Abstract
The coronavirus disease 2019 (COVID-19) epidemic has brought major challenges to the global health system, and influenza is also a problem that cannot be ignored. We aimed to explore and compare the clinical characteristics of COVID-19 and influenza to deepen the understanding of these two diseases and provide some guidance for clinicians to make differential diagnoses. We searched PubMed, Embase and Web of Science for articles and performed a meta-analysis using Stata 14.0 with a random-effects model. This meta-analysis was conducted in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines. One hundred articles involving 226,913 COVID-19 patients and 201,617 influenza patients were included, and all the articles included patients with these two diseases as experimental and control groups. Compared to influenza, COVID-19 was more common among men (OR = 1.46, 95% CI: 1.23–1.74) and people with a higher body mass index (MD = 1.43, 95% CI: 1.09–1.77). The proportion of current smokers among COVID-19 patients was lower than that among influenza patients (OR = 0.25, 95% CI: 0.18–0.33). Patients with COVID-19 had longer stays in the hospital (MD = 3.20, 95% CI: 2.58–3.82) and ICU (MD = 3.10, 95% CI: 1.44–4.76), required mechanical ventilation more frequently (OR = 2.30, 95% CI: 1.77–3.00), and had higher mortality (OR = 2.22, 95% CI: 1.93–2.55). We also found significant differences in some blood parameters between the two groups of patients. Upper respiratory symptoms were more obvious in influenza patients, and the proportion of comorbidities was higher than that among COVID-19 patients. There are some differences in the major characteristics, symptoms, laboratory findings and comorbidities between COVID-19 patients and influenza patients. COVID-19 patients often require more medical resources and have worse clinical outcomes.
Similar content being viewed by others
Introduction
In February 2020, the World Health Organization (WHO) named the disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) coronavirus disease 2019 (COVID-19). The first case occurred in Wuhan, China, in December 2019, and COVID-19 quickly spread worldwide, posing great challenges to the health systems of various countries1,2. SARS-CoV-2 shares a 79.6% nucleotide identity with SARS-CoV and 51.8% identity with Middle East respiratory syndrome coronavirus (MERS-CoV). The latter two pathogens caused regional outbreaks of acute respiratory diseases in 2002–2003 and 2012, respectively, and each disease caused hundreds of deaths3,4,5. Influenza virus is an RNA virus that can cause influenza in humans and animals and belongs to the Orthomyxoviridae family. This family has four genera; however, only genera A and B are clinically relevant to humans6. Influenza has obvious seasonality7. The WHO estimates that the annual influenza season usually leads to approximately 3–5 million critical cases and 290,000 to 650,000 deaths8. Although influenza often leads to significant mortality and morbidity, the public usually thinks that influenza is a trivial disease, similar to the common cold9. In the early stage of the pandemic, COVID-19 was downplayed as only a ‘little flu’ by some people10. There are many similarities between SARS-CoV-2 and influenza virus in terms of transmission and manifestations, but compared with influenza virus, SARS-CoV-2 is more contagious11, and influenza makes diagnosing COVID-19 difficult12.
We aimed to explore and compare the clinical characteristics of COVID-19 and influenza to deepen the understanding of these two diseases and provide some guidance for clinicians to make differential diagnoses. In November 2021, the Omicron variant was first detected and quickly became the main pathogenic strain worldwide, which was quite different from those of previous strains13. The virulence of the Omicron variant has significantly decreased with a lower rate of severe cases of disease, hospitalizations, and deaths, but it is more infectious and transmissible14,15. The non-Omicron strains primarily affects the lungs, whereas the Omicron variant primarily affects the upper respiratory tract16. A study by the University of Hong Kong found that the Omicron variant infects human bronchi more than 70 times faster than the Alpha variant and has a higher replication speed. In contrast, the original strain infects the lung more than 10 times faster than the Omicron variant, and the replication speed is also faster than the latter17. We think that it is not rigorous enough to put the Omicron variant and the non-Omicron strains in the same group when comparing the clinical characteristics of COVID-19 and influenza. Therefore, patients infected with the Omicron variant were not included in our study.
Materials and methods
This meta-analysis was conducted in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (PROSPERO registration number: CRD42023397835).
Eligibility criteria
Articles that met the following criteria were included in this meta-analysis: 1) articles with controls; 2) the experimental group and the control group were COVID-19 and influenza patients, respectively; and 3) the data collection time of COVID-19 patients had to be before November 2021, that is, before the outbreak of the Omicron variant of SARS-CoV-213.
The exclusion criteria were as follows: 1) animal studies, 2) case reports, 3) reviews and comments, 4) duplicate data, 5) samples from children only, and 6) sample size less than five.
Information sources and search strategy
We searched PubMed, Embase and Web of Science for articles published before November 25, 2023. There was no restriction on the language of the article to collect useful information on a global scale. The search strategy in PubMed was as follows: ((((((COVID-19[Title]) OR (2019-nCoV[Title])) OR (Coronavirus Disease 2019[Title])) OR (SARS-CoV-2[Title])) OR (2019 Novel Coronavirus Disease[Title])) OR (2019 Novel Coronavirus Infection[Title])) AND ((((Flu[Title]) OR (Influenza[Title])) OR (Influenzas[Title])) OR (Grippe[Title])).
Study selection process
All articles obtained from PubMed, Embase and Web of Science were imported into NoteExpress software. We first deleted duplicate articles through the matching function of the software and then conducted a preliminary screening by reading titles and abstracts to exclude articles that were not related to our study. Finally, through full-text reading, selected articles were further screened to determine which articles could be included in our meta-analysis.
Data selection process and items
Data extraction was performed independently by two authors to ensure the accuracy of the data. Disagreements were resolved by discussion and referred to a third author for a final decision.
The following information on COVID-19 and influenza patients was included in this meta-analysis: 1) major characteristics, including age, sex, body mass index (BMI), length of hospital stay, length of intensive care unit (ICU) stay, number of current smokers, number of patients admitted to the ICU, number of patients who received mechanical ventilation, number of patients who received extracorporeal membrane oxygenation (ECMO) and number of deaths; 2) symptoms, including fever, cough, expectoration, dyspnea, rhinorrhea, sore throat, headache, chest pain, myalgia, nausea, vomiting, diarrhea and abdominal pain; 3) laboratory findings, including lymphocytes, white blood cells, neutrophils, C-reactive protein, creatinine, hemoglobin, platelets, procalcitonin, aspartate aminotransferase (AST), alanine aminotransferase (ALT), lactate dehydrogenase (LDH) and creatine kinase; and 4) comorbidities, including diabetes, hypertension, heart failure, coronary heart disease, chronic renal failure, stroke, cancer, chronic liver disease, asthma, chronic obstructive pulmonary disease (COPD) and immunodepression.
