1064 (2015).
Mortality, G. B. D. & Causes of Death, C. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 388, 1459–1544 (2016).
Pan, H. et al. Genome-wide association study using whole-genome sequencing identifies risk loci for Parkinson’s disease in Chinese population. NPJ Parkinsons Dis. 9, 22 (2023).
Blauwendraat, C. et al. Parkinson’s disease age at onset genome-wide association study: defining heritability, genetic loci, and alpha-synuclein mechanisms. Mov. Disord. 34, 866–875 (2019).
Pang, S. Y. et al. The interplay of aging, genetics and environmental factors in the pathogenesis of Parkinson’s disease. Transl. Neurodegener. 8, 23 (2019).
Shao, Y. & Le, W. Recent advances and perspectives of metabolomics-based investigations in Parkinson’s disease. Mol. Neurodegener. 14, 3 (2019).
Le, W. D. et al. Mutations in NR4A2 associated with familial Parkinson disease. Nat. Genet. 33, 85–89 (2003).
Kun-Rodrigues, C. et al. A systematic screening to identify de novo mutations causing sporadic early-onset Parkinson’s disease. Hum. Mol. Genet. 24, 6711–6720 (2015).
Guo, J. F. et al. Coding mutations in NUS1 contribute to Parkinson’s disease. Proc. Natl. Acad. Sci. USA 115, 11567–11572 (2018).
Nalls, M. A. et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 18, 1091–1102 (2019).
Stenson, P. D. et al. The Human Gene Mutation Database (HGMD((R))): optimizing its use in a clinical diagnostic or research setting. Hum. Genet. 139, 1197–1207 (2020).
Chai, C. & Lim, K. L. Genetic insights into sporadic Parkinson’s disease pathogenesis. Curr. Genomics 14, 486–501 (2013).
Charvin, D., Medori, R., Hauser, R. A. & Rascol, O. Therapeutic strategies for Parkinson disease: beyond dopaminergic drugs. Nat. Rev. Drug Discov. 17, 844 (2018).
Fink, A. L. The aggregation and fibrillation of alpha-synuclein. Acc. Chem. Res. 39, 628–634 (2006).
Lashuel, H. A., Overk, C. R., Oueslati, A. & Masliah, E. The many faces of alpha-synuclein: from structure and toxicity to therapeutic target. Nat. Rev. Neurosci. 14, 38–48 (2013).
Mishra, S. K. et al. Role of the ubiquitin-like protein Hub1 in splice-site usage and alternative splicing. Nature 474, 173–178 (2011).
Saelens, W., Cannoodt, R. & Saeys, Y. A comprehensive evaluation of module detection methods for gene expression data. Nat. Commun. 9, 1090 (2018).
Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinforma. 9, 559 (2008).
Tesson, B. M., Breitling, R. & Jansen, R. C. DiffCoEx: a simple and sensitive method to find differentially coexpressed gene modules. BMC Bioinforma. 11, 497 (2010).
Hu, Y., Pan, Z., Hu, Y., Zhang, L. & Wang, J. Network and Pathway-based analyses of genes associated with Parkinson’s disease. Mol. Neurobiol. 54, 4452–4465 (2017).
Fu, L. M. & Fu, K. A. Analysis of Parkinson’s disease pathophysiology using an integrated genomics-bioinformatics approach. Pathophysiology 22, 15–29 (2015).
Godini, R., Fallahi, H. & Ebrahimie, E. A comparative system-level analysis of the neurodegenerative diseases. J. Cell Physiol. 234, 5215–5229 (2019).
Nabirotchkin, S. et al. Next-generation drug repurposing using human genetics and network biology. Curr. Opin. Pharm. 51, 78–92 (2020).
Pushpakom, S. et al. Drug repurposing: progress, challenges and recommendations. Nat. Rev. Drug Discov. 18, 41–58 (2019).
MacArthur, D. G. et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science 335, 823–828 (2012).
Reay, W. R. & Cairns, M. J. Advancing the use of genome-wide association studies for drug repurposing. Nat. Rev. Genet. 22, 658–671 (2021).
Langston, J. W., Ballard, P., Tetrud, J. W. & Irwin, I. Chronic Parkinsonism in humans due to a product of meperidine-analog synthesis. Science 219, 979–980 (1983).
Tawara, T. et al. Effects of paraquat on mitochondrial electron transport system and catecholamine contents in rat brain. Arch. Toxicol. 70, 585–589 (1996).
