Abstract
Many Parkinson’s disease (PD) patients manifest complications related to treatment called levodopa-induced dyskinesia (LID). Preventing the onset of LID is crucial to the management of PD, but the reasons why some patients develop LID are unclear. The ability to prognosticate predisposition to LID would be valuable for the investigation of mitigation strategies. Thirty rats received 6-hydroxydopamine to induce Parkinsonism-like behaviors before treatment with levodopa (2 mg/kg) daily for 22 days. Fourteen developed LID-like behaviors. Fluorodeoxyglucose PET, T2-weighted MRI and cerebral perfusion imaging were collected before treatment. Support vector machines were trained to classify prospective LID vs. non-LID animals from treatment-naïve baseline imaging. Volumetric perfusion imaging performed best overall with 86.16% area-under-curve, 86.67% accuracy, 92.86% sensitivity, and 81.25% specificity for classifying animals with LID vs. non-LID in leave-one-out cross-validation. We have demonstrated proof-of-concept for imaging-based classification of susceptibility to LID of a Parkinsonian rat model using perfusion-based imaging and a machine learning model.
Similar content being viewed by others
Introduction
Levodopa (dopamine precursor) is the gold standard for PD treatment (>90% of PD patients are treated with levodopa)1. Levodopa-induced dyskinesia (LID) is a complication of long-term pharmaceutical treatment of PD. LID typically presents as peak-dose involuntary choreic (jerky/irregular) movements. Nearly half of all PD patients treated with levodopa develop LID within 5 years, while >90% of all patients eventually develop LID after 15 years2,3. LID significantly impairs motor function and contradicts the effect of levodopa for managing the motor symptoms of PD, which affects quality of life during peak-dose times. Management options are limited but include adjusting the levodopa dosage, introducing or increasing dopamine agonists, or prescribing amantadine to help reduce dyskinesia. However, the interventions often provide only temporary relief and do not address the underlying pathophysiological mechanisms of LID, and many patients experience reduced responsiveness to therapy or may require invasive intervention. It was previously recommended that levodopa treatment must be withheld as long as possible to delay the LID development; however, this was later revised because the clinical benefit of levodopa is greater than the potential harm4,5,6. LID is now regarded as a consequence of standard pharmacological PD management in many patients.
There is currently no disease-modifying therapy for PD, and most patients eventually develop LID1. The key mechanism of LID is associated with excessive synaptic dopamine release, but other neurotransmitters (e.g., glutamate, serotonin) and non-neuronal factors (e.g., astrocytic signaling) are also involved7,8,9. One major innovation in LID management is the usage of intestinal gel formulations that enable the slow and sustained release of levodopa10. This helps reduce off-time; however, it does not solve the fundamental problem, since this approach focuses on refining dosage rather than addressing the underlying issues inherent to treatment with levodopa. Deep brain stimulation (DBS) is regarded as the most effective treatment option for LID; however, less than 10% of PD patients are eligible for DBS due to many contraindications (e.g., old age, cognitive impairment)11. DBS is also an invasive neurosurgical procedure that requires specialized resources and personnel. Even with marked improvements in drug delivery and the use of brain stimulation technology, LID is still an unavoidable reality for many PD patients.
An alternative approach would be preventive treatment. Less attention has been paid to those PD patients who do not develop LID (non-LID or NLID). Protective factors have previously been identified, but they are often limited to demographic variables (e.g., male sex and older age of disease onset) due to the lack of longitudinal data with deep phenotyping and lack of non-invasive investigation due to ethical reasons12. Animal models have greatly improved our understanding. For instance, we have developed a novel low-dose levodopa treatment model that mimics the variability of LID emergence in humans, which demonstrated that vasomotor response to dopamine is significantly altered in Parkinsonian LID rodents13,14.
This previous work in our group generated a neuroimaging dataset of 30 rodents, which were imaged with multiple modalities and time points, including a treatment-naive baseline13. In the present work, we detail a machine learning-based model to identify prospective LID in Parkinsonian rats using pre-treatment baseline neuroimaging data. We investigate the utility of three separate neuroimaging modalities and their multimodal combinations for this purpose: structural magnetic resonance (MR) imaging, cerebral perfusion imaging (flow rate and volumetric data) and fluorodeoxyglucose emission tomography (FDG PET). The proposed animal model will provide a novel window of opportunity for researchers to investigate the brains of animals susceptible to LID in the earliest possible stage, prior to chronic levodopa treatment.