Study risk of bias assessment
The Newcastle–Ottawa quality assessment scale was used to assess the quality and risk of bias of the included articles. A total score of seven or more meant that the article had a low risk of bias and high quality.
Reporting bias assessment
We used funnel plots and Egger’s test to evaluate reporting bias assessment, and a p value < 0.05 indicated the presence of bias.
Statistical analysis
Data were analyzed and evaluated by using odds ratios (ORs), mean differences (MDs) and standardized mean differences (SMDs), where ORs were applied to dichotomous variables, MDs were applied to continuous variables with the same units of measurement, SMDs were applied to continuous variables with different units of measurement, and confidence intervals (CIs) were both set at 95%. For data with only sample size and quartile, we used the transformation formula to calculate the mean and standard deviation18. The I2 statistic was used to quantify heterogeneity: 0–40% may not be important; 30–60% may represent moderate heterogeneity; 50–90% may represent substantial heterogeneity; 75–100% may represent considerable heterogeneity19. Subgroup analysis was used to investigate the source of heterogeneity. A random-effects model was used to estimate the effect value. Stata 14.0 was used for statistical analysis, and a p value of z test < 0.05 indicated that the result was statistically significant.
Results
Study selection
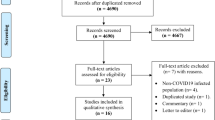
From the three databases, we retrieved a total of 4968 articles and deleted 2398 duplicate articles through NoteExpress software. A total of 2482 articles that were not relevant to our study were excluded by reading the titles and abstracts. Of the remaining 388 articles, 288 were further screened and excluded by reading the full text. If data from multiple articles were from the same research institution and the dates of extraction of patient information overlapped, the data were considered to be at risk of duplication, the article with higher quality was retained, and the rest were deleted. The flow diagram of the article selection process is shown in Fig. 1.
Risk of bias in studies
The Newcastle–Ottawa quality assessment scale is described in Supplementary Table 1. We found that most of the articles included in this meta-analysis were of high quality and had a low risk of bias.
Characteristics and results of individual studies
After screening, a total of 100 articles were included in this meta-analysis (Table 1), including eight prospective studies, 78 retrospective studies, and 14 studies that did not specify the design. Data were collected from 27 regions and included 226,913 COVID-19 patients and 201,617 influenza patients. There were 17 articles that specifically studied patients with influenza A, 41 articles in which the type of influenza was A/B, and the remaining 42 articles did not record the influenza subtype. The earliest data for influenza were from 2006, and all the data for COVID-19 were obtained before the outbreak of the Omicron variant of SARS-CoV-2.
Results of syntheses
Major characteristics (Supplementary Table 2)
The age characteristics20,21,22,23 of four studies, sex characteristics20,21,22,24 of four studies and BMI characteristics23 of one study were screened after matching the experimental group and the control group; thus, their data were not included in this meta-analysis. We found no significant differences in age between patients with COVID-19 and those with influenza (MD = 0.21, 95% CI: −0.99–1.41, I2 = 98.0%, p = 0.730, Supplementary Figure 1). Compared to influenza, COVID-19 was more common among men (OR = 1.46, 95% CI: 1.23–1.74, I2 = 99.0%, p < 0.001, Supplementary Figure 2) and people with a higher BMI (MD = 1.43, 95% CI: 1.09–1.77, I2 = 67.4%, p < 0.001, Supplementary Figure 3). The proportion of current smokers among COVID-19 patients was lower (OR = 0.25, 95% CI: 0.18–0.33, I2 = 55.4%, p < 0.001, Fig. 2) than that among influenza patients. Although the use of ECMO did not differ between the two groups (OR = 1.06, 95% CI: 0.76–1.47, I2 = 62.5%, p = 0.740, Fig. 3), patients with COVID-19 had longer stays in the hospital (MD = 3.20, 95% CI: 2.58–3.82, I2 = 98.2%, p < 0.001, Supplementary Figure 4) and ICU (MD = 3.10, 95% CI: 1.44–4.76, I2 = 96.9%, p < 0.001, Supplementary Figure 5), more patients required mechanical ventilation (OR = 2.30, 95% CI: 1.77–3.00, I2 = 93.7%, p < 0.001, Supplementary Figure 6) and ICU admission (OR = 2.03, 95% CI: 1.72–2.40, I2 = 95.6%, p < 0.001, Supplementary Figure 7), and patients had higher mortality (OR = 2.22, 95% CI: 1.93–2.55, I2 = 94.1%, p < 0.001, Fig. 4).
Symptoms (Supplementary Table 3, Supplementary Figures 8–20)
COVID-19 patients often present with influenza-like symptoms. By comparison, there was no significant difference in the proportion of patients with abdominal pain (OR = 1.06, 95% CI: 0.83–1.36, I2 = 0.0%, p = 0.657), chest pain (OR = 0.92, 95% CI: 0.55–1.53, I2 = 31.8%, p = 0.753), headache (OR = 1.04, 95% CI: 0.72–1.50, I2 = 75.8%, p = 0.835), myalgia (OR = 0.72, 95% CI: 0.49–1.05, I2 = 76.0%, p = 0.09), fever (OR = 0.73, 95% CI: 0.48–1.11, I2 = 85.3%, p = 0.139), nausea (OR = 0.81, 95% CI: 0.40–1.64, I2 = 73.9%, p = 0.551), vomiting (OR = 1.15, 95% CI: 0.51–2.56, I2 = 64.3%, p = 0.739), or dyspnea (OR = 0.76, 95% CI: 0.50–1.17, I2 = 80.2%, p = 0.217) between COVID-19 patients and influenza patients. Patients with COVID-19 were more likely to have diarrhea (OR = 1.65, 95% CI: 1.30–2.08, I2 = 19.6%, p < 0.001), while rhinorrhea (OR = 0.38, 95% CI: 0.19–0.75, I2 = 78.7%, p = 0.005), expectoration (OR = 0.29, 95% CI: 0.22–0.39, I2 = 47.3%, p < 0.001), cough (OR = 0.54, 95% CI: 0.40–0.72, I2 = 78.1%, p < 0.001) and sore throat (OR = 0.64, 95% CI: 0.44–0.93, I2 = 73.0%, p = 0.020) were more common among influenza patients.