Dauer, W. & Przedborski, S. Parkinson’s disease: mechanisms and models. Neuron 39, 889–909 (2003).
Inden, M. et al. Parkinsonian rotenone mouse model: reevaluation of long-term administration of rotenone in C57BL/6 mice. Biol. Pharm. Bull. 34, 92–96 (2011).
Terzioglu, M. & Galter, D. Parkinson’s disease: genetic versus toxin-induced rodent models. FEBS J. 275, 1384–1391 (2008).
Borsche, M., Pereira, S. L., Klein, C. & Grunewald, A. Mitochondria and Parkinson’s disease: clinical, molecular, and translational aspects. J. Parkinsons Dis. 11, 45–60 (2021).
Betarbet, R. et al. Chronic systemic pesticide exposure reproduces features of Parkinson’s disease. Nat. Neurosci. 3, 1301–1306 (2000).
Innos, J. & Hickey, M. A. Using rotenone to model Parkinson’s disease in mice: a review of the role of pharmacokinetics. Chem. Res Toxicol. 34, 1223–1239 (2021).
Ahn, E. H. et al. Initiation of Parkinson’s disease from gut to brain by delta-secretase. Cell Res. 30, 70–87 (2020).
Liu, J. et al. Piperlongumine restores the balance of autophagy and apoptosis by increasing BCL2 phosphorylation in rotenone-induced Parkinson disease models. Autophagy 14, 845–861 (2018).
Wagner, J. et al. Anle138b: a novel oligomer modulator for disease-modifying therapy of neurodegenerative diseases such as prion and Parkinson’s disease. Acta Neuropathol. 125, 795–813 (2013).
Ramasamy, A. et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat. Neurosci. 17, 1418–1428 (2014).
Trabzuni, D. et al. Fine-mapping, gene expression and splicing analysis of the disease associated LRRK2 locus. PLoS One 8, e70724 (2013).
Trabzuni, D. et al. Widespread sex differences in gene expression and splicing in the adult human brain. Nat. Commun. 4, 2771 (2013).
Lamb, J. The Connectivity Map: a new tool for biomedical research. Nat. Rev. Cancer 7, 54–60 (2007).
Lamb, J. et al. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313, 1929–1935 (2006).
Fan, C., Wong, P. -p & Zhao, H. DStruBTarget: integrating binding affinity with structure similarity for ligand-binding protein prediction. J. Chem. Inf. Model.60, 400–409 (2019).
Moran, L. B. et al. Whole genome expression profiling of the medial and lateral substantia nigra in Parkinson’s disease. Neurogenetics 7, 1–11 (2006).
Miyashita, A. et al. Genes associated with the progression of neurofibrillary tangles in Alzheimer’s disease. Transl. Psychiatry 4, e396 (2014).
Botta-Orfila, T. et al. Microarray expression analysis in idiopathic and LRRK2-associated Parkinson’s disease. Neurobiol. Dis. 45, 462–468 (2012).
Botta-Orfila, T. et al. Brain transcriptomic profiling in idiopathic and LRRK2-associated Parkinson’s disease. Brain Res. 1466, 152–157 (2012).
Fernandez-Santiago, R. et al. Aberrant epigenome in iPSC-derived dopaminergic neurons from Parkinson’s disease patients. EMBO Mol. Med 7, 1529–1546 (2015).
Shehadeh, L. A. et al. SRRM2, a potential blood biomarker revealing high alternative splicing in Parkinson’s disease. PLoS One 5, e9104 (2010).
Langfelder, P., Luo, R., Oldham, M. C. & Horvath, S. Is my network module preserved and reproducible?. PLoS Comput. Biol. 7, e1001057 (2011).
Chen, G. et al. UNC5C receptor proteolytic cleavage by active AEP promotes dopaminergic neuronal degeneration in Parkinson’s disease. Adv. Sci.9, e2103396 (2022).
Wu, Z. et al. C/EBPbeta/delta-secretase signaling mediates Parkinson’s disease pathogenesis via regulating transcription and proteolytic cleavage of alpha-synuclein and MAOB. Mol. Psychiatry 26, 568–585 (2021).
Hawkins, N. T., Maldaver, M., Yannakopoulos, A., Guare, L. A. & Krishnan, A. Systematic tissue annotations of genomics samples by modeling unstructured metadata. Nat. Commun. 13, 6736 (2022).
Kudo, S., Umehara, K., Abe, Y., Furukawa, M. & Odomi, M. Intracerebral penetration of carteolol hydrochloride in rats. Psychopharmacology131, 388–393 (1997).