Results
The purpose of the present study was limited to developing a machine learning model that classified LID vs. NLID animals using baseline neuroimaging data. Our previous study demonstrated that approximately 50% of the treated animals develop LID-like behaviors13. In the present study, 14 of 30 animals developed LID.
Modality-based performance comparison
Classification of LID vs. NLID status by support vector machine (SVM) was evaluated on 4 separate neuroimaging modalities (anatomical T2, cerebral blood flow, cerebral blood volume and FDG PET) for our rodent model (nLID = 14 and nNLID = 16) and their multimodal combinations. The performance of each was estimated by examining accuracy (ACC), area-under-curve (AUC), sensitivity (SEN), specificity (SPE), and both positive and negative predictive values (PPV/NPV) in leave-one-out cross-validation (LOOCV) over the entire subject set. All results are compiled in Table 1.
SVM performance varied considerably depending on the neuroimaging modality and modality combinations used for the classification of LID versus NLID subjects. Among individual modalities, models trained on perfusion data demonstrated very high performance. The SVM trained on CBV imaging achieved 86.67% ACC and 86.16% AUC. It also demonstrated strong performance with balanced SEN and SPE at 92.86% and 81.25% respectively. Similarly, the CBF model performed comparably well, with 88.84% AUC, 83.33% ACC, 62.49% SEN, and 100% SPE.
In contrast, models trained with structural and metabolic imaging performed less effectively. The model trained on T2-weighted MR images achieved 70.00% ACC and 60.27% AUC with a notably high 92.86% SEN (tied with CBV imaging) but a much lower 50.00% SPE. FDG PET imaging models performed similarly, with 70.00% ACC, 64.29% AUC, 64.29% SEN, and 75.00% SPE.
Multimodal combinations of imaging modalities were explored to potentially take advantage of complementary information and increase model performance. The combination of CBF and CBV outperformed all other multimodal methods with an ACC of 86.67%, 90.62% AUC, 100% SEN, and 75.00% SPE. On the other end of the spectrum, the lowest-performing multimodal combination was FDG PET combined with CBV, which produced a model with 66.67%, 70.09% AUC, 64.29% SEN, and 68.75% SPE.
Other multimodal combinations allowed models to accurately classify rodents with AUCs ranging from 80.36% to 83.93%. ACCs from 76.67% to 83.33% and reasonably balanced performance. Combining all modalities did not significantly improve performance beyond that of the best-performing unimodal models, with 83.33% ACC and 81.70% AUC, 85.71% SPE, and 81.25% SPE.
PPV and NPV also varied based on the modality. The CBV model had 78.79% PPV and 92.31% NPV, indicating strong overall prediction quality. The CBF model achieved 100% PPV and 76.19% NPV. By contrast, T2-weighted imaging models had a lower performance with 61.90% PPV and 88.89% NPV. FDG PET models performed at a very similar level with 69.23% PPV and 70.59% NPV. Other multimodal combinations yielded PPVs ranging from 73.68% to 100% and NPVs between 76.19% and 92.31%.
Though the combined cerebral perfusion model performed best in terms of AUC and tied with unimodal CBV on accuracy, further investigation was carried out on CBV-based modeling alone for its substantially better balance between positive and negative subjects.
Hyperplane visualization and permutation testing
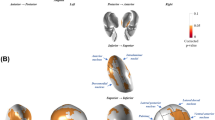
The hyperplane for the CBV-based SVM model was visualized on the Schwarz T2 intracranial template. Beta values were z-scored after extraction for ease of visual interpretation. Positive and negative values of the hyperplane (visualized in red and blue, respectively) reflect those features (brain voxels) that are strongly associated with classification in the positive (LID) or negative (NLID) class. To discern only the most significant voxels, permutation testing was performed. Subject labels were randomly permuted over 10,000 iterations. The voxel weights from each iteration’s hyperplane were compared to those from the original model and used to compute a significance map of p-values over the brain data volume. A logical mask of all voxels where p < 0.05 was created, and all other voxels were excluded in a refined hyperplane (Fig. 1).