Laboratory findings (Supplementary Table 4, Supplementary Figures 21–32)
Our study showed that platelet counts (SMD = 0.42, 95% CI: 0.28–0.56, I2 = 92.2%, p < 0.001) and hemoglobin (SMD = 0.24, 95% CI: 0.07–0.41, I2 = 91.5%, p = 0.006) and ALT (SMD = 0.35, 95% CI: 0.17–0.54, I2 = 88.6%, p < 0.001) levels were higher in COVID-19 patients than in influenza patients, while creatinine levels (SMD = −0.14, 95% CI:−0.03–−0.25, I2 = 75.3%, p = 0.012), procalcitonin levels (SMD = −0.47, 95% CI: −0.73–−0.22, I2 = 81.5%, p < 0.001), neutrophil levels (SMD = −0.48, 95% CI: −0.79–−0.17, I2 = 97.0%, p = 0.003) and white blood cell counts (SMD = −0.26, 95% CI: −0.37–−0.16, I2 = 88.5%, p < 0.001) were significantly lower than in influenza patients. There were no significant differences in lymphocyte counts (SMD = 0.14, 95% CI: 0.00–0.27, I2 = 90.1%, p = 0.05) or C-reactive protein (SMD = −0.05, 95% CI: −0.28–0.18, I2 = 95.6%, p = 0.664), LDH (SMD = 0.05, 95% CI: −0.23–0.33, I2 = 95.4%, p = 0.773), AST (SMD = 0.09, 95% CI: −0.20–0.37, I2 = 94.3%, p = 0.551) or creatine kinase (SMD = −0.09, 95% CI: −0.31–0.14, I2 = 81.9%, p = 0.454) levels between the two groups.
Comorbidities (Supplementary Table 5, Supplementary Figures 33–43)
We compared 11 comorbidities between patients with COVID-19 and those with influenza and found that patients with COVID-19 were more likely to develop diabetes (OR = 1.09, 95% CI: 1.01 –1.17, I2 = 86.2%, p = 0.028). Patients with COVID-19 were less likely to have most of the comorbidities than patients with influenza (heart failure (OR = 0.63, 95% CI: 0.53–0.74, I2 = 96.6%, p < 0.001), coronary heart disease (OR = 0.59, 95% CI: 0.76–0.99, I2 = 0.0%, p = 0.039), stroke (OR = 0.82, 95% CI: 0.69–0.96, I2 = 76.7%, p = 0.015), cancer (OR = 0.62, 95% CI: 0.51–0.75, I2 = 94.0%, p < 0.001), chronic renal failure (OR = 0.70, 95% CI: 0.51–0.98, I2 = 70.3%, p = 0.039), chronic liver disease (OR = 0.62, 95% CI: 0.39–0.98, I2 = 48.0%, p = 0.043), COPD (OR = 0.42, 95% CI: 0.36–0.49, I2 = 82.9%, p < 0.001) and immunodepression (OR = 0.43, 95% CI: 0.33–046, I2 = 93.8%, p < 0.001)); however, there was no significant difference in the prevalence of hypertension (OR = 0.97, 95% CI: 0.89–1.07, I2 = 92.2%, p = 0.571) or asthma (OR = 0.75, 95% CI: 0.55–1.01, I2 = 75.4%, p = 0.058).
Reporting biases
Funnel plots and Egger’s test were used for reporting bias analysis, and we did not find reporting bias in most of the studies (Supplementary Figures 44–131).
Heterogeneity
Some outcomes in our study might have substantial or considerable heterogeneity, and we attempted to perform a subgroup analysis by using the region or influenza subtype as the basis for classification, but unfortunately, we did not find an accurate source of heterogeneity.
Discussion
The COVID-19 pandemic has been the worst global health crisis since the 1918 H1N1 influenza pandemic, and its impact on the global health care system is much greater than that of influenza25. The characteristics of this new respiratory disease are inevitably compared to seasonal influenza because of their similar symptoms and infectious nature. Angiotensin-converting enzyme 2 (ACE2), a functional receptor of SARS-CoV-2, is crucial for the fusion of virus and cell membranes, and its expression and activity directly mediate SARS-CoV-2 infection. ACE2 is highly expressed not only in lung alveolar type 2 cells and gland cells but also in the ileum and colon and less so on the surface of nasopharyngeal cells26,27. The influenza virus mainly binds to alpha2,6-linked cell receptors via sialic acid-linked glycoproteins. The distribution of sialic acid on the cell surface is one of the determinants of host tropism, and these receptors are expressed particularly in the respiratory tract, from the nasopharynx and trachea to the bronchi, with the exception of the alveoli28. Therefore, it is not surprising that patients with influenza in our study were more likely to present with upper respiratory symptoms than patients with COVID-19, and COVID-19 patients experienced a greater occurrence of diarrhea.
In earlier reports, COVID-19 was more common among men, and our study further indicated that patients with COVID-19 had a higher proportion of men than patients with influenza, which may be associated with higher levels of ACE2 expression in men29. To minimize the spread of SARS-CoV-2, maintaining social distance is recommended in many areas, which may increase loneliness, depression, and psychological pressure in some people, thereby adversely affecting dietary habits and reducing exercise, which can lead to an increase in BMI. Obese individuals are more susceptible to SARS-CoV-2, and obesity is an independent risk factor for COVID-1930. It is well known that smokers are mostly men31. Although the proportion of men among patients with COVID-19 was higher, we found that the proportion of current smokers among COVID-19 patients was significantly lower than that among influenza patients. Previous studies have noted a lower rate of SARS-CoV-2 infection in countries with a high rate of smoking32. Farsalinos et al. even proposed a controversial idea of nicotine as a therapeutic option for COVID-1933. Nicotine acts in a similar fashion as the naturally occurring neurotransmitter acetylcholine on nicotinic acetylcholine receptors and may have anti-inflammatory effects34. Patients with COVID-19 spent longer in the hospital and ICU than patients with influenza and were more frequently admitted to the ICU, reflecting the fact that COVID-19 patients require more medical resources and have a more serious medical condition. Moreover, a greater use of mechanical ventilation also suggested that SARS-CoV-2 was more likely to cause lower respiratory symptoms than influenza virus. Comparing the mortality of these two groups of patients worldwide is always challenging. The main reason is that influenza deaths are usually based on estimates, whereas for COVID-19, although there is underreporting, deaths are currently reported as direct counts rather than estimates35. We included clinical data from multiple regions worldwide before the outbreak of Omicron and found that mortality among patients with COVID-19 was much higher than that among patients with influenza.