Fan, C., Wong, P. P. & Zhao, H. DStruBTarget: integrating binding affinity with structure similarity for ligand-binding protein prediction. J. Chem. Inf. Model 60, 400–409 (2020).
Mishina, M. et al. Function of sigma1 receptors in Parkinson’s disease. Acta Neurol. Scand. 112, 103–107 (2005).
Yang, X. D. et al. Expression of the gene coading for PGC-1alpha in peripheral blood leukocytes and related gene variants in patients with Parkinson’s disease. Parkinsonism Relat. Disord. 51, 30–35 (2018).
Dipanwita, S., Arindam, B., Atanu, B., Kunal, R. & Jharna, R. Genetic polymorphisms in DRD4 and Risk for Parkinson’s disease among eastern Indians. Neurol. India 70, 729–732 (2022).
Barrero, F. J. et al. Depression in Parkinson’s disease is related to a genetic polymorphism of the cannabinoid receptor gene (CNR1). Pharmacogenomics J. 5, 135–141 (2005).
Yu, H. et al. The neuroprotective effects of the CB2 agonist GW842166x in the 6-OHDA mouse model of Parkinson’s disease. Cells. https://doi.org/10.3390/cells10123548 (2021).
Shukla, R. et al. Modulation of GPCR receptors common to gut inflammatory diseases and neuronal disorders, Alzheimer’s and Parkinson’s diseases as druggable targets through Withania somnifera bioactives: an in silico study. J. Biomol. Struct. Dyn. 41, 4485–4503 (2023).
Cummings, J. et al. Pimavanserin for patients with Parkinson’s disease psychosis: a randomised, placebo-controlled phase 3 trial. Lancet 383, 533–540 (2014).
Senatorov, V. V., Ren, M., Kanai, H., Wei, H. & Chuang, D. M. Short-term lithium treatment promotes neuronal survival and proliferation in rat striatum infused with quinolinic acid, an excitotoxic model of Huntington’s disease. Mol. Psychiatry 9, 371–385 (2004).
Gage, M., Gard, M. & Thippeswamy, T. Characterization of cortical glial scars in the diisopropylfluorophosphate (DFP) rat model of epilepsy. Front Cell Dev. Biol. 10, 867949 (2022).
Ohm, D. T. et al. Accumulation of neurofibrillary tangles and activated microglia is associated with lower neuron densities in the aphasic variant of Alzheimer’s disease. Brain Pathol. 31, 189–204 (2021).
Grishagin, I. V. Automatic cell counting with ImageJ. Anal. Biochem 473, 63–65 (2015).
Ellenbroek, B. & Youn, J. Rodent models in neuroscience research: Is it a rat race?. Dis. Model Mech. 9, 1079–1087 (2016).
Porsolt, R. D., Anton, G., Blavet, N. & Jalfre, M. Behavioural despair in rats: a new model sensitive to antidepressant treatments. Eur. J. Pharm. 47, 379–391 (1978).
Steru, L., Chermat, R., Thierry, B. & Simon, P. The tail suspension test: a new method for screening antidepressants in mice. Psychopharmacology85, 367–370 (1985).
Tarren, J. R., Lester, H. A., Belmer, A. & Bartlett, S. E. Acute ethanol administration upregulates synaptic alpha4-subunit of neuronal nicotinic acetylcholine receptors within the nucleus accumbens and amygdala. Front. Mol. Neurosci. 10, 338 (2017).
Friedman, L. G. et al. Cadherin-8 expression, synaptic localization, and molecular control of neuronal form in prefrontal corticostriatal circuits. J. Comp. Neurol. 523, 75–92 (2015).
Zhu, G. et al. Rats harboring S284L Chrna4 mutation show attenuation of synaptic and extrasynaptic GABAergic transmission and exhibit the nocturnal frontal lobe epilepsy phenotype. J. Neurosci. 28, 12465–12476 (2008).
Shimojo, M. et al. Synaptotagmin-11 mediates a vesicle trafficking pathway that is essential for development and synaptic plasticity. Genes Dev. 33, 365–376 (2019).
Russell, T. L. et al. Medullary respiratory circuit is reorganized by a seasonally-induced program in preparation for hibernation. Front. Neurosci. 13, 376 (2019).
Seet, R. C. et al. Oxidative damage in Parkinson disease: Measurement using accurate biomarkers. Free Radic. Biol. Med. 48, 560–566 (2010).