Anatomical overlap of the model hyperplane
After permutation testing over 10,000 iterations and cluster size thresholding (50 voxels or larger), there remained 6 significant clusters of voxels within the SVM hyperplane. These clusters are visualized in Fig. 2. Positive contributions toward LID classification highlight the roles of the bilateral striatum (clusters 1 and 3) and right piriform cortex (cluster 4). Negative contributions are centered over the right globus pallidus, internal capsule and striatum (cluster 2), right insular cortex (cluster 5), left somatosensory cortex (cluster 6) and right piriform cortex (cluster 5). Note that the striatum showed mixed contributions on the right-hand side rostrocaudally, as did the piriform cortex mediolaterally. Table 2 contains a breakdown of the cluster properties and coordinates.
Clusters are visualized upon the Schwarz intracranial rat template MRI. Crosshairs indicate the center of mass for each cluster. Coordinates of the center of mass and extent over all three spatial dimensions are given in millimeters from the origin in real space. Clusters are shown in order of descending volume.
Discussion
The present work is a proof of concept for using pre-treatment perfusion imaging to classify susceptibility to LID in a Parkinsonian rodent model. Prior to this study, there had not been any known project attempting to classify Parkinsonian rodents by their susceptibility to LID with machine learning and multiple neuroimaging modalities. Among those tested, perfusion-based imaging (CBV and CBF) performed best for predicting LID susceptibility. They also indicate that structural (T2-weighted MR imaging) or metabolic (FDG PET) modalities alone may be insufficient for distinguishing between LID and NLID rodents for timeframes within the duration of study. An image-guided predictive model could assist investigators in noninvasively identifying rodent subjects at heightened risk for LID before symptoms emerge. Researchers could then develop experimental designs in which rodents identified as likely to develop LID can be specifically targeted for intervention.
Our findings align with emerging evidence from animal and human neuroimaging studies that suggest vascular abnormalities, specifically abnormal neurovascular coupling, play a central role in the pathogenesis of LID. For instance, Jourdain et al. have previously demonstrated a dissociation between levodopa-induced increases in striatal blood flow and stable glucose metabolism, but also found resting hyperperfusion in the putamen and globus pallidus of dyskinetic patients without corresponding changes in dopaminergic function15,16. These studies suggest that dysregulated cerebral perfusion is mechanistically linked to LID. Previous work in our group showed that animals with LID have dysregulated blood vessel response to dopaminergic stimulation. This disregulation results in increased blood flow to the striatum and midbrain structures at peak levodopa dose when vascular hyperreactivity causes exaggerated and transient surges in blood flow and dopamine influx13.
In addition to subcortical structures, our model identified the sensorimotor and insular cortices as contributors to LID classification. The involvement of the sensorimotor cortex is consistent with prior studies reporting increased connectivity and perfusion in this region during dyskinetic states and highlights the potential role of maladaptive cortical-striatal plasticity in LID vulnerability15,16. The insular cortex is important for integrating internal states, salience and affective signals with motor planning. It is tempting to explain its perfusion-based contributions to the model as evidence that insular regions may also participate in dyskinesia, either through neurovascular mechanisms or broader network-level dysregulation17. Together, these cortical features support a growing view that LID is not solely a basal ganglia disorder but emerges from distributed dysfunction across motor and non-motor domains. It is therefore plausible that the success of perfusion imaging in classifying future LID vs. non-LID status stems from capturing early, baseline differences that exist prior to levodopa exposure. Even before LID emerges behaviorally, subtle differences in cerebrovascular dynamics may be detectable through imaging metrics of CBV or CBF. In contrast, neither T2-weighted MR imaging nor FDG PET metabolic imaging revealed comparable predictive power at this stage. While metabolic and structural adaptations certainly evolve over time in PD, they may not reflect the acute or pre-symptomatic vasomotor predisposition that triggers dyskinesia once treatment with levodopa begins.
