Abstract
mRNA vaccines have shown great efficacy against SARS-CoV-2, yet challenges remain in optimizing vaccine components to achieve enhanced immune response and vaccine stability. In this study, we developed CPVax-CoV, a new lyophilized mRNA vaccine that features novel thiolactone-based ionizable lipids and newly designed untranslated regions (UTRs) for enhanced expression. Incorporation of these optimized components into our vaccine candidate CPVax-CoV significantly improved immune responses in mice compared to commercially available mRNA vaccines. Moreover, lyophilized CPVax-CoV has proven to be thermostable, maintaining its biological activity for up to one year at 4 °C and 25 °C after lyophilization, overcoming the cold-chain limitations of current mRNA vaccines. This vaccine demonstrates protective efficacy against ancestral SARS-CoV-2 and the Omicron XBB variant, offering a scalable solution for global distribution and pandemic preparedness. These findings underscore the potential of this platform for future next-generation mRNA vaccine development.
Similar content being viewed by others

Introduction
The outbreak of coronavirus disease 2019 (COVID-19) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has significantly influenced global health, social dynamics, and economic activity. Vaccination remains the most potent strategy for preventing infection and controlling virus spread. Consequently, considerable efforts have been directed towards the rapid development and approval of various vaccines worldwide1.
Among these, mRNA technology has led the race of anti-SARS-CoV-2 vaccines and emerged as a promising vaccine platform due to its rapid development, manufacturing versatility, safety profile, and ability to elicit broad immune responses. Unlike traditional platforms, mRNA vaccines offer advantages such as rapid design based solely on sequence information, bypassing the need for virus culture or recombinant protein production. They also circumvent issues like insertional mutagenesis and pre-existing immunity that may hinder other vaccine types, such as DNA or viral vector vaccines2,3. Approved mRNA vaccines, including BNT162b2/Comirnaty (Pfizer/BioNTech)4 and mRNA-1273/SpikeVax (Moderna)5, have validated the potential of mRNA technology, demonstrating significant success in mitigating viral spread, reducing hospitalizations, and lowering mortality rates since their emergency authorization in 20206. Additionally, bivalent mRNA vaccines targeting specific SARS-CoV-2 variants have been approved, providing enhanced protection against emerging strains7, and ongoing research is focused on the development of combined vaccines that address both COVID-19 and seasonal influenza8.
Despite their proven efficacy and rapid development, there is still considerable room for improvement in the field of mRNA vaccine components and stability to enhance protective efficacy against emerging variants or new pathogens and to ensure equal distribution and accessibility worldwide9,10,11,12,13. Here we present a new mRNA vaccine platform that incorporates novel optimizations on mRNA sequence design and lipid formulation that is suitable for lyophilization and stable for long-term storage.
Optimization of mRNA sequence, comprising antigen-coding sequence and non-coding regulatory elements, directly impacts protein expression and vaccine efficacy14. mRNA vaccines typically comprise a codon-optimized nucleotide sequence encoding the antigen of interest15 along with other stabilizing elements, including two non-coding regulatory sequences (untranslated regions, UTRs) flanking the coding sequence (5’UTR and 3’UTR). UTRs contain multiple regulatory elements and are essential for mRNA stability and translation efficiency. Here, we have incorporated novel UTR sequences optimized for efficient protein expression after intramuscular injection16 into Spike coding mRNA.
Along with mRNA sequence optimization, efficient delivery of mRNA into the cytoplasm of target cells is crucial to exert its therapeutic function. Lipid nanoparticles (LNPs) are currently the leading delivery system for mRNA vaccines. LNPs are commonly composed of 4 lipid components: an ionizable lipid, a helper phospholipid, cholesterol, and a PEGylated lipid17. These lipids encapsulate the antigen-coding mRNA and protect the nucleic acid from degradation. The ionizable lipid is the most critical component of LNPs, as it determines the efficacy of transfection and endosomal escape. They typically feature a tertiary amine that remains deprotonated at physiological pH, improving the biocompatibility of LNPs, but becomes positively charged at lower pH conditions. Protonation of ionizable lipids at the endosomal acidic pH promotes membrane destabilization and delivery of nucleic acid cargo into the cytosol of target cells18. Furthermore, ionizable lipid impacts vaccine adjuvanticity, which directly affects the therapeutic effect of mRNA vaccines19. Currently, there are only two ionizable lipids (SM-102 and ALC-0315) approved for commercial use as part of mRNA vaccines20,21. We recently developed a new library of rationally designed ionizable lipids able to induce high protein expression levels in mice inoculated by the intramuscular route22,23, the most common route for administration of mRNA vaccines. However, their applicability as part of mRNA vaccines and their ability to induce immunity have not been addressed yet. In this study, we have characterized this new family of ionizable lipids as part of mRNA vaccines against SARS-CoV-2, showing a strong potential for their use in mRNA-LNPs-based vaccines.
A critical limitation in the global distribution of current mRNA vaccines is their dependence on cold-chain logistics to preserve mRNA integrity and maintain the physicochemical properties of LNPs24,25. The need for ultra-cold storage conditions poses a major barrier to ensuring higher coverage in resource-limited settings, which is essential for ensuring equitable protection11. To address this challenge, lyophilization, or freeze-drying, has long been utilized to stabilize pharmaceuticals and vaccines, prolonging their shelf life and simplifying storage and distribution logistics. Applying this technique to mRNA vaccines holds immense potential to overcome the cold chain requirements associated with traditional liquid formulations, thereby enabling broader global access to advanced immunization technologies, since cryogenic transport and storage are needed for the current licensed mRNA vaccines26. In this study, we developed a lyophilized formulation of our candidate mRNA vaccine, CPVax-CoV, which maintains LNP physicochemical properties and biological activity, showing comparable results to the standard liquid formulation.
COVID-19 crisis underscored the critical need for novel vaccine platforms able to provide rapid and effective solutions in response to future outbreaks and emerging pathogens. Here we have applied innovative optimizations on delivery systems, mRNA sequence design, and vaccine stability to develop CPVax-CoV, a novel SARS-CoV-2 mRNA-based vaccine that is thermostable for long-term storage in lyophilized format and elicits robust protective immunity compared to commercially available SARS-CoV-2 vaccines. Importantly, lyophilized CPVax-CoV induced protective immunity in mice challenged with SARS-CoV-2 even after one year of storage at 4 °C and 25 °C, while maintaining intact LNP physicochemical properties. This platform has been validated against ancestral and Omicron SARS-CoV-2 variants of concern (VOCs), and in a recently developed mouse model using a mouse-adapted SARS-CoV-2 virus that resembles human COVID1927. Our findings support the efficacy of CPVax-CoV as a robust and flexible mRNA vaccine platform and provide significant impact on pandemic preparedness, highlighting the potential of novel ionizable lipids, UTR sequences, and lyophilization technology to be transferred to future mRNA vaccines against infectious diseases.
Results
Novel ionizable lipid-based mRNA vaccines induce a robust anti-SARS-CoV-2 immune response
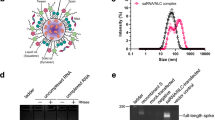
We have recently reported the development of an extensive library of novel thiolactone-based ionizable lipids proven to be safe and effective for mRNA expression in vivo, showing enhanced results compared to other approved lipid-based LNPs benchmarks22,23. Remarkably, from these previous studies, CP-LC-0729 emerged as the top-performing lipid, demonstrating high in vivo efficiency in delivering firefly luciferase mRNA. In this work, we selected CP-LC-0729 along with several promising novel ionizable lipids (structures shown in Supplementary Fig. 1) in terms of protein expression after intramuscular administration (Supplementary Fig. 2) to evaluate their potential to induce immunity for mRNA vaccine applications.
They were synthesized and characterized using HPLC-MS and NMR, and we incorporated these lipids into LNP formulations to construct various mRNA vaccine candidates, all using the Spike-coding mRNA sequence from the commercial Comirnaty vaccine. Both the mRNA and LNPs were prepared in-house via in vitro transcription and microfluidic mixing, respectively. These vaccine candidates were compared to a control LNP formulation, which replicated the Comirnaty vaccine using ALC-0315 as the ionizable lipid. Thus, ALC-0315 was formulated using Comirnaty lipid components at molar lipid ratios of 46.6:9.4:42.7:1.6 (ALC-0315: DSPC: cholesterol: ALC-0159), and the evaluated candidate lipids were formulated at molar ratios of 50:10:38.5:1.5 (ionizable lipid:DOPE:cholesterol:DMG-PEG2000). Physicochemical characterization of the resulting LNPs showed optimal values of diameter and encapsulation efficiency with no significant differences between the various candidates (Supplementary Table 1).