Diabetes leads to a functional immune deficiency and increased ACE2 expression in humans, leading to an increase in susceptibility to SARS-CoV-2 and exacerbation of the disease36. The above may explain why we observed a higher prevalence of diabetes among patients with COVID-19 than among patients with influenza. However, in our findings, patients with influenza tended to have more comorbidities than patients with COVID-19, which seemed to suggest that SARS-CoV-2 was more likely to infect healthy people. This may be because people with comorbidities were more cautious during the COVID-19 pandemic and were more active in wearing masks and avoiding social contact and thus had a lower incidence of COVID-19 compared to influenza. Oronasal entry of respiratory viruses results in their direct infection in the pulmonary system with the opportunity to enter the blood and then come into contact with extrapulmonary organs through the bloodstream, so patients with COVID-19 or influenza often have some abnormalities in blood parameters37. In the results of our meta-analysis, there were significant differences in some laboratory findings between patients with COVID-19 and patients with influenza, but the relative increase or decrease in the indices could not represent the severity of organ injury or disease, which only provided a certain reference for distinguishing the two viral infections. At present, the gold standard for distinguishing the two diseases is still reverse transcription-polymerase chain reaction (RT‒PCR)38.
In the early stage of the COVID-19 pandemic, the global circulation of influenza virus also decreased. This may be due to the following reasons: 1) To curb the spread of SARS-CoV-2, countries adopted various social isolation measures and strictly enforced social distancing, which also prevented the spread of influenza virus39. 2) During the pandemic, doctors prioritized the detection of SARS-CoV-2 and reduced efforts to detect influenza virus40. 3) SARS-CoV-2 infection may provide some cross-protection against influenza through nonspecific mechanisms, which stimulate antiviral defenses or trigger an adaptive response to secondary infection with influenza41. However, specific policies to control the COVID-19 epidemic, such as banning social gatherings and travel restrictions, are unlikely to be implemented every year. During the COVID-19 epidemic, low levels of influenza activity may have reduced people’s immunity, and cross-reactive antibodies against influenza infection often expire within one year42.
In the future, COVID-19 is likely to become a seasonal epidemic similar to influenza43. It is also very important to prevent the coinfection of these two diseases because this increases the severity of infection. A study by Public Health England showed that the risk of death of patients infected with both diseases was approximately two times higher than that of patients with COVID-19 alone44. Adherence to vaccination against both viruses is the most effective means of reducing morbidity, mortality and economic impact. Although SARS-CoV-2 infection may occur in a fully vaccinated population, vaccinated individuals are still much less likely to become infected, die, or transmit the disease to others than unvaccinated individuals45.
Limitations
This meta-analysis had the following limitations. Most of the articles included in this study were retrospective, and of the 100 articles, only 1822,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62 explicitly noted that the sample did not contain patients with SARS-CoV-2 and influenza virus coinfection, and the coinfection was not described in detail in other articles, so we were unable to determine the absence of coinfection in all samples. Because we could not extract enough data from the included studies, we could not perform subgroup analysis according to SARS-CoV-2 variant or vaccination status of COVID-19 patients. We did not include the data of Omicron patients in this study, and intend to focus on this field in future studies.
Conclusions
There are some differences in the major characteristics, symptoms, laboratory findings and comorbidities between COVID-19 patients and influenza patients. COVID-19 patients often require more medical resources and have worse clinical outcomes.
Data Availability
No datasets were generated or analysed during the current study.
References
Naming the coronavirus disease and the virus that causes it; Technical guidance; World Health Organization; Available online at: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it (Accessed November 25, 2023).
Sim, M. R. The COVID-19 pandemic: major risks to healthcare and other workers on the front line. Occup. Env. Med. 77, 281–282 (2020).
Zhou, P. et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 579, 270–273 (2020).
Ren, L. L. et al. Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study. Chin. Med. J. 133, 1015–1024 (2020).
Hosseiny, M., Kooraki, S., Gholamrezanezhad, A., Reddy, S. & Myers, L. Radiology perspective of coronavirus disease 2019 (COVID-19): lessons from severe acute respiratory syndrome and middle east respiratory syndrome. AJR Am. J. Roentgenol. 214, 1078–1082 (2020).
Venkatram, S., Alapati, A., Dileep, A. & Diaz-Fuentes, G. Change in patterns of hospitalization for influenza during COVID-19 surges. Influenza Other Respir. Viruses 16, 72–78 (2022).
Tan, M. P. et al. Dearth of influenza among older adults admitted with respiratory symptoms in Malaysia during the coronavirus disease 2019 pandemic in 2021. Front. Med. 9, 977614 (2022).
World Health Organization. Influenza (Seasonal). Available online: https://www.who.int/news-room/fact-sheets/detail/influenza-(seasonal) (Accessed November 25, 2023).
Bekkat-Berkani, R. & Romano-Mazzotti, L. Understanding the unique characteristics of seasonal influenza illness to improve vaccine uptake in the US. Vaccine. 36, 7276–7285 (2018).
Picheta, R., Cotovio, V. & Darlington, S. As hospitals in Brazil teeter on the brink of collapse, Bolsonaro does pushups with supporters. CNN;May 18, 2020. Available online at: https://edition.cnn.com/2020/05/18/americas/brazil-coronavirus-sao-paulo-bolsonaro-rally-intl/index.html (Accessed November 25, 2023).
Olsen, S. J. et al. Decreased influenza activity during the COVID-19 pandemic-United States, Australia, Chile, and South Africa, 2020. Am. J. Transplant. 20, 3681–3685 (2020).
Wu, X. J. et al. Co‐infection with SARS‐CoV‐2 and influenza A virus in patient with pneumonia, China. Emerg. Infect. Dis. 26, 1324–1326 (2020).
Chen, J., Wang, R., Gilby, N. B. & Wei, G. W. Omicron variant (B.1.1.529): infectivity, vaccine breakthrough, and antibody resistance. J. Chem. Inf. Model. 62, 412–422 (2022).
Radhakrishnan, N. et al. Comparison of the clinical characteristics of SARS-CoV-2 Delta (B.1.617.2) and Omicron (B.1.1.529) infected patients from a single hospitalist service. BMC Infect. Dis. 23, 747 (2023).
Pingping, Z. et al. Comparison between original SARS-CoV-2 strain and omicron variant on thin-section chest CT imaging of COVID-19 pneumonia. Radiologie 63, 55–63 (2023).