Our analysis and classification models face some limitations. First, although our sample size is within a typical range for rodent imaging studies, any machine learning approach in a small dataset raises concerns of overfitting, even with robust validation procedures like LOOCV. A larger study is warranted to ensure the generalizability of the method. Second, our low-dose levodopa regimen is designed to replicate variable LID onset in rats but might not mirror the diversity of clinical presentation in humans. Furthermore, it is possible that some of the NLID rodents would have eventually developed dyskinesia with extended observation periods and/or higher dosage of levodopa, implying that the NLID label in this work may reflect only a delayed onset rather than true resistance. All our animal subjects were females, making generalizability to male rodents a priority for any future studies. Finally, while our results suggest baseline differences detectable by CBV imaging and SVM, these are not necessarily causative and may in fact be epiphenomena of another yet unknown physiological mechanism contributing to the eventual onset of LID.
The findings in the present work demonstrate that perfusion-based imaging is a strong predictor of dyskinesia risk in a Parkinsonian rodent model, supporting the view that abnormal neurovascular coupling is a critical and potentially modifiable component of LID pathophysiology. If further validated, early detection of perfusion anomalies could guide preclinical research in testing strategies aimed at preventing or delaying LID, for example, by targeting astrocyte–vascular interactions. As LID remains a major challenge in long-term levodopa therapy for PD, studies like ours underscore the need to move beyond dopamine-centric models and incorporate the vascular dimension into both mechanistic and therapeutic considerations.
Methods
Methodological details and ethical approval
Previous research in our group led to the development of a novel Parkinsonian rodent model of LID that was used in the present work13,14. A validated rodent model of LID was developed using unilateral 6-hydroxydopamine (6-OHDA) lesions to induce dopaminergic denervation. This model was used to examine neurovascular and astrocytic calcium responses to dopamine in the dyskinetic state. Full details, including animal housing conditions, surgical procedures and humane endpoints are described in Booth et al.13,14. For the purposes of the present work, a summary of imaging and injection timepoints is given in Fig. 3.
All experiments received approval from the University of Manitoba’s Animal Care Committee and were conducted in compliance with the guidelines of the Canadian Council on Animal Care.
Levodopa dosage, behavioral testing, imaging schedule and acquisition parameters
Female Sprague–Dawley rats were housed under standard conditions (12-hour light/dark cycle, ad libitum food and water). Thirty rats underwent unilateral 6-OHDA lesioning to the right medial forebrain bundle (12.5 µg in 2.5 µL of 0.2% ascorbic acid/saline, infused over 5 min). To minimize extrasynaptic degradation, pargyline hydrochloride (10 mg/kg, intraperitoneal) was administered 30 min prior to surgery. After a 2-week recovery period, animals were screened for hemiparkinsonian-like behaviors using the cylinder test.
Following lesion validation, animals received daily levodopa (2 mg/kg subcutaneous) co-administered with benserazide (15 mg/kg) for 22 days. A control group (n = 5) received isotonic saline. Dyskinesia severity was assessed on days 1, 11, and 22 using the abnormal involuntary movements (AIM) test, scoring axial, orolingual and limb movements on a 0–4 scale. Animals with AIM scores ≥2 in at least one category by day 22 were classified as LID18. To confirm lesion integrity, immunohistochemical staining was performed on striatal and substantia nigra (SN) sections. First, tyrosine hydroxylase immunostaining quantified dopaminergic cell loss in the SN. Animals with <90% dopaminergic depletion in the substantia nigra, as determined by tyrosine hydroxylase staining, were excluded. This threshold was determined by prior work demonstrating that insufficient lesioning fails to produce consistent motor deficits or susceptibility to LID13,14.
Animals were scanned with PET-MR both OFF and ON levodopa at two timepoints: t1, when levodopa treatment first began and LID symptoms had not yet appeared (OFF1 and ON1 studies were performed on consecutive days; at OFF1, animals had never received levodopa treatment), and t2, when levodopa medication had been administered for 21 days (OFF2 and ON2 studies were also performed on consecutive days). On the day of OFF2, animals were administered levodopa immediately after scanning sessions. Dyskinesia was evaluated using three abnormal involuntary movement (AIMS) tests on days 1, 11, and 23. AIMS refers to uncoordinated, uncontrollable movements that occur without conscious effort and are reflected as side effects of PD. The details of the AIMS test scoring are described elsewhere18. The animals were sacrificed by perfusion fixation the day following the last AIMs test, and their brain tissue was removed.
