Abstract
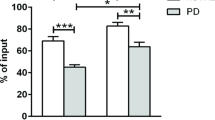
Parkinson’s disease (PD) is characterised by α-synuclein aggregation, dopaminergic neuron loss and chronic neuroinflammation. Disruption of the blood-brain barrier enables immune cell infiltration, including dendritic cells (DCs) and CD4+ T-cells, contributing to disease progression. To explore peripheral immune mechanisms in PD, we isolated DCs and CD4+ T-cells from the blood of 17 PD patients and 10 controls using magnetic separation, followed by flow cytometry and single-cell RNA sequencing. Cell-type annotation identified CD4+ T-cell and DC subtypes, including rare DC3 cells. PD patients showed reduced circulating DCs, with no change in CD4+ T-cell levels. Differential gene expression and pathway analysis suggest CD4+ effector memory T-cells (TEMs) and cDC2s as important mediators of immune responses in PD, enriched for immune-related pathways including T-cell activation and antigen presentation. Our findings implicate specific immune subsets in PD-associated neuroinflammation, suggesting cDC2s and CD4+ TEMs as potential targets for immunomodulatory strategies.
Similar content being viewed by others
Data availability
The single-cell RNA sequencing data obtained during this study will be made publicly available in the GitHub repository [https://github.com/PKatarina/PD_SingleCell] at the publication date of this article. All other data supporting this study are available from the corresponding authors upon reasonable request.
References
Ou, Z. et al. Global trends in the incidence, prevalence, and years lived with disability of Parkinson’s disease in 204 countries/territories from 1990 to 2019. Front. Public Health 9, 776847 (2021).
Ben-Shlomo, Y. et al. The epidemiology of Parkinson’s disease. Lancet 403, 283–292 (2024).
Savica, R. et al. Medical records documentation of constipation preceding Parkinson disease: a case-control study. Neurology 73, 1752–1758 (2009).
Bovenzi, R. et al. Shaping the course of early-onset Parkinson’s disease: insights from a longitudinal cohort. Neurol. Sci. 44, 3151–3159 (2023).
Jia, F., Fellner, A. & Kumar, K. R. Monogenic Parkinson’s disease: genotype, phenotype, pathophysiology, and genetic testing. Genes 13, 471 (2022).
Islam, M. S. et al. Pesticides and Parkinson’s disease: current and future perspective. J. Chem. Neuroanat. 115, 101966 (2021).
Zhao, J., Yu, S., Zheng, Y., Yang, H. & Zhang, J. Oxidative modification and its implications for the neurodegeneration of Parkinson’s disease. Mol. Neurobiol. 54, 1404–1418 (2017).
Moon, H. E. & Paek, S. H. Mitochondrial dysfunction in Parkinson’s disease. Exp. Neurobiol. 24, 103–116 (2015).
Ciaramella, A. et al. Blood dendritic cell frequency declines in idiopathic parkinson’s disease and is associated with motor symptom severity. PLoS ONE 8, e65352 (2013).
Williams, G. P. et al. CD4 T cells mediate brain inflammation and neurodegeneration in a mouse model of Parkinson’s disease. Brain 144, 2047–2059 (2021).
Trudler, D. et al. Soluble α-synuclein–antibody complexes activate the NLRP3 inflammasome in hiPSC-derived microglia. Proc. Natl. Acad. Sci. USA 118, e2025847118 (2021).
Takata, F., Nakagawa, S., Matsumoto, J. & Dohgu, S. Blood-brain barrier dysfunction amplifies the development of neuroinflammation: understanding of cellular events in brain microvascular endothelial cells for prevention and treatment of BBB dysfunction. Front. Cell. Neurosci. 15, 661838 (2021).
Al-Bachari, S., Naish, J. H., Parker, G. J. M., Emsley, H. C. A. & Parkes, L. M. Blood–brain barrier leakage is increased in Parkinson’s disease. Front. Physiol. 11, 593026 (2020).
Rock, R. B. et al. Role of microglia in central nervous system infections. Clin. Microbiol. Rev. 17, 942–964 (2004).
Wendimu, M. Y. & Hooks, S. B. Microglia phenotypes in aging and neurodegenerative diseases. Cells 11, 2091 (2022).
Mula, A., Yuan, X. & Lu, J. Dendritic cells in Parkinson’s disease: regulatory role and therapeutic potential. Eur. J. Pharmacol. 976, 176690 (2024).