To evaluate the immune response elicited by the different vaccine formulations, BALB/c mice were intramuscularly immunized with 1 µg of mRNA/mice following a 21-day prime-boost regimen and specific B and T cell responses were analyzed (Fig. 1a). Production of specific anti-RBD IgG antibodies were detected in serum from all the vaccinated animals and they were significantly enhanced after boost immunization. Mice vaccinated with LNPs formulated with our novel ionizable lipids showed comparable antibody levels to mice vaccinated with the control vaccine using ALC-0315 as ionizable lipid. Importantly, antibody levels in mice vaccinated with LNPs containing CP-LC-0729 lipid were significantly higher than in those vaccinated with the control vaccine after boost (Fig. 1b). This enhanced immune response aligns with the previously reported superior protein expression of CP-LC-072923, which may be facilitated by its proposed improved endosomal escape efficiency. T-cell response was evaluated 3 weeks post-boost in splenocytes after specific stimulation with SARS-CoV-2 Spike peptide mix. IFN-γ-producing cells (Fig. 1c) and cytokine secretion of IFN-γ and IL-4 (Fig. 1d, e) were measured by ELISPOT and ELISA, respectively, showing T cell induction in all vaccinated animals with strong IFN-γ and low IL-4 response, indicating a Th1-biased response. Although all groups exhibited elevated T cell responses, IFN-γ production was notably higher in mice vaccinated with CP-LC-0729, CP-LC-0867, CP-LC-0743, or ALC-0315 compared to those vaccinated with CP-LC-0431 or CP-LC-0474-based vaccines.
a Immunization scheme and sample collection schedule. BALB/c mice (n = 5) were immunized intramuscularly at day 0 (prime) and 21(boost) with 1 µg of mRNA/animal. Blood samples were obtained at week 3 (prior to boost) and 6, and specific antibody levels were determined in serum. T-cell response was evaluated in splenocytes 3 weeks post-boost. b Reciprocal endpoint titers of antigen-specific IgG antibodies in serum samples determined by ELISA using RBD recombinant protein from SARS-CoV-2 wild-type variant. c IFN-γ-secreting splenocytes quantification by ELISPOT after O.N. stimulation with SARS-CoV-2 Spike peptide pool (d, e) IFN-γ and IL-4 quantification in supernatants of splenocytes after overnight stimulation with SARS-CoV-2 Spike peptide pool determined by ELISA. Graphs are represented as mean ± SD. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001 as determined by two-way OVA (b) and one-way ANOVA (c, d) with Tukey post-test.
Taken together, these results demonstrate that LNPs formulated with our proprietary novel ionizable lipids exhibit robust induction of both B and T cell responses in vivo, providing evidence for their potential use in next-generation mRNA vaccines against SARS-CoV-2 and, potentially, other pathogens. CP-LC-0729 lipid was chosen to be further characterized in the next assays as it exhibits superior antibody response after boost compared to the evaluated candidates and the control vaccine.
mRNA sequence optimization enhances S-specific B and T-cell responses
Despite their significant impact on therapeutic protein expression, current mRNA vaccine candidates often utilize 5′ and 3′UTRs derived from commonly used human genes with minimal optimization. We have previously explored novel UTR sequences following a semi-rational RNA sequence design approach, leading to the discovery of new RNA motifs that enhance mRNA expression in vivo16. Notably, the optimized 5′ UTR includes two tandem repeats of the sequence 5′-GCCACC-3′ positioned immediately upstream of the start codon, which contributes to enhanced translation efficiency. These optimized UTR sequences, when incorporated into mRNA, significantly improve protein production and outperform UTR designs used in commercially approved mRNA vaccines16. Herein, we designed a codon-optimized mRNA sequence based on the full-length Spike protein of the Wuhan SARS-CoV-2 virus (SARS-CoV-2 wild type (Wuhan-Hu-1), hereinafter referred to as wild type), incorporating our proprietary UTR sequences (Supplementary Fig. 3), and we tested its efficacy as part of a new mRNA vaccine candidate.
First, we evaluated Spike in vitro expression in HEK293T to verify antigen production of our newly developed mRNA Spike sequence based on wild type strain (CP-S) by Western Blot and flow cytometry (Supplementary Fig. 3). Then, we encapsulated Comirnaty or CP-S mRNA in LNPs formulated with CP-LC-0729 lipid at molar lipid ratios of 40.7:34.9:23.3:1.2 (CP-LC-0729:DOPE:cholesterol:DMG-PEG2000). These specific molar ratios were chosen based on our previous optimization studies of the LNP formulation for CP-LC-0729, which demonstrated enhanced intramuscular mRNA expression using these lipid ratios23. In order to evaluate vaccine immunity in vivo, BALB/c mice were immunized in a 21-day prime-boost regimen. Serum antibody responses were analyzed at different time-points, and T-cell response was evaluated in the spleen 3 weeks post-boost (Fig. 2a).
a Immunization scheme and sample collection schedule. Mice (n = 5) were immunized intramuscularly at day 0 (prime) and 21(boost) with 1 µg of mRNA/animal. Blood samples were obtained at week 3 (prior to boost) and 6, and specific antibody levels were determined in serum. T-cell response was evaluated in splenocytes 3 weeks post-boost. b Reciprocal endpoint titers of antigen-specific IgG antibodies in serum samples determined by ELISA using RBD recombinant protein from wild-type variant. c Neutralization titers in serum samples were determined by neutralization assay using Spike-pseudotyped lentivirus and infection in HEK293T-ACE2-TMPRSS2 cells. NT50 titers refer to the dilution of a serum sample at which 50% of the pseudovirus infection is inhibited. d IFN-γ-secreting splenocytes quantification by ELISPOT after O.N. stimulation with SARS-CoV-2 Spike peptide pool. e IFN-γ quantification in supernatants of splenocytes after overnight stimulation with SARS-CoV-2 Spike peptide pool determined by ELISA. f T-lymphocyte CD4 and CD8 cell frequencies in spleen analyzed by flow cytometry. g Tfh and GC B cells analysis. Mice (n = 5) were immunized intramuscularly with 5 µg of mRNA/animal. Inguinal lymph nodes were extracted on day 7. h Tfh (CD45R-, CD4+, CD44hi, CXCR5+, PD-1+) and GC B cell (CD19+, CD95+, GL7+) frequencies were determined by flow cytometry analysis. Graphs are represented as mean ± SD. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001 as determined by two-way ANOVA (a) and one-way ANOVA (c–h) with Tukey post-test.
Anti-RBD total IgG antibody levels in serum were 2.5-fold higher in the animals vaccinated with LNPs formulated with CP-S mRNA compared to the ones encapsulating Comirnaty mRNA or the control vaccine after the first immunization. After the second immunization, these differences increased significantly, showing 5-fold and 4.3-fold higher titers in the CP-S mRNA group (Fig. 2b).
Neutralizing antibodies were analyzed through a pseudovirus-based assay using GFP-expressing lentivirus pseudotyped with SARS-CoV-2 Spike protein from wild-type strain (Supplementary Fig. 4). Neutralizing titer 50 (NT50) post-boost was comparable in all the experimental groups and slightly higher, although not significantly, in serum from mice vaccinated with CP-LC-0729 encapsulating CP-S mRNA, in accordance with the results of total anti-RBD binding IgG titers (Fig. 2c).
Specific T cell response was analyzed in splenocytes at 3 weeks post-boost. Cytokine secretion after specific stimulation showed enhanced production of IFN-γ in mice vaccinated with CP-LC-0729 LNPs compared with the cell samples from animals immunized with the control vaccine. Notably, this difference was significantly higher in CP-LC-0729 LNPs encapsulating CP-S mRNA, both in terms of IFN-γ secreting splenocytes (Fig. 2d) and total IFN-γ secretion (Fig. 2e). T cell populations inthe spleen were evaluated by flow cytometry, showing higher levels of total CD4 T cells in mice vaccinated with CP-LC-0729 LNPs encapsulating CP-S mRNA. No significant differences were found in total CD8 T cell levels between the evaluated vaccines (Fig. 2f).
To further characterize the immune response triggered by these mRNA vaccine candidates, we evaluated early germinal center (GC) B cell and T follicular helper (Tfh) cell responses in the draining lymph nodes. Tfh are critical for GC formation, which directly impacts B-cell differentiation and long-lived plasma cell expansion. Early Tfh and GC responses have been previously related to protective humoral immunity in mRNA vaccines28. BALB/c mice were immunized with a single dose of 5 µg mRNA/mouse. Administering a single, higher mRNA dose was intended to ensure a robust immune signal at this early timepoint, facilitating accurate detection and characterization of these cellular populations. One-week post-prime Tfh and GC responses were evaluated in the draining lymph nodes by flow cytometry (Supplementary Fig. 5). As shown in Fig. 2h, significant expansion of GC B cells and cell populations was observed in the vaccinated animals compared to the non-vaccinated controls. This result correlates with a robust antibody-specific response previously observed in the serum analysis (Fig. 2b).
In summary, these data indicate that our optimized mRNA sequence enhances the immune response induced by our novel lipid-based vaccine, underscoring the importance of UTR optimization in the development of more effective mRNA vaccines. Based on these results, we defined our final vaccine candidate, CPVax-CoV, that incorporates novel ionizable lipid CP-LC-0729 and CP-S mRNA sequence. Our results indicate that this novel vaccine candidate induces robust immune B and T cell responses and outperforms the Comirnaty vaccine.