Yang, N. et al. Clinical and pulmonary CT characteristics of patients infected with the SARS-CoV-2 omicron variant compared with those of patients infected with the alpha viral strain. Front. Public. Health. 10, 931480 (2022).
HKUMed Finds Omicron SARS-CoV-2 Can Infect Faster and Better Than Delta in Human Bronchus but With Less Severe Infection in Lung. Available online at: https://www.med.hku.hk/en/news/press/20211215-omicron-SARS-CoV-2-infection (Access January 11, 2025).
Wan, X., Wang, W., Liu, J. & Tong, T. Estimating the sample mean and standard deviation from the sample size, median, range and/or interquartile range. BMC Med. Res. Methodol. 14, 135 (2014).
Cochrane Handbook for Systematic Reviews of Interventions. Available online at: https://training.cochrane.org/handbook (Access January 11, 2025).
Wallemacq, S. et al. A comparative analysis of the outcomes of patients with influenza or COVID-19 in a tertiary hospital in Belgium. J. Infect. Chemother. 28, 1489–1493 (2022).
Mizera, L. et al. COVID-19 versus seasonal influenza: myocardial injury and prognostic importance. BMC Infect. Dis. 22, 539 (2022).
Kim, S. H., Wi, Y. M., Lim, S., Han, K. T. & Bae, I. G. Differences in clinical characteristics and chest images between coronavirus disease 2019 and influenza-associated pneumonia. Diagnostics 11, 261 (2021).
Kronibus, N. et al. Respiratory physiology of COVID-19 and influenza associated acute respiratory distress syndrome. J. Clin. Med. 11, 6237 (2022).
Seligman, B. et al. 30-day mortality following COVID-19 and influenza hospitalization among US veterans aged 65 and older. J. Am. Geriatr. Soc. 70, 2542–2551 (2022).
Kaye, A. D. et al. Economic impact of COVID-19 pandemic on healthcare facilities and systems: In-ternational perspectives. Best. Pract. Res. Clin. Anaesthesiol. 35, 293–306 (2021).
Hamming, I. et al. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 203, 631–637 (2004).
Lukassen, S. et al. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 39, e105114 (2020).
Osman, M., Klopfenstein, T., Belfeki, N., Gendrin, V. & Zayet, S. A comparative systematic review of COVID-19 and influenza. Viruses. 13, 452 (2021).
Qu, J. et al. Clinical characteristics of COVID-19 and its comparison with influenza pneumonia. Acta Clin. Belg. 75, 348–356 (2020).
Ho, F. K. et al. Modifiable and non-modifiable risk factors for COVID-19, and comparison to risk factors for influenza and pneumonia: results from a UK Biobank prospective cohort study. BMJ Open. 10, e040402 (2020).
Cai, H. Sex difference and smoking predisposition in patients with COVID-19. Lancet Respir. Med. 8, e20 (2020).
Gonzalez-Rubio, J. et al. A systematic review and meta-analysis of hospitalised current smokers and COVID-19. Int. J. Env. Res. Public. Health 17, 7394 (2020).
Farsalinos, K., Barbouni, A. & Niaura, R. Systematic review of the prevalence of current smoking among hospitalized COVID-19 patients in China: could nicotine be a therapeutic option? Intern. Emerg. Med. 15, 845–852 (2020).
Piao, W. H. et al. Nicotine and inflammatory neurological disorders. Acta Pharmacol. Sin. 30, 715–722 (2009).
Faust, J. S. & Rio, C. D. Assessment of deaths from COVID-19 and from seasonal influenza. JAMA Intern. Med. 180, 1045–1046 (2020).
Larsson, E., Eriksson, J., Eriksson, M. & Oldner, A. Critical influenza and COVID-19—A comparative nationwide case-control study. Crit. Care Explor. 4, e0705 (2022).
Zhang, J., Huang, X. & Tao, Z. Correlation of clinical characteristics between patients with seasonal influenza and patients infected by the wild type or delta variant of SARS-CoV-2. Front. Public. Health 10, 981233 (2020).
Li, Y. et al. High-resolution computed tomography in the differential diagnosis between imported COVID-19 and seasonal influenza pneumonia. Iran. Red. Crescent MedJ 22, e109 (2020).
Caterson, H. et al. Higher mortality in extracorporealmembrane-oxygenation patients with COVID-19 compared with H1N1-influenza. Respirology. 27, SUPPL 1 (207-) (2022).
Agca, H. et al. Changing epidemiology of influenza and other respiratory viruses in the first year of COVID-19 pandemic. J. Infect. Public. Health 14, 1186–1190 (2021).
Karlsson, E. A. et al. Review of global influenza circulation, late 2019 to 2020, and the impact of the COVID-19 pandemic on influenza circulation. Releve Epidemiologique Hebd. 96, 241–264 (2021).
Kucharski, A. J., Lessler, J., Cummings, D. A. T., Riley, S. & Rowland-Jones, S. Timescales of influenza A/H3N2 antibody dynamics. PLoS Biol. 16, e2004974 (2018).
Nichols, L. K., Maki, S. B., Szpunar, S. M., Bhargava, A. & Saravolatz, L. D. A comparison of coronavirus disease 2019 (COVID-19) versus influenza during the pandemic: can we distinguish COVID-19 from flu? Infect. Control. Hosp. Epidemiol. 43, 1265–1268 (2022).
Stowe, J. et al. Interactions between SARS-CoV-2 and influenza, and the impact of coinfection on disease severity: a test-negative design. Int. J. Epidemiol. 50, 1124–1133 (2021).
Ashraf, M., Rajaram, S. & English, P. M. How the COVID 19 pandemic will shape influenza public health initiatives: the UK experience. Hum. Vaccin. Immunother. 18, 2056399 (2022).
Yin, Z. et al. A comparison of clinical and chest CT findings in patients with influenza A (H1N1) virus infection and coronavirus disease (COVID-19). AJR Am. J. Roentgenol. 215, 1065–1071 (2020).
Marcoux, D. et al. A retrospective, monocentric study comparing Co and secondary infections in critically Ill COVID-19 and influenza patients. Antibiotics 11, 704 (2022).
Maamar, A. et al. Acquired agitation in acute respiratory distress syndrome with COVID-19 compared to influenza patients: a propensity score matching observational study. Virol. J. 19, 145 (2022).
Strohbehn, I. A. et al. Acute kidney injury incidence, recovery, and long-term kidney outcomes among hospitalized patients With COVID-19 and influenza. Kidney Int. Rep. 6, 2565–2574 (2021).