Meena, M. et al. Transmigration across a steady-state blood–brain barrier induces activation of circulating dendritic cells partly mediated by actin cytoskeletal reorganization. Membranes 11, 700 (2021).
Brochard, V. et al. Infiltration of CD4+ lymphocytes into the brain contributes to neurodegeneration in a mouse model of Parkinson disease. J. Clin. Investig. 119, 182–192 (2009).
Luckheeram, R. V., Zhou, R., Verma, A. D. & Xia, B. CD4+ T cells: differentiation and functions. Clin. Dev. Immunol. 2012, 1–12 (2012).
Ahmadi, A., Gispert, J. D., Navarro, A., Vilor-Tejedor, N. & Sadeghi, I. Single-cell transcriptional changes in neurodegenerative diseases. Neuroscience 479, 192–205 (2021).
Jia, Q., Chu, H., Jin, Z., Long, H. & Zhu, B. High-throughput single-сell sequencing in cancer research. Signal Transduct. Target. Ther. 7, 145 (2022).
Zheng, G. X. Y. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017).
Pozojevic, J. & Spielmann, M. Single-cell sequencing in neurodegenerative disorders. Mol. Diagn. Ther. 27, 553–561 (2023).
Clarke, Z. A. et al. Tutorial: guidelines for annotating single-cell transcriptomic maps using automated and manual methods. Nat. Protoc. 16, 2749–2764 (2021).
Sulzer, D. et al. T cells from patients with Parkinson’s disease recognize α-synuclein peptides. Nature 546, 656–661 (2017).
Wang, P. et al. Single-cell transcriptome and TCR profiling reveal activated and expanded T cell populations in Parkinson’s disease. Cell Discov. 7, 52 (2021).
Xiong, L.-L. et al. Single-cell RNA sequencing reveals peripheral immunological features in Parkinson’s disease. Npj Park. Dis. 10, 185 (2024).
Freuchet, A. et al. Differential memory enrichment of cytotoxic CD4 T cells in Parkinson’s disease patients reactive to α-synuclein. Npj Park. Dis. 11, 127 (2025).
Rickenbach, C. et al. Altered T-cell reactivity in the early stages of Alzheimer’s disease. Brain 148, 3364–3378 (2025).
Wang, Y. et al. Characteristics of T cells in single-cell datasets of peripheral blood and cerebrospinal fluid in Alzheimer’s disease patients. J. Alzheimer’s. Dis. 99, S265–S280 (2024).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021).
Dutertre, C.-A. et al. Single-cell analysis of human mononuclear phagocytes reveals subset-defining markers and identifies circulating inflammatory dendritic cells. Immunity 51, 573–589.e8 (2019).
Xu, H. & Jia, J. Single-cell RNA sequencing of peripheral blood reveals immune cell signatures in Alzheimer’s disease. Front. Immunol. 12, 645666 (2021).
Mair, F. & Liechti, T. Comprehensive phenotyping of human dendritic cells and monocytes. Cytometry A 99, 231–242 (2021).
Wilk, A. J. et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 26, 1070–1076 (2020).
Poch, T. et al. Single-cell atlas of hepatic T cells reveals expansion of liver-resident naive-like CD4+ T cells in primary sclerosing cholangitis. J. Hepatol. 75, 414–423 (2021).
Szabo, P. A. et al. Single-cell transcriptomics of human T cells reveals tissue and activation signatures in health and disease. Nat. Commun. 10, 4706 (2019).
Liu, X., Zhu, Z. & Wang, X. Specificity and function of T cell subset identities using single-cell sequencing. Clin. Transl. Discov. 3, e199 (2023).
Saichi, M. et al. Single-cell RNA sequencing of blood antigen-presenting cells in severe COVID-19 reveals multi-process defects in antiviral immunity. Nat. Cell Biol. 23, 538–551 (2021).
Villani, A.-C. et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356, eaah4573 (2017).
Ashburner, M. et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 (2000).
Kanehisa, M., Sato, Y., Furumichi, M., Morishima, K. & Tanabe, M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 47, D590–D595 (2019).
Fabregat, A. et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 46, D649–D655 (2018).
Jin, S. et al. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 12, 1088 (2021).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019).
Scherzer, C. R. et al. Molecular markers of early Parkinson’s disease based on gene expression in blood. Proc. Natl. Acad. Sci. USA 104, 955–960 (2007).