The administration protocol differed for each imaging session. For ON sessions, animals were given 2 mg/kg levodopa with 15 mg/kg benserazide. An equivalent volume of isotonic saline was given prior to scanning for OFF sessions. After twenty minutes, the animal was placed in an induction chamber with 5% isoflurane and 0.5 L/min oxygen flow to induce anesthesia. The animals were shifted to a nasal cone with an oxygen flow rate of 0.5 l per minute and an isoflurane concentration of 1.5–2%. To administer tracer during the scan, a 23-gauge needle was used to cannulate the tail vein before loading onto the scanning bed. Rats were placed face down in the anesthetic system’s nasal cone. Throughout the imaging procedure, body temperature and respiration were recorded and kept constant at 37 ± 0.5 °C and 60–80 breaths per minute, respectively.
Studies were acquired with an MR Solutions 7.0 T drymag scanner with the clip-on PET module. The PET and MRI imaging were performed sequentially, and a rat head RF coil was used for all MR imaging.
Structural MR, dynamic susceptibility contrast (DSC) MR (gadolinium contrast enhancement) and FDG PET studies were collected. Anatomical images were acquired using a fast spin echo T2-weighted pulse sequence (26 ×1 mm coronal slices, TR = 4000 ms, TE = 45 ms, matrix size = 250 × 256 and 3 averages) over an acquisition time of 12 min. DSC perfusion maps were acquired using a gradient-echo planar imaging sequence (TR = 333 ms, TE = 20 ms, field of view = 30 × 30 mm, matrix size = 64 × 64, slice gap = 0.2 mm, slice thickness = 1.5 mm, temporal resolution = 0.333 s) such that the entire rostral striatum and the majority of caudal striatum were included in the acquisition. In total, 180 acquisitions were taken over 60 s, and 0.3 mmol/kg of Gadovist (gadobutrol-based contrast agent) in isotonic saline was injected through tail vein cannulation. From these, relative cerebral blood perfusion flow (CBF) images were generated using custom scripts in the MATLAB 2023a environment (MathWorks Inc., Natick, MA, USA) by voxel-wise calculation of the transverse relaxation rate curve maximum,
where S(t) is signal intensity at time t, S0 is the pre-contrast baseline intensity and TE is the echo time of the pulse sequence. Cerebral blood perfusion volume (CBV) images were generated by integrating over the perfusion flow volume. PET images were acquired immediately after DSC acquisition, and the bed position of the subject was moved to the PET coil for a 15-min acquisition. PET images were reconstructed using filtered back projection.
Neuroimaging data preprocessing
Preprocessing operations were performed using the Statistical Parametric Mapping (SPM12) software package (http://www.fil.ion.ucl.ac.uk/spm/ Wellcome Department of Cognitive Neurology, London, UK) within the MATLAB R2023a environment. All images were resized by ×10, then manually re-oriented to align with the Schwartz rat T2 template, which was also resized by ×10. The Schwartz template is based on 97 anatomical MR studies from Sprague Dawley rats, volumetrically reconstructed into Paxinos and Watson space19,20. All functional images (PET, CBF, and CBV) were coregistered to T2-MRI using SPM12’s default coregistration function and were visually inspected. T2-weighted structural images were spatially normalized into template space using the “Old Normalize” function in SPM12, with no affine regularization, a nonlinear frequency cutoff of 25 and 16 iterations. The same deformations were applied to write the coregistered functional images into template space. Resulting images were generated with the template image’s bounding box ([−95, −168, −138; 95, 70, 52]) and voxel size (2 × 2 × 2 mm). All functional images (FDG PET, CBF, and CBV) were smoothed with a Gaussian kernel with a full width at half maximum of 8 × 8 × 8 mm. Each image was proportionally scaled to the mean signal intensity of the entire volume.