Picazo, O., Becerril-Montes, A., Huidobro-Perez, D. & Garcia-Segura, L. M. Neuroprotective actions of the synthetic estrogen 17alpha-ethynylestradiol in the hippocampus. Cell Mol. Neurobiol. 30, 675–682 (2010).
Matsuda, N. et al. Diverse effects of pathogenic mutations of Parkin that catalyze multiple monoubiquitylation in vitro. J. Biol. Chem. 281, 3204–3209 (2006).
Hey, C. et al. Inhibition of arginase in rat and rabbit alveolar macrophages by N omega-hydroxy-D,L-indospicine, effects on L-arginine utilization by nitric oxide synthase. Br. J. Pharm. 121, 395–400 (1997).
Yumino, K., Kawakami, I., Tamura, M., Hayashi, T. & Nakamura, M. Paraquat- and diquat-induced oxygen radical generation and lipid peroxidation in rat brain microsomes. J. Biochem. 131, 565–570 (2002).
Yoshida, T., Tanaka, M., Sotomatsu, A. & Hirai, S. Activated microglia cause superoxide-mediated release of iron from ferritin. Neurosci. Lett. 190, 21–24 (1995).
Hong, D. P., Fink, A. L. & Uversky, V. N. Smoking and Parkinson’s disease: Does nicotine affect alpha-synuclein fibrillation?. Biochim. Biophys. Acta 1794, 282–290 (2009).
Kurako Iu, L. & Volianskii, V. E. New directions in contemporary pharmacotherapy of parkinsonism (review)]. Zh. Nevropatol. Psikhiatr. Im. S S Korsakova 84, 1401–1406 (1984).
Corbett, J. L. Beta adrenergic blockers in tremor. Aust. Fam. Physician 10, 633–635 (1981).
Sandler, M., Fellows, L. E., Calne, D. B. & Findley, L. J. Letter: oxprenolol and levodopa in Parkinsonian patients. Lancet 1, 168 (1975).
von Egidy, H. [What is assured in treatment with beta receptor blockaders?]. Internist13, 481–484 (1972).
Thompson, M. K. Effect of oxprenolol on Parkinsonian tremor. Lancet 2, 388 (1972).
Jackson, R., Crawford, C. O. & Peel, J. S. Oxprenolol in Parkinson’s disease. N. Z. Med. J. 74, 246–247 (1971).
Pang, Z. et al. MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 49, W388–W396 (2021).
Pang, Z., Zhou, G., Chong, J. & Xia, J. Comprehensive meta-analysis of COVID-19 global metabolomics datasets. Metabolites. https://doi.org/10.3390/metabo11010044 (2021).
Vavougios, G. D. Human coronaviruses in idiopathic Parkinson’s disease: implications of SARS-CoV-2’s modulation of the host’s transcriptome. Infect. Genet. Evol. 89, 104733 (2021).
Nitkowska, M., Tomasiuk, R., Czyzyk, M. & Friedman, A. Prolactin and sex hormones levels in males with Parkinson’s disease. Acta Neurol. Scand. 131, 411–416 (2015).
Hilal, T., Puetter, V., Otto, C., Parczyk, K. & Bader, B. A dual estrogen receptor TR-FRET assay for simultaneous measurement of steroid site binding and coactivator recruitment. J. Biomol. Screen 15, 268–278 (2010).
Sekulic-Jablanovic, M., Wright, M. B., Petkovic, V. & Bodmer, D. Pioglitazone ameliorates gentamicin ototoxicity by affecting the TLR and STAT pathways in the early postnatal organ of corti. Front. Cell Neurosci. 14, 566148 (2020).
Jung, J. et al. Discovery of novel dual adenosine A(2A) and A(1) receptor antagonists with 1H-pyrazolo[3,4-d]pyrimidin-6-amine core scaffold as anti-Parkinson’s disease Agents. Pharmaceuticals. https://doi.org/10.3390/ph15080922 (2022).
Xu, X. et al. The gut microbial metabolite phenylacetylglycine protects against cardiac injury caused by ischemia/reperfusion through activating beta2AR. Arch. Biochem. Biophys. 697, 108720 (2021).
Lewis, P. A. & Cookson, M. R. Gene expression in the Parkinson’s disease brain. Brain Res. Bull. 88, 302–312 (2012).
Feleke, R. et al. Cross-platform transcriptional profiling identifies common and distinct molecular pathologies in Lewy body diseases. Acta Neuropathol. 142, 449–474 (2021).