Delhommeau, G. et al. Bacterial pulmonary co-infections on ICU admission: comparison in patients with SARS-CoV-2 and influenza acute respiratory failure: a multicentre cohort study. Biomedicines. 10, 2646 (2022).
Zheng, J. et al. Clinical and virological impact of single and dual infections with influenza A (H1N1) and SARS-CoV-2 in adult inpatients. PLoS Negl. Trop. Dis. 15, e0009997 (2021).
Hedberg, P. et al. Clinical phenotypes and outcomes of SARS-CoV-2, influenza, RSV and seven other respiratory viruses: a retrospective study using complete hospital data. Thorax. 77, 154–163 (2022).
Zhao, S. et al. Combining initial chest CT with clinical variables in differentiating coronavirus disease 2019 (COVID-19) pneumonia from influenza pneumonia. Sci. Rep. 11, 6422 (2021).
Shen, C. et al. Comparative analysis of early-stage clinical features between COVID-19 and influenza A H1N1 virus pneumonia. Front. Public. Health 8, 206 (2020).
Oliva, A. et al. Comparison of clinical features and outcomes in COVID-19 and influenza pneumonia patients requiring intensive care unit admission. Infection 49, 965–975 (2021).
Liu, L. et al. Comparison of clinical features and outcomes of medically attended COVID-19 and influenza patients in a defined population in the 2020 respiratory virus season. Front. Public. Health 9, 587425 (2021).
Liu, M. et al. COVID-19 pneumonia: CT findings of 122 patients and differentiation from influenza pneumonia. Eur. Radiol. 30, 5463–5469 (2020).
Godaert, L., Cebille, A., Proye, E. & Drame, M. COVID-19 versus seasonal influenza: comparison of profiles of older adults hospitalized in a short-term geriatric ward in France. Am. J. Trop. Med. Hyg. 106, 108–113 (2021).
Yang, Z. et al. Distinguishing COVID-19 from influenza pneumonia in the early stage through CT imaging and clinical features. Front. Microbiol. 13, 847836 (2022).
Torun, S. et al. Influenza viruses and SARS-CoV-2 in adult: ‘Similarities and differences’. Tuberk. Toraks 69, 458–468 (2021).
Zhang, J., Huang, X., Ding, D. & Tao, Z. Platelet-driven coagulopathy in COVID-19 patients: in comparison to seasonal influenza cases. Exp. Hematol. Oncol. 10, 34 (2021).
Wong, J. M. et al. Clinical features of COVID-19, dengue, and influenza among adults presenting to emergency departments and urgent care Clinics-Puerto Rico, 2012-2021. Am. J. Trop. Med. Hyg. 108, 107–114 (2022).
Desai, A., Rosenzvit, C., Nauka, P., Gong, M. & Chen, T. Comparison of acute respiratory failure between SARS-CoV-2 and influenza. Crit. Care Med. 50, 139 (2022).
Gjurasin, B. et al. A retrospective comparison between influenza and COVID-19-associated ARDS in a Croatian tertiary care center. Wien. Klin. Wochenschr. 133, 406–411 (2022).
Marignan, D. et al. A retrospective comparison of COVID-19 and seasonal influenza mortality and outcomes in the ICUs of a French university hospital. Eur. J. Anaesthesiol. 39, 427–435 (2022).
Shafran, N. et al. Abnormal liver tests in patients with SARS-CoV-2 or influenza–prognostic similarities and temporal disparities. JHEP Rep. 3, 100258 (2021).
Bhasin, B. et al. AKI in hospitalized patients with COVID-19 and seasonal influenza: a comparative analysis. Kidney360 2, 619–628 (2021).
Martinez, A., Cardona, L. & Ricci, N. Antibiotic prescribing trends in hospitalized influenza versus COVID-19 patients at a community-based health system. Open. Forum Infect. Dis. 8, S248 (2021). SUPPL 1.
Shimol, A. B. et al. Can laboratory evaluation differentiate between coronavirus disease-2019, influenza, and respiratory syncytial virus infections? A retrospective cohort study. Croat. Med. J. 62, 623–629 (2021).
Wang, H., Wei, R., Rao, G., Zhu, J. & Song, B. Characteristic CT findings distinguishing 2019 novel coronavirus disease (COVID-19) from influenza pneumonia. Eur. Radiol. 30, 4910–4917 (2020).
Verma, A. A. et al. Characteristics and outcomes of hospital admissions for COVID-19 and influenza in the Toronto area. CMAJ. 193, E410–E418 (2021).
Herbstreit, F. et al. Characteristics of critically Ill patients with COVID-19 compared to patients with influenza-A single center experience. J. Clin. Med. 10, 2056 (2021).
Sieber, P. et al. Characteristics of patients with Coronavirus Disease 2019 (COVID-19) and seasonal influenza at time of hospital admission: a single center comparative study. BMC Infect. Dis. 21, 271 (2021).
Choreno-Parra, J. A. et al. Clinical and immunological factors that distinguish COVID-19 from pandemic influenza A(H1N1). Front. Immunol. 12, 593595 (2021).
Zayet, S. et al. Clinical features of COVID-19 and influenza: a comparative study on Nord Franche-Comte cluster. Microbes Infect. 22, 481–488 (2020).
Ludwig, M., Jacob, J., Basedow, F., Andersohn, F. & Walker, J. Clinical outcomes and characteristics of patients hospitalized for Influenza or COVID-19 in Germany. Int. J. Infect. Dis. 103, 316–322 (2021).
Tsukada, N., Inamura, J., Igarashi, S. & Sato, K. Clinical outcomes of coronavirus disease 2019 and seasonal influenza in patients with hematological disorders: a retrospective study. J. Rural. Med. 17, 158–165 (2022).
Lv, Y. et al. Comparative analysis of elderly hospitalized patients with coronavirus disease 2019 or influenza A H1N1 virus infections. Int. J. Infect. Dis. 125, 278–284 (2022).
Biasco, L. et al. Comparative frequency and prognostic impact of myocardial injury in hospitalized patients with COVID-19 and Influenza. Eur. Heart J. Open. 1, oeab025 (2021).
Raff, L. A. et al. Comparative outcomes between COVID-19 and influenza patients placed on veno-venous extracorporeal membrane oxygenation for severe ARDS. Am. J. Surg. 223, 388–394 (2022).
Kong, J. et al. Comparative study of hematological and radiological feature of severe/critically ill patients with COVID-19, influenza A H7N9, and H1N1 pneumonia. J. Clin. Lab. Anal. 35, e24100 (2021).