Comparison of neuroimaging modalities with a support vector machine
An SVM is a very robust machine intelligence model that is commonly used for the purpose of binary classification. It functions on the principle of maximizing the margin between classes in an extremely high-dimensional feature space21. A series of SVMs (a priori defined hyperparameters: linear kernel, 5% outlier fraction and iterative single-data algorithm as optimizer) was trained to classify LID vs. NLID subjects, and modality-specific performance between models (T2-weighted MRI, CBV, CBF, and FDG PET) was compared by AUC, SEN, SPE, ACC, and both PPV and NPV. Multimodal combinations of neuroimaging data were also investigated as a possible method of increasing performance through complementary information.
Visualizing model decisions
The hyperplane of the best-performing model was visualized and underwent permutation testing. Study labels (LID vs. NLID) were permuted randomly, and an SVM was trained on this permuted data. The original hyperplane weights were ranked in the permuted distribution of hyperplane weights at each voxel, and the p-value was determined. Weights were deemed significant if they were ranked within the top 500 of 10,000 iterations (p < 0.05). Only the hyperplane weights of significant voxels were visualized and carried forward for further anatomical analysis under the assumption that these voxels are meaningful for LID vs NLID classification. The original hyperplane was z-scored relative to the intracranial voxel weights and then masked by the statistically significant voxels of the permuted hyperplane. Clusters of voxels were then characterized using MarsBaR and a pre-determined size threshold of 50 voxels22. The anatomy was identified by volume-of-interest analysis with the Schwarz atlas found in the Small Animal Molecular Imaging Toolbox (SAMIT)23.
Validation
Due to the modest sample size of our rodent population, we employed leave-one-out cross-validation (LOOCV) to estimate the generalizability of our models. This also allowed us to infer the effect of overfitting upon base models trained on all available data.
Data availability
Data may be shared by reasonable request to the corresponding author.
Code availability
Models and MATLAB scripts may be shared upon reasonable request to the corresponding author.
References
Turcano, P. et al. Levodopa-induced dyskinesia in Parkinson disease: a population-based cohort study. Neurology 91, e2238–e2243 (2018).
Ahlskog, J. E. & Muenter, M. D. Frequency of levodopa-related dyskinesias and motor fluctuations as estimated from the cumulative literature. Mov. Disord. 16, 448–458 (2001).
Hely, M. A., Morris, J. G., Reid, W. G. & Trafficante, R. Sydney Multicenter Study of Parkinson’s disease: non-L-dopa-responsive problems dominate at 15 years. Mov. Disord. 20, 190–199 (2005).
Fahn, S. A new look at levodopa based on the ELLDOPA study. J. Neural Transm. Suppl. 70, 419–426 (2006).
Fahn, S. Is levodopa toxic?. Neurology 47, S184–S195 (1996).
Cilia, R. et al. The modern pre-levodopa era of Parkinson’s disease: insights into motor complications from sub-Saharan Africa. Brain 137, 2731–2742 (2014).
Pavese, N. et al. Clinical correlates of levodopa-induced dopamine release in Parkinson disease: a PET study. Neurology 67, 1612–1617 (2006).
Jenner, P. Molecular mechanisms of L-DOPA-induced dyskinesia. Nat. Rev. Neurosci. 9, 665–677 (2008).
Ko, J. H., Lerner, R. P. & Eidelberg, D. Effects of levodopa on regional cerebral metabolism and blood flow. Mov. Disord. 30, 54–63 (2015).
Nyholm, D. Duodopa® treatment for advanced Parkinson’s disease: a review of efficacy and safety. Parkinsonism Relat. Disord. 18, 916–929 (2012).
Munhoz, R. P. et al. Eligibility criteria for deep brain stimulation in Parkinson’s disease, tremor, and dystonia. Can. J. Neurol. Sci.43, 462–471 (2016).
Tran, T. N., Vo, T. N. N., Frei, K. & Truong, D. D. Levodopa-induced dyskinesia: clinical features, incidence, and risk factors. J. Neural Transm. (Vienna) 125, 1109–1117 (2018).
Booth, S. et al. The vasomotor response to dopamine is altered in the rat model of l-dopa-induced dyskinesia. Mov. Disord. 36, 938–947 (2021).