CPVax-CoV biodistribution and preliminary safety evaluation
Safety is an essential requisite for any newly developed vaccine. LNPs, especially ionizable lipids, are frequently identified as a safe concern in mRNA vaccines, since they exhibit adjuvant properties and are determinant for the pharmacokinetics (PK) and biodistribution profile of mRNA vaccines29,30. Consequently, special attention needs to be addressed to the tolerability and safety profile of novel ionizable lipids as part of LNPs-based therapies. We have preliminarily evaluated our ionizable lipid candidate CP-LC-0729 in mice after systemic administration of a high dose of FLuc mRNA-LNPs by intravenous injection, showing a good safety profile with no signs of hepatic or systemic toxicity23.
To further characterize the safety and PK profile of the CPVax-CoV vaccine, we carried out a biodistribution study to track LNPs accumulation and elimination in vivo. LNPs were labeled with indium-111 oxinate and intramuscularly injected in male and female BALB/c mice. Images were acquired using microSPECT/CT at 1 h, 3 h, and 6 h and 1, 2, 3, 4, and 7 days post-injection. The visual analysis of the images showed a strong signal at the injection site, but the LNPs signal was also found in the sacral ganglion chain, bone marrow (long bone heads/iliac), and kidneys (elimination pathway) (Fig. 3a).
BALB/c mice (male n = 3, female n = 3) were immunized intramuscularly with 5 µg of mRNA/animal encapsulated in Indium-111 labeled LNPs. a In vivo SPECT/CT representative images of coronal and axial sections acquired at different time points post-immunization for up to one week. The lower image shows the signal progression in the presacral lymph node (red arrow) with an adjusted threshold. b Signal quantification at the injection site at different points post-vaccination using standardized uptake value (SUV) units. c Signal quantification in the presacral lymph node at different points post-vaccination. d Ex vivo signal quantification at day 7 post-immunization, represented as injected dose per gram normalized to brain signal. e Serum biochemical analysis of hepatic enzymes 24 h post-immunization. Graphs are represented as mean ± SD. ns non-significant as determined by two-way ANOVA with Tukey post-test (e).
Maximum signal intensity at the injection site decreased over time, but persisted for at least 4 days, reflecting a primarily distribution of LNPs at the injection site within the first 24 h post-administration (Fig. 3b). Draining through the retroperitoneal and medial sacral lymph nodes was observed in all animals starting from 1 h post-administration, with signal intensity peaking at 6 h and gradually decreasing thereafter (Fig. 3c). At 6 h post-administration, the appearance of signal in the bone marrow was visually detected, peaking approximately at 1–2 days post-injection (Supplementary Fig. 6). Ex vivo analysis on day 7 supported the in vivo imaging data, showing clear drainage through the lymph nodes when normalized by organ weight (Fig. 3d).
Migration to draining lymph nodes is compatible with the observed Tfh cell expansion, GC formation, and immune response elicited by vaccination, the signal observed in kidneys is compatible with urine excretion, and no accumulation in liver or other organs was detected.
To corroborate vaccine tolerability, hepatic enzyme levels in serum were evaluated 24 h post-vaccination. No increment in AST and ALT levels was observed compared to the serum analysis data pre-vaccination (Fig. 3e). These data provide preliminary evidence of vaccine safety and support CPVax-CoV for further evaluation as an mRNA vaccine.
CPVax-CoV induces protection against SARS-CoV-2 challenge
To evaluate vaccine-induced protection of CPVax-CoV against SARS-CoV-2, we conducted protection experiments in vivo and validated the results in two different mouse infection models: one using a mouse-adapted SARS-CoV-2 strain and the other using hACE2-K18 transgenic mice.
First, a mouse-adapted SARS-CoV-2 strain (MA20) was used to infect wild-type BALB/c mice. MA20 was obtained from a clinical isolate of the SARS-CoV-2 Alpha variant (Pango lineage Nomenclature B.1.1.7) after 20 serial passages in one-year-old C57BL/6 mice. MA20 is able to induce severe lung pathology in mice and recapitulates key pathological features of COVID-19 in humans, including higher severity in males, lymphodepletion, and an increase in inflammatory cytokine response and acute respiratory distress syndrome (ARDS)27. BALB/c female mice were immunized in a 21-day prime-boost regimen with 1 µg of mRNA/animal of CPVax-CoV and the control vaccine. Four weeks post-boost, mice were intranasally infected with MA20. Survival, viral load and cytokine profile in lungs were determined (Fig. 4a). All vaccinated animals survived over a 25-day period after virus challenge (endpoint of the experiment) without showing weight loss or clinical signs of disease (Fig. 4b). In contrast, unvaccinated control mice showed severe weight loss and were euthanized on day 4 post-infection according to previously established human endpoint criteria (Fig. 4b, c). In accordance with survival outcomes, the infecting virus was detected by TCID50 assay in lungs from unvaccinated mice but not in lungs from vaccinated mice at day 2 or 4 post-challenge, except for one mouse in the control vaccine group, in which a low viral load was detected (Fig. 4d).
a Immunization scheme and sample collection schedule of the mouse-adapted infection model. BALB/c mice were immunized intramuscularly at day 0 (prime) and 21 (boost) with 1 µg of mRNA/animal. At week 7, mice were challenged with 1 × 104 TCID50 of the mouse-adapted strain MA20. At days 2 and 4 post-challenge, lung viral load and cytokine profile were evaluated (n = 5). Clinical signs and survival were monitored during 25 days after challenge in an independent experimental group of each condition (n = 10). b Body weight monitored over 25 days after challenge, calculated as a percentage of the initial weight (pre-challenge). c Survival rate over 25 days after challenge. d Lung viral load determined by virus titration assay in VERO E6 cells at day 2 and 4 post-infection. e Cytokine and chemokine profile in lung homogenates collected at day 2 and 4 post-challenge and analyzed by Luminex assay. Graphs are represented as mean ± SD. ns non-significant; **p < 0.01; ****p < 0.0001 as determined by two-way ANOVA with Tukey post-test.
Inflammatory virus-associated cytokine and chemokine markers were analyzed in lung homogenates using a multiplex assay as a complementary indicator of vaccine-induced protection. These cytokines are typically associated with virus-induced inflammation, as well as lung damage, and correlate with disease progression31. Cytokine and chemokine levels were significantly increased in unvaccinated compared to vaccinated animals, especially at day 2 post-infection. These cytokines were not upregulated in the lungs from the vaccinated groups in any of the evaluated time-points after virus challenge (Fig. 4e).
The results obtained with the virus adapted strain were validated in a transgenic C57BL/6 hACE2-K18 mouse model infected with an Alpha variant SARS-CoV-2 strain (Fig. 5a). The K18-hACE2 model of SARS-CoV-2 infection has been previously described as closely mimicking many aspects of severe COVID-19 and has been widely used in the evaluation of vaccines and antiviral therapies32. In accordance with the results for the mouse-adapted strain, no weight loss or clinical signs of disease were observed in vaccinated animals during the course of the experiment, in contrast with the non-vaccinated controls (Fig. 5b). Lung viral load was not detected in lungs from any of the vaccinated groups at day 4 post-infection (Fig. 5c). In addition, no significant increment in lung inflammatory cytokines and chemokines was detected in the vaccinated mice, in contrast with the non-vaccinated controls (Fig. 5d).
a Immunization scheme and challenge schedule of the K18 infection model. C57BL/6-hACE2 K18 mice (n = 5) were immunized intramuscularly at day 0 (prime) and 21(boost) with 1 µg of mRNA/animal. At week 7, mice were challenged with 1 × 105 TCID50 of SARS-CoV-2 alpha strain. At day 3 post-challenge, lung viral load was evaluated. b Body weight monitoring after 4 days of infection was calculated as a percentage of the initial weight (pre-challenge). c Lung viral load determined by virus titration assay in VERO E6 cells at day 4 post-infection. d Cytokine and chemokine profile in lung homogenates collected at day 4 post-challenge and analyzed by Luminex assay. Graphs are represented as mean ± SD. ns non-significant; ***p < 0.001; ****p < 0.0001 as determined by two-way ANOVA (b) and one-way ANOVA (c) with Tukey post-test.
Collectively, these data reveal that CPVax-CoV exhibits a strong vaccine-induced protection against viral SARS-CoV-2 challenge in wild-type and ACE-2 humanized mouse models.
CPVax-CoV platform validation for SARS-CoV-2 VOCs
Despite the remarkable efficacy demonstrated by SARS-CoV-2 vaccines, the emergence of novel variants poses a persistent challenge, compromising the level of protection conferred by current immunization strategies. As the virus continues to evolve, these variants exhibit diverse genetic mutations that may impact vaccine effectiveness. This underscores the need for continuous surveillance and adaptation of vaccination strategies to address the evolving challenge posed by these variants. To validate the versatility of the CPVax-CoV vaccine platform to be adapted to emerging VOC, we developed a vaccine candidate against the Omicron XBB.1.5 variant using CPVax-CoV technology. We chose the XBB lineage since it was one of the most recently emerged and prevalent variants at the time these experiments were conducted33,34. Moreover, XBB.1.5 sublineage exhibited superior growth advantages over most omicron mutants and has shown a strong evasive ability against the neutralization of plasma and serum from vaccinated or convalescent individuals35,36.