Borrageiro, G., Haylett, W., Seedat, S., Kuivaniemi, H. & Bardien, S. A review of genome-wide transcriptomics studies in Parkinson’s disease. Eur. J. Neurosci. 47, 1–16 (2018).
Soldner, F. et al. Parkinson-associated risk variant in distal enhancer of alpha-synuclein modulates target gene expression. Nature 533, 95–99 (2016).
Pierce, S. E., Tyson, T., Booms, A., Prahl, J. & Coetzee, G. A. Parkinson’s disease genetic risk in a midbrain neuronal cell line. Neurobiol. Dis. 114, 53–64 (2018).
McGregor, M. M. & Nelson, A. B. Circuit mechanisms of Parkinson’s disease. Neuron 101, 1042–1056 (2019).
Surmeier, D. J., Obeso, J. A. & Halliday, G. M. Selective neuronal vulnerability in Parkinson disease. Nat. Rev. Neurosci. 18, 101–113 (2017).
Poewe, W. et al. Parkinson disease. Nat. Rev. Dis. Prim. 3, 17013 (2017).
Hijaz, B. A. & Volpicelli-Daley, L. A. Initiation and propagation of alpha-synuclein aggregation in the nervous system. Mol. Neurodegener. 15, 19 (2020).
Das, T., Hwang, J. J. & Poston, K. L. Episodic recognition memory and the hippocampus in Parkinson’s disease: A review. Cortex 113, 191–209 (2019).
Boonpraman, N., Yoon, S., Kim, C. Y., Moon, J. S. & Yi, S. S. NOX4 as a critical effector mediating neuroinflammatory cytokines, myeloperoxidase and osteopontin, specifically in astrocytes in the hippocampus in Parkinson’s disease. Redox Biol. 62, 102698 (2023).
Dorsey, E. R., Sherer, T., Okun, M. S. & Bloem, B. R. The emerging evidence of the Parkinson pandemic. J. Parkinsons Dis. 8, S3–S8 (2018).
Zou, J. J. et al. Bioequivalence and pharmacokinetic comparison of a single 200-mg dose of meclofenoxate hydrochloride capsule and tablet formulations in healthy Chinese adult male volunteers: a randomized sequence, open-label, two-period crossover study. Clin. Ther. 30, 1651–1657 (2008).
Liao, Y., Wang, R. & Tang, X. C. Centrophenoxine improves chronic cerebral ischemia induced cognitive deficit and neuronal degeneration in rats. Acta Pharm. Sin. 25, 1590–1596 (2004).
Wang, S. et al. Chemical compensation of mitochondrial phospholipid depletion in yeast and animal models of Parkinson’s disease. PLoS One 11, e0164465 (2016).
Nehru, B., Verma, R., Khanna, P. & Sharma, S. K. Behavioral alterations in rotenone model of Parkinson’s disease: attenuation by co-treatment of centrophenoxine. Brain Res. 1201, 122–127 (2008).
Zhao, Z. et al. Fecal microbiota transplantation protects rotenone-induced Parkinson’s disease mice via suppressing inflammation mediated by the lipopolysaccharide-TLR4 signaling pathway through the microbiota-gut-brain axis. Microbiome 9, 226 (2021).
Sonia Angeline, M., Chaterjee, P., Anand, K., Ambasta, R. K. & Kumar, P. Rotenone-induced Parkinsonism elicits behavioral impairments and differential expression of parkin, heat shock proteins and caspases in the rat. Neuroscience 220, 291–301 (2012).
Zhao, X. et al. Baicalein alleviates depression-like behavior in rotenone-induced Parkinson’s disease model in mice through activating the BDNF/TrkB/CREB pathway. Biomed. Pharmacother. 140, 111556 (2021).
Oliver, J. E. & Restell, M. Serial testing in assessing the effect of meclofenoxate on patients with memory defects. Br. J. Psychiatry 113, 219–222 (1967).
Marcer, D. & Hopkins, S. M. The differential effects of meclofenoxate on memory loss in the elderly. Age Ageing 6, 123–131 (1977).
Platel, A. & Porsolt, R. D. Habituation of exploratory activity in mice: a screening test for memory enhancing drugs. Psychopharmacology78, 346–352 (1982).
Nehru, B., Bhalla, P. & Garg, A. Evidence for centrophenoxine as a protective drug in aluminium induced behavioral and biochemical alteration in rat brain. Mol. Cell Biochem. 290, 33–42 (2006).