Zhang, X. et al. Region-specific protein abundance changes in the brain of MPTP-induced Parkinson’s disease mouse model. J. Proteome Res. 9, 1496–1509 (2010).
Lei, K. et al. Immune-associated biomarkers for early diagnosis of Parkinson’s disease based on hematological lncRNA–mRNA co-expression. Biosci. Rep. 40, BSR20202921 (2020).
Liu, P., Li, P. & Burke, S. Critical roles of Bcl11b in T-cell development and maintenance of T-cell identity. Immunol. Rev. 238, 138–149 (2010).
Koga, T. Understanding the pathogenic significance of altered calcium-calmodulin signaling in T cells in autoimmune diseases. Clin. Immunol. 262, 110177 (2024).
Ferretti, A. P., Bhargava, R., Dahan, S., Tsokos, M. G. & Tsokos, G. C. Calcium/calmodulin kinase IV controls the function of both T cells and kidney resident cells. Front. Immunol. 9, 2113 (2018).
Moquin-Beaudry, G. et al. Mapping the peripheral immune landscape of Parkinson’s disease patients with single-cell sequencing. Brain 148, 2847–2860 (2025).
Kustrimovic, N. et al. Parkinson’s disease patients have a complex phenotypic and functional Th1 bias: cross-sectional studies of CD4+ Th1/Th2/T17 and Treg in drug-naïve and drug-treated patients. J. Neuroinflammation 15, 205 (2018).
Jiang, S., Gao, H., Luo, Q., Wang, P. & Yang, X. The correlation of lymphocyte subsets, natural killer cell, and Parkinson’s disease: a meta-analysis. Neurol. Sci. 38, 1373–1380 (2017).
Stevens, C. H. et al. Reduced T helper and B lymphocytes in Parkinson’s disease. J. Neuroimmunol. 252, 95–99 (2012).
Chen, Y. et al. Clinical correlation of peripheral CD4+-cell sub-sets, their imbalance and Parkinson’s disease. Mol. Med. Rep. 12, 6105–6111 (2015).
Lindestam Arlehamn, C. S. et al. α-Synuclein-specific T cell reactivity is associated with preclinical and early Parkinson’s disease. Nat. Commun. 11, 1875 (2020).
Geginat, J., Sallusto, F. & Lanzavecchia, A. Cytokine-driven proliferation and differentiation of human naive, central memory, and effector memory CD4ϩ T Cells. J. Exp. Med. 194, 1711–1719 (2001).
Serre-Miranda, C. et al. Effector memory CD4+ T cells are associated with cognitive performance in a senior population. Neurol. Neuroimmunol. Neuroinflammation 2, e54 (2015).
Liu, J., Zhang, X., Cheng, Y. & Cao, X. Dendritic cell migration in inflammation and immunity. Cell. Mol. Immunol. 18, 2461–2471 (2021).
Saito, Y., Komori, S., Kotani, T., Murata, Y. & Matozaki, T. The Role of type-2 conventional dendritic cells in the regulation of tumor immunity. Cancers 14, 1976 (2022).
Yin, X. et al. Human blood CD1c+ dendritic cells encompass CD5high and CD5low subsets that differ significantly in phenotype, gene expression, and functions. J. Immunol. 198, 1553–1564 (2017).
Bourdely, P. et al. Transcriptional and functional analysis of CD1c+ human dendritic cells identifies a CD163+ subset priming CD8+CD103+ T cells. Immunity 53, 335–352.e8 (2020).
Leal Rojas, I. M. et al. Human blood CD1c+ dendritic cells promote Th1 and Th17 effector function in memory CD4+ T cells. Front. Immunol. 8, 971 (2017).
Kleiveland, C. R. Peripheral blood mononuclear cells. in (eds. Verhoeckx, K. et al.) The Impact of Food Bioactives on Health, 161–167 (Springer International Publishing, 2015).
Zhang, X. et al. CD4 T cells with effector memory phenotype and function develop in the sterile environment of the fetus. Sci. Transl. Med. 6, 238ra72 (2014).
Dzamko, N. Cytokine activity in Parkinson’s disease. Neuronal Signal 7, NS20220063 (2023).
Di Lazzaro, G. et al. Differential profiles of serum cytokines in Parkinson’s disease according to disease duration. Neurobiol. Dis. 190, 106371 (2024).