We first verified antigen expression in vitro of the new XBB.1.5 mRNA after HEK293T transfection by Western Blot and Flow Cytometry, and we confirmed that Spike XBB.1.5 mRNA was correctly translated in eukaryotic cells (Supplementary Fig. 3). Then, XBB.1.5 mRNA was formulated into LNPs with CPVax-CoV lipid composition, resulting in CPVax-CoV-XBB vaccine, which exhibited comparable physicochemical properties to that of CPVax-CoV (Supplementary Table 1).
Immunity of these vaccines against ancestral and XBB strains was evaluated in BALB/c mice following a 21-day prime-boost vaccination (Fig. 6a). To evaluate variant-specific antibody neutralizing ability, an XBB.1.5 Spike pseudovirus expressing GFP was constructed. Infection ability of XBB-Pseudovirus (XBB-PSV) was confirmed in HEK293T-ACE2-TMPRSS2 cells (Supplementary Fig. 4). Serum neutralization was determined after boost immunization against both wild-type and XBB.1.5 variants, and neutralization titers were found to be variant-specific. No neutralization was observed in serum from CPVax-CoV-XBB against the ancestral wild-type variant. Conversely, NT50 of wild-type Spike-based vaccines were not detected against XBB.1.5 variant (Fig. 6b). Spleen-specific T-cell response was also evaluated at week 6 post-prime. Variant selectivity was lower for the specific T-cell response than that observed for the antibody response. IFN-y secreting cells were detected in splenocytes from all vaccinated animals after stimulation with specific peptide mixes of wild-type or XBB Spike variants (Fig. 6c). A strong cross-reactive T cell response against various SARS-CoV-2 VOCs has been previously reported, highlighting the importance of robust T-cell induction in SARS-CoV-2 immunization strategies37.
a Timeline of CPVax-CoV-XBB immunity evaluation. BALB/c mice (n = 5) were immunized intramuscularly at day 0 (prime) and 21(boost) with 1 µg of mRNA/animal. Blood samples were obtained at week 6, and serum neutralizing ability was determined. T-cell response was evaluated in splenocytes 3 weeks post-boost. b NT50 neutralization titers in serum of vaccinated mice determined at 3 weeks post-boost by Spike pseudovirus-neutralization assay using wild type and XBB.1.5 Spike variants. c IFNγ-secreting splenocytes quantification by ELISPOT after O.N. stimulation with SARS-CoV-2 Spike peptide pools of wild type and XBB.1.5 variants. d Immunization scheme and challenge schedule of the K18 infection model with the XBB strain. C57BL/6-hACE2 K18 mice (n = 5) were immunized intramuscularly at day 0 (prime) and 21(boost) with 1 µg of mRNA/animal. At week 7, mice were challenged with 5 × 103 TCID50 of SARS-CoV-2 XBB.1.5 strain. At day 3 post-challenge, lung viral load was evaluated. e NT50 neutralization titers in the serum of mice vaccinated were determined at 3 weeks post-boost by Spike pseudovirus-neutralization assay using wild type and XBB.1.5 Spike variants. f Lung viral load determined by virus titration assay in VERO E6 cells at day 3 post-infection. Graphs are represented as mean ± SD. ns non-significant; *p < 0.05; **p < 0.01; as determined by two-way ANOVA (c), unpaired t-test (e), and one-way ANOVA (f) with Tukey post-test.
Finally, we evaluated vaccine-induced protection of our XBB vaccine candidate. The infection kinetics of the XBB strain in hACE2-k18 and BALB/c mouse models were first evaluated to choose the optimal infection model for the vaccine efficacy study. In contrast to ancestral strains, no weight loss or clinical signs of disease were detected over the first week of infection (Supplementary Fig. 7). However, infecting viral particles were present in lungs of infected mice in both mouse models, although virus titers were higher in hACE2-K18 mice (Supplementary Fig. 7), which was selected as infection model for XBB strain. These pathological outcomes are in accordance with previous results observed in rodent infection models of SARS-CoV-2 for XBB strains38,39.
K18 mice were immunized with CPVax-CoV and CPVax-CoV-XBB following a 21-day prime-boost regimen (Fig. 6d). Serum neutralization titers against XBB-PSV were analyzed before infection. Neutralization against XBB-PSV was not detected for the CPVax-CoV (Fig. 6e), showing comparable results to those obtained in the BALB/c mouse model (Fig. 6b). Protective efficacy was evaluated by determination of viral load in lungs. Despite the differences observed between wild type and XBB.1.5 Spike-based vaccines in serum neutralizing ability, no infecting viruses were detected in lungs from any vaccinated mice (Fig. 6f), further underscoring the critical role of T-cell responses in mediating protection against SARS-CoV-2. Accordingly, no weight loss was observed in any of the vaccinated groups after challenge (Supplementary Fig. 8).
These results demonstrate the potential of the CP vaccine platform to be effectively used with different antigen sequences for the development of SARS-CoV-2 vaccines against VOCs. Notably, the CPVax-CoV wild-type vaccine also provided significant protection against the XBB variant, highlighting its broad efficacy.
Lyophilized CPVax-CoV exhibits long-term thermostability
Thermostability and storage requirements are major factors limiting universal access to RNA vaccines. The two currently licensed mRNA vaccines, Comirnaty and Spikevax, require cryogenic transport and storage at −80 °C to −60 °C and −20 °C, respectively40, with a significant loss of quality under suboptimal conditions due to mRNA hydrolysis and LNP destabilization41,42. Consequently, developing thermostable mRNA vaccines is crucial to ensure accessibility in medium and low-income settings.
To address this challenge, we have developed a lyophilized formulation for the CPVax-CoV vaccine that maintains LNPs' physicochemical properties and effectively preserves vaccine-induced immune response after long-term storage at 4 °C and 25 °C. To this end, we employed an optimized lyophilization method based on a previously described protocol43. Furthermore, a long-term stability study was conducted for up to one year, demonstrating that the lyophilization process effectively preserved the characteristics and in vivo functionality of LNPs under both refrigerated and ambient temperature conditions throughout the study period, without the need for freezing.
To evaluate the long-term thermostability and biological efficacy of our lyophilized CPVax-CoV formulation, lyophilized CPVax-CoV LNPs from the same production batch were stored at 4 °C and 25 °C for up to one year. Non-lyophilized CPVax-CoV and Comirnaty vaccines stored at −80 °C were included as experimental controls, as the standard conditions used for the long-term storage of LNP-based vaccines44. At various time points throughout this time period, we assessed the physicochemical properties of the LNPs, including particle size, zeta potential, polydispersity index (PDI), and encapsulation efficiency, alongside their in vivo biological activity. BALB/c mice were immunized via a single intramuscular injection at each indicated time point using the same dosage as in the non-lyophilized vaccine formulation (1 µg of mRNA per animal). Serum samples were collected 21 days post-vaccination for specific antibody quantification. After 12 months of storage, a protective efficacy study was also conducted to assess the vaccine’s ability to confer protection against SARS-CoV-2 challenge in the mouse model (Fig. 7a).
a Timeline of the stability study. Lyophilized CPVax-CoV was stored at 4 °C and 25 °C, and standard liquid vaccines were stored at −80 °C as controls. Aliquots of each condition were reconstituted at the indicated time-points to characterize their physicochemical properties and to evaluate their in vivo activity. At 0, 3, 6, and 9 months, production of specific antibodies in serum was analyzed by ELISA after a single immunization of BALB/c mice with 1 µg of mRNA/animal. At 12 months, an efficacy study was conducted. After prime-boost immunization, animals were infected with mouse-adapted SARS-CoV-2, and neutralization, viral load and cytokines in lungs were evaluated. b Determination of diameter, PDI, Z potential, and encapsulation efficacy of liquid and lyophilized LNPs at the indicated time-points of storage. c Reciprocal endpoint titers of anti-RBD IgG antibodies in serum samples collected at day 21 post-immunization were determined by ELISA after the indicated time-points of storage. d Determination of diameter, PDI, Z potential, and encapsulation efficacy at different time-points after resuspension and storage at 4 °C of CPVax-CoV. e Reciprocal endpoint titers of anti-RBD IgG antibodies in serum samples collected at day 21 post-immunization were determined by ELISA at different time-points after resuspension and storage at 4 °C of CPVax-CoV. Graphs are represented as mean ± SD. ns non-significant; as determined by one-way ANOVA with Tukey post-test. wks weeks.
As shown in Fig. 7b, the size of the LNPs increased slightly after lyophilization, a result that is consistent with previous reports in the literature45. PDI, RNA encapsulation efficiency, and zeta potential showed no significant deviations between the lyophilized and non-lyophilized LNPs. Over the course of the one-year storage period, these parameters remained largely constant under the tested conditions. Lyophilized CPVax-CoV stored at 4 °C maintained their initial particle size for up to one year after lyophilization, while LNPs stored at 25 °C exhibited a slight increase in size after six months, which then stabilized and remained consistent for up to one year (Supplementary Table 2). In line with the physicochemical characterization results, the lyophilized CPVax-CoV vaccine maintained robust in vivo biological activity throughout the entire storage period, irrespective of the storage temperature. IgG anti-RBD levels remained consistently high across all experimental groups at each evaluated time point. Importantly, no significant differences were observed between the controls stored at −80 °C and the lyophilized CPVax-CoV stored at 4 °C or 25 °C. Notably, the lyophilized vaccine stored at 25 °C successfully induced specific antibody levels that were comparable to those of the −80 °C stored controls after one year, demonstrating outstanding preservation of immunogenicity (Fig. 7c). These results underscore the long-term thermostability of our lyophilized CPVax-CoV formulation, highlighting its potential for effective deployment in diverse temperature conditions without compromising efficacy.