Rolls, E. T. The cingulate cortex and limbic systems for emotion, action, and memory. Brain Struct. Funct. 224, 3001–3018 (2019).
Kim, M. et al. Extra-basal ganglia iron content and non-motor symptoms in drug-naive, early Parkinson’s disease. Neurol. Sci. 42, 5297–5304 (2021).
Calabresi, P., Castrioto, A., Di Filippo, M. & Picconi, B. New experimental and clinical links between the hippocampus and the dopaminergic system in Parkinson’s disease. Lancet Neurol. 12, 811–821 (2013).
Geschwind, D. H. & Konopka, G. Neuroscience in the era of functional genomics and systems biology. Nature 461, 908–915 (2009).
Breuer, M. E., Willems, P. H., Russel, F. G., Koopman, W. J. & Smeitink, J. A. Modeling mitochondrial dysfunctions in the brain: from mice to men. J. Inherit. Metab. Dis. 35, 193–210 (2012).
Ghosh, A. et al. Mitochondrial pyruvate carrier regulates autophagy, inflammation, and neurodegeneration in experimental models of Parkinson’s disease. Sci. Transl. Med. 8, 368ra174 (2016).
Rak, M. & Rustin, P. Supernumerary subunits NDUFA3, NDUFA5 and NDUFA12 are required for the formation of the extramembrane arm of human mitochondrial complex I. FEBS Lett. 588, 1832–1838 (2014).
Pereira, B., Videira, A. & Duarte, M. Novel insights into the role of Neurospora crassa NDUFAF2, an evolutionarily conserved mitochondrial complex I assembly factor. Mol. Cell Biol. 33, 2623–2634 (2013).
Lu, J. et al. Polymorphic variation in cytochrome oxidase subunit genes. J. Alzheimers Dis. 21, 141–154 (2010).
Dunnick, J. et al. Critical pathways in heart function: bis(2-chloroethoxy)methane-induced heart gene transcript change in F344 rats. Toxicol. Pathol. 34, 348–356 (2006).
Liu, J. et al. Integration of epidemiologic, pharmacologic, genetic and gut microbiome data in a drug-metabolite atlas. Nat. Med 26, 110–117 (2020).
Jakkamsetti, V. et al. Brain metabolism modulates neuronal excitability in a mouse model of pyruvate dehydrogenase deficiency. Sci. Transl. Med. https://doi.org/10.1126/scitranslmed.aan0457 (2019).
Giuliano, C. et al. Neuroprotective and symptomatic effects of cannabidiol in an animal model of Parkinson’s disease. Int. J. Mol. Sci. https://doi.org/10.3390/ijms22168920 (2021).
Wi, R., Chung, Y. C. & Jin, B. K. Functional crosstalk between CB and TRPV1 receptors protects nigrostriatal dopaminergic neurons in the MPTP model of Parkinson’s disease. J. Immunol. Res. 2020, 5093493 (2020).
Gugliandolo, A., Pollastro, F., Bramanti, P. & Mazzon, E. Cannabidiol exerts protective effects in an in vitro model of Parkinson’s disease activating AKT/mTOR pathway. Fitoterapia 143, 104553 (2020).
Udovin, L. et al. Role of astrocytic dysfunction in the pathogenesis of Parkinson’s disease animal models from a molecular signaling perspective. Neural Plast. 2020, 1859431 (2020).
Li, M. et al. Sensation of TRPV1 via 5-hydroxytryptamine signaling modulates pain hypersensitivity in a 6-hydroxydopamine induced mice model of Parkinson’s disease. Biochem. Biophys. Res. Commun. 521, 868–873 (2020).
Crivelaro do Nascimento, G. et al. Cannabidiol increases the nociceptive threshold in a preclinical model of Parkinson’s disease. Neuropharmacology 163, 107808 (2020).
Kang, B., Li, N., Liu, S., Qi, S. & Mu, S. Protective effect of isopulegol in alleviating neuroinflammation in lipopolysaccharide-induced BV-2 cells and in Parkinson disease model induced with MPTP. J. Environ. Pathol. Toxicol. Oncol. 40, 75–85 (2021).
Du, Z. R. et al. GPER and IGF-1R mediate the anti-inflammatory effect of genistein against lipopolysaccharide (LPS)-induced nigrostriatal injury in rats. J. Steroid Biochem. Mol. Biol. 214, 105989 (2021).