Benaim, A. R. et al. Comparing COVID-19 and Influenza Presentation and Trajectory. Front. Med. 8, 656405 (2021).
Pandey, M. et al. Comparative incidence of early and late bloodstream and respiratory tract co-infection in patients admitted to ICU with COVID-19 pneumonia versus Influenza A or B pneumonia versus no viral pneumonia: wales multicentre ICU cohort study. Crit. Car 26, 158 (2022).
Charlton, M. et al. Comparing SARS-CoV-2 and influenza A(H1N1)pdm09-infected patients requiring ECMO–A single-centre, retrospective observational cohort experience. J. Infect. 82, 84–123 (2021).
Donnino, M. W. et al. Comparison between Patients Hospitalized with Influenza and COVID-19 at a Tertiary Care Center. J. Gen. Intern. Med. 36, 1689–1695 (2021).
Ding, L. et al. Comparison of acute respiratory distress syndrome in patients with COVID-19 and influenza A (H7N9) virus infection. Int. J. Infect. Dis. 122, 593–598 (2022).
Hattatoglu, D. G. & Yildiz, B. P. Comparison of clinical and biochemical features of hospitalized COVID‐19 and influenza pneumonia patients. J. Med. Virol. 93, 6619–6627 (2021).
Brehm, T. T. et al. Comparison of clinical characteristics and disease outcome of COVID-19 and seasonal influenza. Sci. Rep. 11, 5803 (2021).
Nasir, N. et al. Comparison of clinical characteristics and outcomes between COVID-19 pneumonia and H1N1 influenza. Adv. Respir. Med. 89, 254–261 (2021).
Cobb, N. L. et al. Comparison of clinical features and outcomes in critically Ill patients hospitalized with COVID-19 versus influenza. Ann. Am. Thorac. Soc. 18, 632–640 (2021).
Marois, C. et al. Comparison of clinical profiles and mortality outcomes between influenza and COVID-19 patients invasively ventilated in the ICU: a retrospective study from all Paris public hospitals from 2016 to 2021. Crit. Care Explor. 4, e0737 (2022).
Neto, A. S. et al. Comparison of critical care occupancy and outcomes of critically Ill patients during the 2020 COVID-19 winter surge and 2009 H1N1 influenza pandemic in Australia. Ann. Am. Thorac. Soc. 18, 1380–1389 (2021).
Yildirim, M. et al. Comparison of critically ill COVID-19 and influenza patients with acute respiratory failure. Acute Crit. Care 37, 168–176 (2022).
Montesinos, I. L. et al. Comparison of hospitalized coronavirus disease 2019 and influenza patients requiring supplemental oxygen in a cohort study: clinical impact and resource consumption. Clin. Infect. Dis. 75, 2225–2238 (2022).
Taniguchi, Y. et al. Comparison of patient characteristics and in-hospital mortality between patients with COVID-19 in 2020 and those with influenza in 2017-2020: a multicenter, retrospective cohort study in Japan. Lancet Reg. Health West. Pac. 20, 100365 (2022).
Lee, J. et al. Comparison of short-term mortality between mechanically ventilated patients with COVID-19 and influenza in a setting of sustainable healthcare system. J. Infect. 81, e76–e78 (2020).
Piroth, L. et al. Comparison of the characteristics, morbidity, and mortality of COVID-19 and seasonal influenza: a nationwide, population-based retrospective cohort study. Lancet Respir. Med. 9, 251–259 (2021).
Kuang, P. D. et al. Comparison of the clinical and CT features between COVID-19 and H1N1 influenza pneumonia patients in Zhejiang, China. Eur. Rev. Med. Pharmacol. Sci. 25, 1135–1145 (2021).
Hernandez-Cardenas, C., Lugo-Goytia, G., Hernandez-Garcia, D. & Perez-Padilla, R. Comparison of the clinical characteristics and mortality in acute respiratory distress syndrome due to COVID-19 versus due to Influenza A-H1N1pdm09. Med. Intensiva 46, 345–347 (2022).
Auvinen, R. et al. Comparison of the clinical characteristics and outcomes of hospitalized adult COVID-19 and influenza patients-a prospective observational study. Infect. Dis. 53, 111–121 (2021).
Fukui, S., Inui, A., Saita, M., Kobayashi, D. & Naito, T. Comparison of the clinical parameters of patients with COVID-19 and influenza using blood test data: a retrospective cross-sectional survey. J. Int. Med. Res. 50, 3000605221083751 (2022).
Talbot, H. K. et al. Coronavirus disease 2019 (COVID-19) versus influenza in hospitalized adult patients in the United States: differences in demographic and severity indicators. Clin. Infect. Dis. 73, 2240–2247 (2021).
Pawelka, E. et al. COVID-19 is not “just another flu”: a real-life comparison of severe COVID-19 and influenza in hospitalized patients in Vienna, Austria. Infection. 49, 907–916 (2021).
Zarei, F. et al. Differentiation of Chest CT findings between influenza pneumonia and COVID-19: interobserver agreement between radiologists. Acad. Radiol. 28, 1331–1338 (2021).
Frohlich, G. M. et al. Hospital outcomes of community-acquired COVID-19 versus influenza: Insights from the Swiss hospital-based surveillance of influenza and COVID-19. Euro Surveill. 27, 2001848 (2022).
Lyons, P. G. et al. Hospital trajectories and early predictors of clinical outcomes differ between SARS-CoV-2 and influenza pneumonia. EBioMedicine. 85, 104295 (2022).
Begum, H. et al. Surveillance for severe influenza and COVID-19 in patients admitted to sentinel Australian hospitals in 2020: the Influenza Complications Alert Network (FluCAN). Commun. Dis. Intell. (2018) 28, 46 (2022).
Rouze, A. et al. Invasive pulmonary aspergillosis among intubated patients with SARS-CoV-2 or influenza pneumonia: a European multicenter comparative cohort study. Crit. Care 26, 11 (2022).
Beatty, K., Hamilton, V. & Kavanagh, P. M. Just a bad flu? Tackling the “infodemic” in Ireland through a comparative analysis of hospitalised cases of COVID-19 and influenza. Public. Health 194, 19–24 (2021).
Fjelltveit, E. et al. Lower antibiotic prescription rates in hospitalized COVID-19 patients than influenza patients, a prospective study. Infect. Dis. 54, 79–89 (2022).