In addition to assessing the long-term stability of the lyophilized CPVax-CoV, we evaluated the short-term stability of the resuspended lyophilized vaccine, a critical aspect for practical medical applications. To this end, the reconstituted CPVax-CoV was stored at 4 °C for up to 96 h, during which we monitored the physicochemical properties of the LNPs and their in vivo biological activity. Our analysis revealed that the LNP size and encapsulation efficiency remained consistent throughout the storage period, showing no significant deviations (Fig. 7d). Furthermore, the vaccine’s ability to induce specific antibody production in mice was unaffected, with no significant differences observed between the freshly resuspended vaccine and the one stored for 96 h (Fig. 7e). These findings underscore the potential utility of lyophilized CPVax-CoV in clinical settings.
Lyophilized CPVax-CoV maintains protective efficacy after one year at 25 °C
Protection induced by lyophilized CPVax-CoV after one year of storage at 4 °C and 25 °C was evaluated in the mouse model. BALB/c mice were vaccinated in a prime-boost regimen and challenged with the MA20 mouse-adapted SARS-CoV-2 strain27. In addition to the one-year stored lyophilized and control vaccines, freshly prepared CPVax-CoV and Comirnaty vaccines were included in this study as additional controls. Serum neutralization was evaluated at 3 weeks post-boost, and viral load and cytokine production were evaluated in lungs at day 3 post-infection (Fig. 8a).
a–e Protection of one-year stored lyophilized CPVax-CoV at 4 °C and 25 °C in BALB/c mice infected with a viral dose of 1 × 104 TCID50 of the MA20 strain. a Immunization scheme and challenge schedule of the MA20 infection model. b NT50 neutralization titers in the serum of vaccinated mice were determined at 3 weeks post-boost by Spike pseudovirus-neutralization assay. c Body weight monitoring during the 3 days after infection was calculated as a percentage of the initial weight (pre-challenge). d Lung viral load was determined by virus titration assay in VERO E6 cells at day 3 post-infection. e Cytokine and chemokine profile in lung homogenates collected at day 3 post-challenge and analyzed by Luminex assay. f Protective efficacy of lyophilized CPVax-CoV and CPVax-CoV-XBB in C57BL/6 humanized ACE2 model against 1 × 105 or 5 × 103 TCID50 of alpha or XBB infection, respectively. Lung viral load determined by virus titration assay in VERO E6 cells on day 3 post-infection. Graphs are represented as mean ± SD. ns non-significant; **p < 0.01; ****p < 0.0001 as determined by one-way ANOVA (b, d, and f) and two-way ANOVA (c) with Tukey post-test.
Prior to virus challenge, anti-RBD IgG antibodies (Supplementary Fig. 9) and serum neutralization were evaluated (Fig. 8b). No significant differences were observed in anti-RBD binding antibody titers between lyophilized vaccines stored at 4 °C and 25 °C and freshly formulated. Neutralizing antibody titers were higher in mice vaccinated with CPVax-Cov lyophilized and stored at 4 °C, while no significant differences were found between lyophilized vaccines stored at 25 °C and freshly formulated LNPs.
No weight loss or clinical signs of disease progression were observed in any of the vaccinated groups (Fig. 8c). Accordingly, infecting virus in lungs were only detected in the non-vaccinated animals in the TCID50 assay (Fig. 8d). These results are consistent with the cytokine profile observed in lung homogenates, since the inflammatory cytokines and chemokines evaluated were significantly higher in lungs from unvaccinated mice and no significant differences in cytokine levels were found between mice vaccinated with controls and 4 and 25 °C lyophilized CPVax-CoV (Fig. 8e).
Finally, we validated our lyophilization technology to be applied to different mRNA payloads and developed a lyophilized CPVax-CoV-XBB. Physicochemical parameters of the lyophilized CPVax-CoV-XBB were comparable to those obtained with CPVax-CoV (Supplementary Table 2). Lyophilized CPVax-CoV-XBB and CPVax-CoV protective efficacy was evaluated in the humanized K18 mouse model after infection with XBB or alpha SARS-CoV-2 strains, respectively (Fig. 8f). In line with the results obtained in the mouse-adapted infection model vaccinated with the wild-type-based Spike sequence, no viral load in lungs was detected in K18 mice immunized with lyophilized CPVax-CoV-XBB or CPVax-CoV at day 3 after infection with their respective viral strains (Fig. 8f).
Collectively, these findings demonstrate that lyophilized CPVax-CoV, stored at both 4 °C and 25 °C for up to one year, provides robust and consistent protection against SARS-CoV-2 infection in mice. Furthermore, this lyophilization technology proved to be effective in maintaining the efficacy of our LNP formulations with different mRNA spike sequences, demonstrating its adaptability to other mRNA sequences from SARS-CoV-2 and potentially other pathogens.
Discussion
The rapid development of mRNA vaccines has proven to be a transformative approach to combating the COVID-19 pandemic46. Despite their success, challenges remain in enhancing vaccine stability and optimizing the delivery systems to ensure easy deployment and broader accessibility. This study addresses these challenges by developing an mRNA vaccine platform that incorporates optimized UTR sequences and rationally designed ionizable lipids in a novel LNP formulation that can be successfully lyophilized for long-term storage to overcome the limitations of existing mRNA vaccine formulations.
LNPs are the leading delivery system for mRNA vaccines, and the ionizable lipids play a critical role in mRNA transfection efficiency and endosomal escape18. Although many ionizable lipids have already been described, very few have been approved for clinical use and are currently licensed for mRNA vaccine applications4,5,47. We have recently described a new library of ionizable lipids developed using a high-throughput screening approach using a multicomponent thiolactone-based synthesis platform that allowed us to synthesize many lipid candidates in a cost-effective, scalable, and time-efficient manner. This screening process enabled us to establish critical structure-function relationships, particularly related to the hydrophobic tails and functional groups that influence endosomal escape and membrane-disruption activity. As a result, we identified several lipid candidates with enhanced in vivo mRNA delivery than gold standard ionizable lipids, as evidenced by superior performance in luciferase protein expression after intramuscular injection. Additionally, the use of our thiolactone-based lipids offers significant advantages due to an optimized synthetic method, featuring a one-pot synthesis with short reaction times (2 h) and room temperature23, in contrast to lipids such as SM-102 or ALC-031548,49. Our goal in selecting these lipids was not only to use potential effective candidates in terms of protein expression but also to achieve broad chemical structural diversity, incorporating variations in polar head groups (both cyclic and linear), hydrophobic tail lengths, branching structures, and the presence of unsaturations.
In the present study, we first evaluated the immunogenic potential of some of our top-performing ionizable lipids in the context of a SARS-CoV-2 vaccination model, hypothesizing that the increased protein expression observed after intramuscular administration would lead to a stronger antigen presentation, thereby enhancing robust immune responses. Our results indicate that all the ionizable lipids tested here efficiently encapsulated the Spike mRNA in a standard LNP formulation and were capable of inducing both humoral and cellular immune responses. Although a strong Th1 polarized immune response was observed across all the experimental groups, lipids CP-LC-0431 and CP-LC-0474 were excluded due to their comparatively lower induction of IFNγ secretion compared to the other lipids and the commercial vaccine formulation. CP-LC-0729 was selected as the lead candidate as it induced one of the strongest immune responses, which correlates with our previous observations for intramuscular protein expression23. Furthermore, CP-LC-0729 has been extensively characterized in our previous work and has proven to be safe and well tolerated after systemic injection23.
Once the ionizable lipid CP-LC-0729 was selected, we incorporated an optimized mRNA sequence encoding the SARS-CoV-2 Spike protein, featuring proprietary UTRs designed to enhance translation efficiency16. The incorporation of this optimized sequence significantly improved the immune response associated with the LNP formulated with CP-LC-0729 in terms of B and T cell responses. Notably, the optimization of UTRs can lead to increased protein expression, allowing for a reduction in the required vaccine dose without compromising immunogenicity. For instance, the Comirnaty vaccine uses 30 μg of mRNA per dose, while the Spikevax vaccine requires higher doses of mRNA—100 µg for the primary series and 50 µg for booster doses. These differences in mRNA dosage likely reflect variations in the efficiency of their respective UTRs and LNP formulations, with Comirnaty’s UTRs potentially offering more efficient translation at a lower mRNA concentration21. Our findings reinforce the critical role of UTR optimization, along with the synergy between mRNA design and LNP delivery systems, in producing robust and durable immune responses with potentially lower mRNA doses. Based on these data, we defined our final vaccine candidate, CPVax-CoV, which combines the ionizable lipid CP-LC-0729 with our optimized mRNA sequence. CPVax-CoV demonstrated complete protection against SARS-CoV-2 infection in both mouse-adapted and humanized murine infection models, as it was able to fully protect vaccinated mice from productive SARS-CoV-2 infection and virus-associated inflammatory response in the lungs. Additionally, the biodistribution study of CPVax-CoV revealed that the vaccine primarily localizes at the injection site and in the draining lymph nodes after intramuscular injection, which is likely contributing to its strong induced immune response. Although more detailed studies are needed to fully characterize the pharmacokinetic profile of CPVax-CoV, renal excretion was observed, and no accumulation of the vaccine was detected in major organs after 7 days, preliminarily indicating a favorable tolerability profile.