Hou, Y. et al. Neuroprotective effects of short-chain fatty acids in MPTP induced mice model of Parkinson’s disease. Exp. Gerontol. 150, 111376 (2021).
Picconi, B., Piccoli, G. & Calabresi, P. Synaptic dysfunction in Parkinson’s disease. Adv. Exp. Med. Biol. 970, 553–572 (2012).
Wood, W. G., Gorka, C., Armbrecht, H. J., Williamson, L. S. & Strong, R. Fluidizing effects of centrophenoxine in vitro on brain and liver membranes from different age groups of mice. Life Sci. 39, 2089–2095 (1986).
Bertoni-Freddari, C., Giuli, C. & Pieri, C. The effect of acute and chronic centrophenoxine treatment on the synaptic plasticity of old rats. Arch. Gerontol. Geriatr. 1, 365–373 (1982).
Todd, R. D. & Neuman, R. J. Gene-environment interactions in the development of combined type ADHD: evidence for a synapse-based model. Am. J. Med. Genet. B Neuropsychiatr. Genet. 144B, 971–975 (2007).
Yeo, H. et al. Developmental expression and subcellular distribution of synaptotagmin 11 in rat hippocampus. Neuroscience 225, 35–43 (2012).
Bekirov, I. H., Nagy, V., Svoronos, A., Huntley, G. W. & Benson, D. L. Cadherin-8 and N-cadherin differentially regulate pre- and postsynaptic development of the hippocampal mossy fiber pathway. Hippocampus 18, 349–363 (2008).
Duan, X., Krishnaswamy, A., De la Huerta, I. & Sanes, J. R. Type II cadherins guide assembly of a direction-selective retinal circuit. Cell 158, 793–807 (2014).
Siddiqui, T. & Bhatt, L. K. Targeting sigma-1 receptor: a promising strategy in the treatment of Parkinson’s disease. Neurochem Res. 48, 2925–2935 (2023).
Malar, D. S. et al. Targeting sigma receptors for the treatment of neurodegenerative and neurodevelopmental disorders. CNS Drugs 37, 399–440 (2023).
Walker, J. M. et al. Sigma receptors: biology and function. Pharm. Rev. 42, 355–402 (1990).
Ronsisvalle, G. et al. Synthesis and binding affinity of cis-(-)- and cis-(+)-N-ethyleneamino-N-nordeoxymetazocine and cis-(-)-N-normetazocine analogues at sigma1, sigma2 and kappa opioid receptors. Eur. J. Pharm. Sci. 12, 277–284 (2001).
Maurice, T., Urani, A., Phan, V. L. & Romieu, P. The interaction between neuroactive steroids and the sigma1 receptor function: behavioral consequences and therapeutic opportunities. Brain Res. Brain Res. Rev. 37, 116–132 (2001).
Booth, R. G. & Baldessarini, R. J. +)-6,7-benzomorphan sigma ligands stimulate dopamine synthesis in rat corpus striatum tissue. Brain Res. 557, 349–352 (1991).
Chaki, S., Okuyama, S., Ogawa, S. & Tomisawa, K. Regulation of NMDA-induced [3H]dopamine release from rat hippocampal slices through sigma-1 binding sites. Neurochem Int. 33, 29–34 (1998).
Nuwayhid, S. J. & Werling, L. L. Sigma1 receptor agonist-mediated regulation of N-methyl-D-aspartate-stimulated [3H]dopamine release is dependent upon protein kinase C. J. Pharm. Exp. Ther. 304, 364–369 (2003).
Gao, J., Jian, J., Jiang, Z. & Van Schepdael, A. Screening assays for tyrosine kinase inhibitors: a review. J. Pharm. Biomed. Anal. 223, 115166 (2023).
Carrick, T. et al. Development of a scintillation proximity assay binding method for the human 5-hydroxytryptamine 6 receptor using intact cells. Anal. Biochem. 381, 27–32 (2008).
Nomura, D. K. et al. A brain detoxifying enzyme for organophosphorus nerve poisons. Proc. Natl. Acad. Sci. USA 102, 6195–6200 (2005).
Greenamyre, J. T., Cannon, J. R., Drolet, R. & Mastroberardino, P.-G. Lessons from the rotenone model of Parkinson’s disease. Trends Pharmacol. Sci. 31, 141 (2010).
Barrett, T. et al. NCBI GEO: archive for functional genomics data sets-update. Nucleic Acids Res. 41, D991–D995 (2013).