Northam, K. A., Murray, B. P., Fischer, W. A. & Charles, A. G. Major bleeding and thrombosis events in COVID-19 versus influenza patients requiring extracorporeal membrane oxygenation. ASAIO J. 68, 779–784 (2022).
Faury, H. et al. Medical features of COVID-19 and influenza infection: a comparative study in Paris, France. J. Infect. 82, e36–e39 (2021).
Kodde, C. et al. Mortality in cancer patients with SARS-CoV-2 or seasonal influenza: an observational cohort study from a German-wide hospital network. Infection. 51, 119–127 (2023).
Iosifescu, A. L., Hoogenboom, W. S., Buczek, A. J., Fleysher, R. & Duong, T. Q. New-onset and persistent neurological and psychiatric sequelae of COVID-19 compared to influenza: a retrospective cohort study in a large New York City healthcare network. Int. J. Methods Psychiatr. Res. 31, e1914 (2022).
Jackel, M. et al. Outcome of acute respiratory distress syndrome requiring extracorporeal membrane oxygenation in Covid-19 or influenza: a single-center registry study. Artif. Organs 45, 593–601 (2021).
Blazoski, C. M., Baram, M., Yang, Q. & Hirose, H. Outcomes of extracorporeal membrane oxygenation in influenza versus COVID-19 during the first wave of COVID-19. J. Card. Surg. 36, 3740–3746 (2021).
Shih, E. et al. Outcomes of extracorporeal membrane oxygenation in patients with severe acute respiratory distress syndrome caused by COVID-19 versus influenza. Ann. Thorac. Surg. 113, 1445–1451 (2022).
Woodcock, T., Greenfield, G., Lalvani, A., Majeed, A. & Aylin, P. Patient outcomes following emergency admission to hospital for COVID-19 compared with influenza: retrospective cohort study. Thorax 78, 706–712 (2022).
Hamilton, M. et al. Predictors of all-cause mortality among patients hospitalized with influenza, respiratory syncytial virus, or SARS-CoV-2. Influenza Other Respir. Viruses 16, 1072–1081 (2022).
Merkler, A. E. et al. Risk of ischemic stroke in patients with coronavirus disease 2019 (COVID-19) vs patients with influenza. JAMA Neurol. 77, 1–7 (2020).
Dabaja-Younis, H. et al. SARS-CoV-2 and seasonal influenza: similarities and disparities. Arch. Virol. 167, 2761–2765 (2022).
Roda, S. et al. Sars-Cov-2 versus Flu: ECMO-associated bloodstream infections. N. Microbiol. 45, 35–39 (2022).
Cousin, N. et al. SARS-CoV-2 versus influenza-associated acute respiratory distress syndrome requiring veno-venous extracorporeal membrane oxygenation support. ASAIO J. 67, 125–131 (2021).
Qian, F. et al. Specific dynamic variations in the peripheral blood lymphocyte subsets in COVID-19 and severe influenza A patients: a retrospective observational study. BMC Infect. Dis. 20, 910 (2020).
Boyd, S. et al. The incidence of venous thromboembolism in critically Ill patients with SARS-CoV-2 infection compared with critically Ill influenza and community-acquired pneumonia patients: a retrospective chart review. Med. Sci. 10, 30 (2022).
Kazancioglu, S., Bastug, A., Ozbay, B. O., Kemirtlek, N. & Bodur, H. The role of haematological parameters in patients with COVID-19 and influenza virus infection. Epidemiol. Infect. 148, e272 (2020).
Nersesjan, V., Amiri, M., Christensen, H. K., Benros, M. E. & Kondziella, D. Thirty-day mortality and morbidity in COVID-19 positive vs. COVID-19 negative individuals and vs. individuals tested for influenza A/B: a population-based study. Front. Med. 7, 598272 (2020).
Lachant, D. et al. Venous thromboembolism associates with SARS-CoV-2 more than seasonal influenza. Thromb. Res. 205, 40–43 (2021).
Zhang, M. et al. Initial chest CT features between COVID-19 and influenza A virus-induced pneumonia: a comparative study. Chin. J. Med. Imaging 28, 891–895 (2020).
Mendoza, M. A. et al. Difference between SARS-CoV-2, seasonal coronavirus, influenza, and respiratory syncytial virus infection in solid organ transplant recipients. Transpl. Infect. Dis. 25, e13998 (2023).
Setayeshgar, S. et al. Comparison of influenza and COVID-19 hospitalisations in British Columbia, Canada: a population-based study. BMJ Open. Respir. Res. 10, e001567 (2023).
Volkov, L. et al. Clinical outcomes and characteristics of critically ill patients with influenza- and COVID-19-induced ARDS: a retrospective, matched cohort study. Front. Med. 9, 1027984 (2023).
Gu, B., Yao, L., Zhu, X., Tang, P. & Chen, C. Comparison of hospitalized patients with severe pneumonia caused by COVID-19 and influenza A (H7N9 and H1N1): a retrospective study from a designated hospital. Open. Med. 17, 1965–1972 (2022).
Luyt, C. E. et al. Herpesviridae lung reactivation and infection in patients with severe COVID-19 or influenza virus pneumonia: a comparative study. Ann. Intensive Care 12, 87 (2022).
Dupont, A. et al. Outcomes and risk factors with COVID-19 or influenza in hospitalized asthma patients. Respir. Res. 23, 342 (2022).
Kristofi, R. et al. Patients with type 1 and type 2 diabetes hospitalized with COVID-19 in comparison with influenza: mortality and cardiorenal complications assessed by nationwide Swedish registry data. Cardiovasc. Diabetol. 21, 282 (2022).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Writing-original draft: Z.Z. and Y.H.; Data curation: X.L., Z.Z. and Y.H.; Writing-review and editing: J.G.; Conceptualization: J.G.; Methodology: X.L. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Han, Y., Guo, J., Li, X. et al. Differences in clinical characteristics between coronavirus disease 2019 (COVID-19) and influenza: a systematic review and meta-analysis. npj Prim. Care Respir. Med. 35, 8 (2025). https://doi.org/10.1038/s41533-025-00414-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41533-025-00414-0
This article is cited by
-
Sensitivity analysis and numerical simulations of a poly-infection model with COVID-19, pneumonia, and influenza
Quality & Quantity (2026)
-
Prevalence, clinical features, and seasonal patterns of SARS-CoV-2, influenza, and HRSV in patients with a history of kidney transplantation in Iran
BMC Infectious Diseases (2025)
-
COVID-19 infection associated with increased risk of new-onset vascular dementia in adults ≥50 years
npj Dementia (2025)