One of the most significant achievements of this study is the successful lyophilization of CPVax-CoV and the demonstration of its long-term stability and protective efficacy. While previous reports have explored lyophilized mRNA vaccines, most have been limited to shorter storage periods and typically assessed physicochemical parameters, in vitro transfection, or immunogenicity only50,51,52,53,54,55,56. Immune responses of lyophilized mRNA-LNP vaccines have been reported after storage of up to 6 months at 4 °C or 25 °C45,50,51, but no data beyond this time frame have been published. Notably, Ai et al. demonstrated in vivo protection using freshly lyophilized formulations50, but no previous study has assessed protective efficacy following extended storage of lyophilized mRNA vaccines. To our knowledge, this is the first report to demonstrate vaccine-induced protection in a relevant in vivo infection model after one year of storage at 25 °C, representing the longest duration of confirmed in vivo biological activity for a lyophilized mRNA-LNP vaccine to date.
We have previously described an optimized lyophilization protocol for LNPs encapsulating luciferase-coding mRNA using SM102 as the ionizable lipid, which successfully preserved LNP physicochemical properties and maintained protein expression in vivo after extended storage at 4 °C, compared to their liquid counterparts43. Applying this optimized protocol to CPVax-CoV, we demonstrated that our recently described ionizable lipids are compatible with lyophilization, and the LNPs retain their integrity and their ability to induce an immune response after prolonged storage.
The demonstration that the lyophilized LNPs remain stable after reconstitution in liquid buffer for at least 96 h at 4 °C further enhances the practicality of this platform in clinical settings, allowing for more flexibility in vaccine administration and in diverse medical environments. It is important to note that this assessment was conducted with freshly prepared lyophilized LNPs, and further studies are needed to evaluate the stability of the reconstituted LNPs after long-term storage and at different temperatures, including room temperature. Notably, current SARS-CoV-2 mRNA vaccines have a limited recommended timeframe for use once diluted, with usability restricted to up to 6 h for Comirnaty and 12 h for Spikevax57.
The results presented here with mRNA encoding the Spike protein of the XBB.1.5 variant demonstrate that this platform is highly adaptable to other mRNA sequences. Interestingly, despite the lack of detectable neutralizing antibodies against XBB.1.5, CPVax-CoV is able to induce protection in the lungs. Further supporting this finding and highlighting the potential for broad protection against other VOCs, it should be noted that the MA20 strain carries several mutations in the Spike protein and in other viral proteins such as ORF1a and N, as a consequence of mouse adaptation. Despite these genetic changes, our vaccine formulation demonstrated strong protective efficacy. In this regard, immunity studies demonstrated that the vaccine elicited a strong variant-independent T cell response, which likely contributed to the observed protection in mice of CPVax-CoV against XBB challenge. This finding highlights the role of cellular immunity in protecting against viral variants37 and suggests that CPVax-CoV could provide cross-protection against emerging variants. However, further studies of immunity and other correlates of protection against XBB and different VOCs are required to confirm these preliminary findings and to fully understand the mechanisms of protection conferred by CPVax-CoV against VOCs.
The findings presented in this study also open up valuable opportunities for future research. First, further investigation into long-term immunity is essential to fully understand the durability of both humoral and cellular responses, particularly the persistence of T cell-mediated protection against VOCs. Exploring long-term immune memory, mucosal immunity, and viral dissemination to other organs, as well as conducting studies in additional relevant animal models, will provide a deeper understanding of CPVax-CoV’s full protective potential, PK, and safety. Additionally, while the lyophilized formulation of CPVax-CoV has demonstrated stability for one year at 25 °C, further research into stability at higher temperatures and under diverse environmental conditions would expand its applicability. This will be especially important for optimizing its widespread use in various climates and global vaccination campaigns, especially in regions where maintaining cold-chain logistics is challenging.
In summary, CPVax-CoV represents a significant advancement in mRNA vaccine development by addressing key challenges in stability, delivery, and efficacy. The combination of novel ionizable lipids and optimized mRNA sequences, along with successful lyophilization, offers a scalable and accessible solution that can overcome the logistical barriers of cold-chain dependency, making it highly suitable for global deployment in future pandemics. Although further research will be crucial to fully unlock its potential and ensure its adaptability across different contexts and pathogens, our findings provide a solid foundation for further research into the use of this technology for broader applications beyond SARS-CoV-2.
Materials and methods
DNA template design
The DNA template for the Comirnaty vaccine was constructed by cloning the publicly available mRNA vaccine sequence58 into a pUC-based plasmid under the control of a T7 promoter.
For the CPVax-CoV vaccine, the coding sequence of the Spike SARS-CoV-2 protein from Wild wild-type-H1 strain (GISAID59 accession number EPI_ISL_402124) was codon-optimized for human expression. This sequence included the K986P and V987P (2P) stabilizing mutations and the substitution of the RRAR furin cleavage site at residues 682–685 with GSAS. For the CPVax-CoV-XBB vaccine, the DNA template was generated using the XBB.1.5 strain sequence (GISAID accession number EPI_ISL_15851788). The same 2P and RRAR to GSAS modifications were introduced, and the sequence was similarly codon-optimized for human use.
Both CPVax-CoV and CPVax-CoV-XBB, optimized sequences were inserted in-frame immediately downstream of the 5´UTR into a pUC-based plasmid. This plasmid featured the following elements arranged in a 5′ to 3′ orientation: a T7 RNA polymerase promoter, an optimized 5′ UTR from human APOA216, the 3′ UTR from human beta-globin (GenBank: NM_000518), a polyadenylation tail consisting of 100 adenines, and a BspQI restriction enzyme site. The complete 5′ and 3′ UTR sequences are provided in the Supplementary Information (Supplementary Fig. 3).
Gene synthesis, cloning, and plasmid preparations were outsourced to Genscript. The purified plasmids were subsequently used for in vitro transcription.
In vitro transcription and mRNA purification
Each plasmid containing the DNA template sequence of interest was digested with BspQI (ON-124, HONGENE), which cleaved the plasmid immediately after the segment to be transcribed. The linearization reaction was then purified with the Wizard SV Gel and PCR Clean-Up (Promega A7270), in accordance with the manufacturer’s instructions.
The purified linear DNA was subsequently employed for mRNA production by in vitro transcription using T7 RNA polymerase following the manufacturer’s instructions. Briefly, transcription reactions were performed at 37 °C for 3 h using the following materials:
Template linear DNA (50 µg/mL), T7 RNA polymerase (5000 U/mL; HONGENE, ON-004), RNAse inhibitor (1000 U/mL; HONGENE, ON-039), Inorganic Pyrophosphatase (2 U/mL; HONGENE, ON-025), ATP (5 µg /mL; HONGENE, R1331), GTP (5 µg/mL; HONGENE, R2331), CTP (5 µg/mL; HONGENE, R3331), N1-Methylpseudouridine (5 µg/mL, HONGENE, R5-027), CAP AG (4 µg mL, Hongene, ON-134), and RNAse-free double-distilled water.
The generated mRNA transcripts were initially treated using DNaseI incubation (Hongene, ON-109) according to the manufacturer’s instructions.
In order to reduce double-stranded RNA (dsRNA) contaminants, the resulting mRNA was purified through a process involving two sequential chromatography steps, starting with anion exchange chromatography (AEX) followed by affinity chromatography60. Initially, the mRNA sample was purified using the CIMmultus PrimaS Chromatography Column (BIA Separations). IVT mixture was diluted once in sample loading/equilibrate buffer A (20 mM Tris, 20 mM BTP, 20 mM glycine, 50 mM NaCl, 10 mM EDTA, pH = 8.0) and loaded onto the column. After unbound IVT components eluted in flow-through, a wash 1 with equilibration buffer was necessary, followed by a high-salt wash with 50 mM Tris, 3.0 M guanidine-HCl, 20 mM EDTA, pH 8.0. And after a wash 3 with buffer A until UV returns to baseline, a step elution was performed with buffer C (20 mM Tris, 20 mM BTP, 20 mM glycine, 50 mM NaCl, 10 mM EDTA, pH = 11.0). Fractions were neutralized immediately after elution. Following, additional purification was carried out through affinity chromatography using a POROS Oligo (dT)25 column (ThermoFisher). Specifically, the buffers employed were as follows: Buffer A, which contained 50 mM disodium phosphate, 0.5 M NaCl, 5 mM EDTA, pH = 7.0, and Buffer B, which contained 50 mM sodium dihydrogen phosphate, 5 mM EDTA, pH = 7.0. The mRNA samples were initially half-diluted in Buffer A 2x. Following this, the column was equilibrated with 100% Buffer A, loaded with mRNA, washed with Buffer B, and ultimately eluted using double-deionized water. To completely remove Buffer B, mRNA was washed with a 30 KDa Amicon® filter and then equilibrated through a one-tenth dilution in citrate buffer 10× with a pH of 6.5.