Booth, S. Limitations in effective treatment of Parkinson’s Disease: neuroanatomical substrate of L-Dopa induced dyskinesia and cognitive impairment. in Human Anatomy and Cell Sciences (University of Manitoba, Winnipeg, Canada, 2024)
Jourdain, V. A. et al. Increased putamen hypercapnic vasoreactivity in levodopa-induced dyskinesia. JCI Insight 2, e96411 (2017).
Jourdain, V. A. et al. Flow-metabolism dissociation in the pathogenesis of levodopa-induced dyskinesia. JCI Insight 1, e86615 (2016).
Barbero, J. A., Unadkat, P., Choi, Y. Y. & Eidelberg, D. Functional brain networks to evaluate treatment responses in Parkinson’s disease. Neurotherapeutics 20, 1653–1668 (2023).
Cenci, M. A., Lee, C. S. & Björklund, A. L-DOPA-induced dyskinesia in the rat is associated with striatal overexpression of prodynorphin- and glutamic acid decarboxylase mRNA. Eur. J. Neurosci. 10, 2694–2706 (1998).
Paxinos, G. Paxinos & Watson the Rat Brain in Stereotaxic Coordinates: The New Coronal Set (Elsevier, 2005).
Schwarz, A. J. et al. A stereotaxic MRI template set for the rat brain with tissue class distribution maps and co-registered anatomical atlas: application to pharmacological MRI. Neuroimage 32, 538–550 (2006).
Perron, J. & Ko, J. H. Review of quantitative methods for the detection of Alzheimer’s disease with positron emission tomography. Appl. Sci. 12, 11463 (2022).
Brett, M., Anton, J.-L., Valabregue, R. & Poline, J.-B. Region of interest analysis using the MarsBar toolbox for SPM 99. Neuroimage 16, S497 (2002).
Vállez Garcia, D. et al. A standardized method for the construction of tracer specific PET and SPECT rat brain templates: validation and implementation of a toolbox. PLoS ONE 10, e0122363 (2015).
Acknowledgements
The authors acknowledge the animal care staff at the Center for Animal Care at Health Sciences Center (Winnipeg, Canada) and the animals used in this study for their invaluable contributions to our research. Financial disclosures and sources of support: J.P. received funding from the Natural Sciences and Engineering Research Council of Canada (CGS-D-579035-2023) and the Ricard Foundation’s Baxter Bursary. S.K. received no funding. S.B. received funding from the Parkinson Canada Graduate Student Award (GSA-2021-0000000065). J.K. received funding from Research Manitoba New Investigator Operating Grant (#2009), the Natural Sciences and Engineering Research Council of Canada Discovery Grant (RGPIN-2016-05964; RGPIN-2023-04283), Mitacs Accelerate (IT41654), and the University of Manitoba University Research Grants Program (#60473).
Author information
Authors and Affiliations
Contributions
Animal care, S.B. and D.Z.; animal evaluation, S.B. and D.Z.; animal tissue harvest, S.B.; conceptualization, J.P. and J.H.K.; conference presentation, J.P and S.K.; funding acquisition, J.P., S.K., S.B., D.Z., and J.H.K.; investigation, J.P., S.K.; methodology, J.P. and J.H.K.; resources, S.B. and D.Z.; supervision, J.H.K.; visualization, J.P.; writing-editing, J.P. and J.H.K.; writing-original draft preparation, J.P. and S.K.
Corresponding author
Ethics declarations
Competing interests
The authors make the following financial disclosures for the last 12 months. J.P. has none; S.K. was employed by J.K. through funding from a Mitacs Accelerate grant through Cubresa Inc., a manufacturer of imaging equipment for small animal research; S.B. has none; D.Z. has none; J.K. has none. Cubresa Inc. had no role in the design or conduct of the study; in the collection, analysis and interpretation of data; in the preparation of the manuscript; or in the review or approval of the manuscript.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Perron, J., Krak, S., Booth, S. et al. Cerebral perfusion imaging predicts levodopa-induced dyskinesia in Parkinsonian rat model. npj Parkinsons Dis. 11, 278 (2025). https://doi.org/10.1038/s41531-025-01133-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41531-025-01133-6
This article is cited by
-
Cerebrospinal fluid dopamine 3-O-sulfate as a novel biomarker for predicting motor complications in Parkinson’s disease: insights from the PPMI cohort
Journal of Translational Medicine (2026)