Kikuchi, M. et al. Disruption of a RAC1-centred network is associated with Alzheimer’s disease pathology and causes age-dependent neurodegeneration. Hum. Mol. Genet. 29, 817–833 (2020).
Ikonomovic, M. D. et al. Cholinergic plasticity in hippocampus of individuals with mild cognitive impairment: correlation with Alzheimer’s neuropathology. J. Alzheimers Dis. 5, 39–48 (2003).
Bennett, D. A. et al. Neuropathology of older persons without cognitive impairment from two community-based studies. Neurology 66, 1837–1844 (2006).
Hensley, K. et al. Analysis of postmortem ventricular cerebrospinal fluid from patients with and without dementia indicates association of vitamin E with neuritic plaques and specific measures of cognitive performance. J. Alzheimers Dis. 24, 767–774 (2011).
Duke, D. C., Moran, L. B., Pearce, R. K. & Graeber, M. B. The medial and lateral substantia nigra in Parkinson’s disease: mRNA profiles associated with higher brain tissue vulnerability. Neurogenetics 8, 83–94 (2007).
Finucane, H. K. et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 47, 1228–1235 (2015).
Consortium, G. P. A global reference for human genetic variation. Nature 526, 68 (2015).
Langfelder, P. & Horvath, S. Eigengene networks for studying the relationships between co-expression modules. BMC Syst. Biol. 1, 1–17 (2007).
Langfelder, P., Mischel, P. S. & Horvath, S. When is hub gene selection better than standard meta-analysis?. PLoS One 8, e61505 (2013).
Piñero, J. et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. https://doi.org/10.1093/nar/gkw943 (2016).
Piñero, J. et al. DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes. Database 2015, bav028 (2015).
Pinero, J. et al. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 48, D845–D855 (2020).
Smyth, G. K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, Article3 (2004).
Davis, S. & Meltzer, P. S. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 23, 1846–1847 (2007).
Lee, C., Kim, J., Shin, S. G. & Hwang, S. Absolute and relative QPCR quantification of plasmid copy number in Escherichia coli. J. Biotechnol. 123, 273–280 (2006).
Dolga, A. M. et al. Subcellular expression and neuroprotective effects of SK channels in human dopaminergic neurons. Cell Death Dis. 5, e999 (2014).
Gokul, K. Oral supplements of aqueous extract of tomato seeds alleviate motor abnormality, oxidative impairments and neurotoxicity induced by rotenone in mice: relevance to Parkinson’s disease. Neurochem. Res. 39, 1382–1394 (2014).
Taylor, M. J. et al. Association of genetic risk factors for psychiatric disorders and traits of these disorders in a Swedish population twin sample. JAMA Psychiatry 76, 280–289 (2019).
Tsikas, D. Assessment of lipid peroxidation by measuring malondialdehyde (MDA) and relatives in biological samples: analytical and biological challenges. Anal. Biochem. 524, 13–30 (2017).
Acknowledgements
We thank the assistance from the Artificial Intelligence Lab and the Big Data Center of Sun Yat-sen Memorial Hospital, Sun Yat-sen University. The work was funded by the National Key Research and Development Program of China (2023YFF1204900), the Natural Science Foundation of China (82371482), the Natural Science Foundation of Guangdong (2024A1515011363), Medical and Health Science and Technology Plan of Longgang Shenzhen (No. LGKCYLWS2022019), and Guangzhou Science and Technology Research Plan (2023A03J0659).
Author information
Authors and Affiliations
Contributions
H. Zhao, R.H., and Yd. Yang designed the study. H. Zhang and C.F. performed the analyses with assistance from H. Zhao. H. Zhang, L.L., F.L., and L.M. contributed to experimental design, computational analyses, immunohistochemistry experiments, cell culture study, neuronal nuclei isolations, metabolomics experiments, mitochondrial-related experiments, and behavioral analyses in mice. H. Zhang, C.F., Yh. Yang, D.N.C., and H. Zhao wrote the manuscript. Yd. Yang, Yh. Yang, D.N.C., and H. Zhao supervised the study. All authors discussed the results and interpretation, and contributed to the final version of the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, H., Fan, C., Li, L. et al. Repurposing the memory-promoting meclofenoxate hydrochloride as a treatment for Parkinson’s disease through integrative multi-omics analysis. npj Parkinsons Dis. 11, 167 (2025). https://doi.org/10.1038/s41531-025-01027-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41531-025-01027-7