The concentration of mRNA was determined by measuring the optical density at 260 nm, then adjusted to a final concentration of 1 mg/mL, aliquoted, and stored at −80 °C until needed.
For quality assurance, all mRNAs underwent analysis through automated electrophoresis (2100 Bioanalyzer G2938B, Agilent). Subsequently, the mRNA samples were aliquoted and stored at −80 °C until needed.
Synthesis and characterization of ionizable lipids
All intermediate compounds and ionizable lipids were synthesized following protocols based on the Sequential Thiolactone Amine Acrylate Reaction23. Briefly, following the synthesis of thiolactone derivatives, the ionizable lipid was obtained via a multicomponent one-pot reaction. Thus, the corresponding thiolactone derivatives (0.15 mmol, 1 equiv.), acrylate (0.15 mmol, 1 equiv.) amine (0.15 mmol, 1 equiv.) were dissolved in 300 μL of tetrahydrofuran (THF) at room temperature. After stirring for 2 h, the THF was removed under reduced pressure, and the product was purified using a CombiFlash NextGen 300+ (gradient of elution: from 100% dichloromethane to 50% of an 80/20/1 mixture of DCM/MeOH/NH4OH (aq)). All synthesized ionizable lipids were characterized by high-performance liquid chromatography coupled with a charged aerosol detector (HPLC-CAD) and mass spectrometry (ThermoFisher ISQ). Noteworthy, in previous publications, CP-LC-0743 was also referred to as A4B2C1.
Molecular structures are shown in Supplementary Fig. 1. Theoretical and experimental molecular weights (MW):
CP-LC-0729 theoretical [M + H]+ = 740.63, experimental [M + H]+ = 740.85
CP-LC-0867 theoretical [M + H]+ = 684.57, experimental [M + H]+ = 684.74
CP-LC-0431 theoretical [M + H]+ = 766.65, experimental [M + H]+ = 766.85
CP-LC-0474 theoretical [M + H]+ = 768.66, experimental [M + H]+ = 768.76
CP-LC-0743 theoretical [M + H]+ = 766.65, experimental [M + H]+ = 766.81
CP-LC-0729 1H-RMN was obtained using a Bruker 400 MHz NMR spectrometer. 1H NMR (400 MHz, CDCl3) δ: 6.76 (m, 1H); 6.37 (d, J = 7.9 Hz, 1H); 4.60 (dt, J = 7.1 Hz J = 7.1 Hz, 1H); 3.98 (d, J = 5.8 Hz, 2H); 3.36 (dt, J = 5.7 Hz J = 5.7 Hz, 2H); 2.78 (t, J = 6.8 Hz, 2H); 2.59 (m, 4H); 2.48 (t, J = 5.9 Hz, 2H); 2.28 (s, 6H); 2.06 (m, 2H); 1.96 (m, 1H); 1.58 (m, 3H); 1.41 (m, 2H); 1.25 (m, 44H); 0.87 (m, 12H) (full spectrum available in Supplementary Fig. 1).
mRNA encapsulation into LNPs
LNPs formulations were prepared following a previously described method with modifications20. Briefly, the purified mRNAs were initially diluted in 10 mM sodium citrate buffer at pH = 4, reaching a final concentration of 266 µg/mL. Simultaneously, the lipid mixture was prepared in ethanol at the different molar ratios used of 46.6:9.4:42.7:1.6 (Comirnaty), 50:10:38.5:1.5 (CP ionizable lipids formulations in Fig. 1) and 40.7:34.9:23.3:1.2 (CPVax-CoV) for ionizable lipids CP-LC-0729, CP-LC-0867, CP-LC-0431, CP-LC-0743, CP-LC-0474 or ALC-0315 (Merck 586224): helper lipids DSPC (Merck 850365P) or DOPE (Merck 850725P): Cholesterol (Sigma C3045) and DMG-PEG2000 (Cayman 33945-1) or ALC-0159 (Cayman 34336). Aqueous mRNA solution and lipid mixture were combined at a molar N/P ratio of 6:1 (Moderna/Pfizer) or ionizable lipid/RNA weight ratio of 10:1 (CPVax-CoV formulation). Microfluidic technique was used for the synthesis of LNPs, thereby NanoAssemblr® Ignite microfluidic device (Precision Nanosystems) was set at a total flow rate (TFR) of 12 mL/min and a aqueous:ethanol flow rate ratio (FRR) of 3:1. The resulting LNPs were dialyzed (Pur-A-Lyzer™ Midi Dialysis Kit) overnight against Tris buffer containing cryoprotectants. Each resulting LNP solution was then collected and adjusted to a final concentration of mRNA of 100 µg/mL, filtered through a 0.22 mm filter, and stored at −80 °C for further use.
The average size, PDI, and zeta potential of LNPs were determined using a Malvern Zetasizer Advance Lab Blue Label (Malvern Instruments Ltd., UK) with a capillary cell (DTS1070) and diluting the sample (typically 1:100) in KCl 10 mM filtrated solution.
The concentration of mRNA in LNPs was measured using Quant-iT™ RiboGreen™ RNA Assay Kit from Thermo Fisher Scientific following the manufacturer’s protocols. Thus, the % of RNA encapsulated was calculated by comparing the total RNA obtained by the lysis of mRNA-LNPs using 0.5% Triton X-100 and the non-encapsulated RNA obtained when the LNPs are not lysed in the absence of detergent. Fluorescence was quantified in a Fluostar Omega microplate reader (BMG Labtech). Agarose gel electrophoresis was additionally used to determine the encapsulation of mRNA in LNPs. The quantification of encapsulated mRNA was determined by band densitometry using ImageJ software. Samples were loaded in a 1% agarose gel including SYBR-Safe, and the electrophoresis was run at 120 V for 30 min. Gels were visualized with a UV transilluminator iBright™ CL750 imaging system, using adequate exposure times to avoid image saturation.
LNP lyophilization
After adjusting the concentration of mRNA-LNPs to 100 µg/mL, the mRNA-LNPs suspension was aliquoted into glass vials with a volume of 300 µL per vial. Lyophilization was conducted using a Genesis Pilot Freeze Dryer, following a three-stage process: initial freezing, primary drying, and secondary drying. After lyophilization, vials were backfilled with pure nitrogen, capped, and transferred to various temperatures for stability assessments. The lyophilized mRNA-LNPs were stored at either 4 °C or 25 °C for different times and compared with the non-lyophilized solution stored at −80 °C as a control. To reconstitute lyophilized samples, 300 μL of RNase-free water was added to each vial and softly mixed until the solution turned into a homogeneous, slightly white, clear suspension.
Cell lines
HEK293T cells were obtained from the American Type Culture Collection (ATCC, CRL-3216 and CRL-1586). Vero E6 cells were kindly provided by Júlia Vergara from the Centro de Investigación en Sanidad Animal IRTA-CReSA (Barcelona, Spain). HEK293T-ACE2-TMPRSS2 cells were obtained from the National Institute for Biological Standards and Controls (NIBSC, 101008).
Cells were cultured on complete DMEM, which consists of high-glucose Dulbecco’s Modified Eagle’s Medium (Merck, D6429) supplemented with 10% Fetal Bovine Serum (Sigma, F7524), 1% Penicillin-Streptomycin Solution (Gibco, 15140122), and 2 mM Glutamax (Fisher, 35050038). Vero E6 cells were additionally supplemented with 25 mM HEPES (4-(2-hydroxyethyl)-1-piperanzineethanesulfonic acid) (Biowest), and HEK293T-ACE2-TMPRSS2 cultures were additionally supplemented with 1 µg/mL puromycin dihydrochloride (Sigma, P8833).
Animals
All animal experiments were conducted in agreement with European and national directives for the protection of experimental animals, and experimental procedures were approved by the Ethics Committee for Animal Experiments of the University of Zaragoza (PI59/21 and PI36/22) or the University of Navarra (Protocol ref. CEEA026/20). In addition, animal experiments using the SARS-CoV-2 strains were approved by the Biosafety Committee of the University of Zaragoza (Refs: 124/20, 156/21, and 184/22) and the GMO Interministerial Council (A/ES/20/99).
For the immunity studies, BALB/cAnNRj mice were purchased from Janvier Labs. For the viral challenge studies, BALB/cOlaHsd and C57BL/6J K18-hACE2 transgenic mice were purchased from Envigo and Charles River (K18-hACE2 JAX Mice Strain), respectively.
Male and female mice aged 8–10 weeks and weighing 18–28 g were used for all the experiments. All mice underwent an acclimation period lasting 3–7 days to adapt to the experimental conditions upon arrival at the research facilities, and were housed and maintained in specific pathogen–free conditions in the facilities of Centro de Investigaciones Biomédicas de Aragón (Zaragoza, Spain; reference ES 50 297 0012 011) for immunity studies and Centro de Investigación de Encefalopatías y Enfermedades Transmisibles Emergentes (Zaragoza, Spain; reference ES 50 297 0012 009) for challenge studies. Housing conditions were controlled, maintaining a room temperature of 20–24 °C, humidity levels ranging from 50% to 70%, and a light intensity of 60 lux, with a light-dark cycle lasting 12 h. During the studies, all animals were monitored by the animal resources center or laboratory staff daily.