Baron, B. W. & Pytel, P. Expression pattern of the BCL6 and ITM2B proteins in normal human brains and in Alzheimer disease. Appl. Immunohistochem. Mol. Morphol. 25, 489–496 (2017).
Yao, L. et al. Bioinformatic analysis of genetic factors from human blood samples and postmortem brains in Parkinson’s disease. Oxid. Med. Cell. Longev. 2022, 9235358 (2022).
Shaffer, A. L. et al. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity 13, 199–212 (2000).
Zhao, J. et al. CCL5 promotes LFA-1 expression in Th17 cells and induces LCK and ZAP70 activation in a mouse model of Parkinson’s disease. Front. Aging Neurosci. 15, 1250685 (2023).
Feng, D. et al. LncRNA SOX21-AS1 promotes activation of BV2 cells via epigenetical silencing of SOCS3 and aggravates Parkinson’s disease. Gerontology 70, 1063–1073 (2024).
Liu, Y. et al. LncRNA MALAT1 facilitates Parkinson’s disease progression by increasing SOCS3 promoter methylation. Gerontology 70, 1294–1304 (2024).
Semenova, E. I. et al. Analysis of ADORA2A, MTA1, PTGDS, PTGS2, NSF, and HNMT gene expression levels in peripheral blood of patients with early stages of Parkinson’s disease. BioMed. Res. Int. 2023, 9412776 (2023).
Contaldi, E., Magistrelli, L. & Comi, C. T Lymphocytes in Parkinson’s disease. J. Park. Dis. 12, S65–S74 (2022).
Chen, Z., Chen, S. & Liu, J. The role of T cells in the pathogenesis of Parkinson’s disease. Prog. Neurobiol. 169, 1–23 (2018).
Annunziato, F., Cosmi, L., Liotta, F., Maggi, E. & Romagnani, S. Defining the human T helper 17 cell phenotype. Trends Immunol. 33, 505–512 (2012).
Bourguin-Plonquet, A. et al. Severe decrease in peripheral blood dendritic cells in hairy cell leukaemia. Br. J. Haematol. 116, 595–597 (2002).
Mahnke, Y. D., Beddall, M. H. & Roederer, M. OMIP-017: Human CD4+ helper T-cell subsets including follicular helper cells. Cytometry A 83A, 439–440 (2013).
McGinnis, C. S., Murrow, L. M. & Gartner, Z. J. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 8, 329–337.e4 (2019).
Qiu, X. et al. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982 (2017).
Szklarczyk, D. et al. The STRING database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 51, D638–D646 (2023).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Bindea, G. et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093 (2009).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Acknowledgements
We thank nurse Andrea Klokočki from University Hospital Centre Zagreb for her valuable assistance in collecting blood samples and gratefully acknowledge all study participants for their generous contribution to this research. This study was conducted as a part of the ‘Molecular mechanisms of immune response and inflammasome activation in Parkinson's disease’ project (IP-2020-02-8475) funded by the Croatian Science Foundation. Sarah Meglaj Bakrač is funded by the Croatian Science Foundation’s Young Researchers’ Career Development Project, DOK-2021-02-5343. Katarina Mandić is funded by the Croatian Science Foundation project UIP-2020-02-1623. The funder played no role in study design, data collection, analysis and interpretation of data, or the writing of this manuscript.
Author information
Authors and Affiliations
Contributions
S.M.B. and A.B. performed single-cell sequencing experiments, data analysis and visualisation. K.M. conducted bioinformatics analysis, coding and visualisation of single-cell sequencing data. A.Ba. contributed to bioinformatics analysis, conceptualisation and supervision. F.B. selected participants and collected the samples. Z.M.S. and L.C.K. carried out PBMC isolation, magnetic separation, flow cytometry and flow cytometry data analysis and visualisation. F.B. acquired funding, supervised the project and validated the results. S.M.B., K.M., A.B. and Z.M.S. drafted the original manuscript. All authors reviewed, edited and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Meglaj Bakrač, S., Mandić, K., Cvetko Krajinović, L. et al. Single-cell analysis of the peripheral immune landscape in Parkinson’s disease: insights into dendritic cell and CD4+ T-cell transcriptomics. npj Parkinsons Dis. (2026). https://doi.org/10.1038/s41531-026-01283-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41531-026-01283-1