For the procedures requiring anesthesia (SPECT-CT imaging and SARS-CoV-2 challenge), all animals were anesthetized via inhalation using isoflurane (IsoVet) mixed with oxygen at a flow rate of 1 L/min in a rodent-specific anesthesia station. Anesthesia was induced with 5% isoflurane and maintained at 2%. Animals were monitored continuously during anesthesia and observed post-procedure until fully recovered. No anesthesia was used for intramuscular injections, blood collection, and weighing. Mice were humanely euthanized with CO2 asphyxiation followed by cervical dislocation.
Virus strains
The SARS-CoV-2, hCoV-19/Sweden/20-53846/2020, (Lineage B.1.1.7; Alpha variant) was provided by the Public Health Agency of Sweden. The mouse-adapted strain SARS-CoV-2 MA20 was obtained by serial passaging of SARS-CoV-2, hCoV-19/Sweden/20-53846/2020, (Lineage B.1.1.7; Alpha variant) in mice as described in ref. 27. The SARS-CoV-2 hCoV19/USA/CA-Stanford-109_S21/2022 (Lineage XBB; Omicron Variant) (GISAID: EPI_ISL_15509864) was isolated from a human on October 10, 2022, in California and was obtained from BEI resources (NR-58925). All procedures involving infectious viruses, including in vivo experiments, were performed under biosafety level 3 (BSL-3) conditions.
mRNA in vitro transfection
HEK293T cells were seeded in 6-well cell culture plates at a density of 4 × 105 cells/well and incubated overnight. The SARS-CoV-2 Spike-coding mRNAs were transfected using Lipofectamine MessengerMax Transfection Reagent (Invitrogen, 15397974) according to the manufacturer’s protocol. Briefly, a mixture of each mRNA (2.5 µg/well) and Lipofectamine MessengerMAX (5 µL/well) was pre-incubated in OptiMEM media. The mRNA-lipofectamine mixture was added to the corresponding wells in duplicate, resulting in a final mRNA concentration of 2.5 µg/well. The cells were incubated for 24 h at 37 °C and 5% CO2.
For Western Blot, cells were lysed after incubation with 0,1% TritonX and centrifuged at 13,000×g for 10 min. The cell lysates were run under reducing conditions by SDS-PAGE (SurePAGE, 4-12% Genscript, M00653) and transferred to a nitrocellulose PVDF membrane (Bio-Rad). The membranes were blocked with 3% BSA in TBST buffer (Tris-buffered saline + 0.1% Tween 20) and incubated with a rabbit polyclonal SARS-CoV-2 Spike antibody (SinoBiological, 40591-T62) overnight at 4 °C. The membrane was washed and incubated with a goat anti-rabbit Ig, Human ads-HRP (Southern Biotech, 4010-05) as a secondary antibody. Colorimetric detection of the samples was performed with Pierce ECL Western Blotting Substrate (Thermo, 32106), and images were acquired using the iBright CL750 Imaging system (Invitrogen).
For flow cytometry analysis, cells were trypsinized after incubation. Collected cells were incubated with human FcR blocking reagent (Miltenyi, 130-059-901) for 15 min at 4 °C. Cells were washed and incubated with fixation buffer (eBioscience, 88-8824-00) for 30 min at 4 °C, followed by three washing steps with permeabilization buffer (eBioscience, 88-8824-00) and incubation with SARS-CoV-2 Spike S1 Subunit Antibody (R&D Systems, MAB105403) diluted in permeabilization buffer for 1 h at room temperature. Subsequently, cells were washed and incubated with FITC Goat Anti-Mouse IgG/IgM (BD, 555988) for 1 h at room temperature. After final washing steps, labeled cells were resuspended in PBS buffer, and acquisition was performed using a Gallios flow cytometer (Beckman Coulter).
In vivo immunization and blood collection
Mice were immunized with LNPs prepared as described above, receiving an intramuscular injection of 1 µg of the specified mRNA-LNP into the right thigh muscle, diluted in Tris buffer containing 15% sucrose in a final volume of 30 µL, and administered using a 30 G insulin syringe. An equal booster dose was administered on day 21 following the initial immunization. Blood samples were collected from the submandibular vein on days 0, 21, and 42 post-prime. Blood was allowed to clot and then centrifuged at 6500×g and 4 °C for 10 min. The sera were collected and stored at −80 °C for antibody analysis.
Determination of antibody levels in serum
The serum samples from immunized mice were analyzed for RBD-specific IgG antibody titers using an indirect enzyme-linked immunosorbent assay (ELISA). 96-well Nunc MaxiSorp microplates (Thermo Scientific) were coated with 50 ng/well of recombinant RBD (Certest Biotec) diluted in carbonate/bicarbonate buffer pH 9.6 and incubated overnight. The next day, plates were washed with phosphate buffer saline–Tween 20 0.05% (PBST) followed by blocking with 3% BSA (Seqens) in PBST for 1 h. After washing, serially diluted mouse sera were added to the plate and incubated for 1.5 h at 37 °C, followed by washing steps and addition of IgG goat anti-mouse horseradish peroxidase (HRP) antibody (Southern Biotech) at a dilution of 1:10.000. The plates were developed using 3,3’,5,5’-tetramethylbenzidine (TMB) substrate (Abcam) and 0.2 M H2SO4 (aq) to stop the reaction. Finally, the absorbance was measured at 450 and 630 nm using a FLUOstar Omega microplate reader (BMG Labtech).
The reciprocal endpoint titer was defined as the highest dilution at which the optical density (OD 450-630 nm) of the sample reached a predetermined cutoff of 0.1 or greater.
Organ single-cell suspensions
Organs were harvested and placed in RPMI 1640 medium (Fisher, 10379144). Spleens and lymph nodes (LNs) were homogenized using a syringe plunger and filtered through a 70 µm cell strainer. The cell strainer was washed with 10 mL of RPMI, and cells were centrifuged at 450×g for 5 min. Red blood cells (RBCs) in spleens were lysed using 1 mL of eBioscience RBC Lysis Buffer (Thermofisher, 00-4300-54) for 1 min. After lysing, the cells were washed with 10 mL of RPMI and centrifuged at 450×g for 5 min.
Cells from tissues were resuspended in 1 mL of RPMI 1640 medium supplemented with 10% heat-inactivated Fetal Bovine Serum (Sigma F7524), 1% Glutamax (Fisher, 35050038), 1% Penicillin-Streptomycin (Gibco, 15140122), and 0.00035% β-mercaptoethanol (Merck, M3148). The resuspended cells were immediately used for counting, culture, or staining.
ELISPOT assay
Enzyme-linked immunospot (ELISPOT) assay was employed to quantify the frequency of cytokine-secreting splenocytes. ELISpot assays were performed using mouse IFN-γ ELISpot kits (CTL Immunospot) according to the manufacturer’s instructions.
96-well immunospot plates were coated with 60 µL of murine IFN-γ capture solution and incubated at 4 °C overnight. The next day, plates were washed with PBS, and 4 × 105 splenocytes/well were stimulated with 50 µg/mL of a peptide pool covering the immunodominant sequence domains of the S protein (Miltenyi, 130-126-700). Phorbol-12-myristate-13-acetate (PMA) 50 ng/mL (Merck, 524400) and ionomycin 500 ng/mL (Merck, 407953) were used as positive control, while splenocytes without stimulation were utilized as negative controls. Samples were incubated for 20 h at 37 °C and 5% CO2. The following day, plates were washed, and 10 µL of anti-murine IFN-γ detection solution was added. After washing, a tertiary solution containing streptavidin-HRP was used, followed by the addition of a chromogen substrate for enzyme activity detection. Finally, plates were scanned using an ImmunoSpotS6 Ultra-V analyzer (CTL EUROPE GMBH), and spot numbers were assessed using ImmunoSpot 7.0.34.0 Professional Analyzer DC software.
ELISA cytokine assay
The 1 × 106S splenocytesl were seeded in 96-well plates at 1 x106 and stimulated with 50 µg/mL of a peptide pool covering the immunodominant sequence domains of the S protein (Miltenyi, 130-126-700). Phorbol-12-myristate-13-acetate (PMA) 50 ng/mL (Merck, 524400) and ionomycin 500 ng/mL (Merck, 407953) were used as positive controls, while splenocytes without stimulation were utilized as negative controls. Samples were incubated for 20 h at 37 °C and 5% CO2. Supernatants were collected after centrifugation at 1500×g for 5 min, and samples were stored at −80 °C until analysis.
Cytokine quantification was conducted using Mabtech ELISA Flex (HRP) kits following the manufacturer’s instructions. Briefly, Nunc MaxiSorp 96-well plates (Thermo Fisher) were coated with capture antibody and incubated at 4 °C overnight. The next day, plates were washed with PBST and blocked with 0.1% BSA in PBST buffer for 1 h at room temperature. After washing, samples and standards dilutions were added and incubated for 2 h at room temperature, followed by the addition of detection biotinylated antibody and incubation for 1 h. Streptavidin-HRP was added and incubated for 1 h at RT. Finally, the plates were washed, and TMB substrate (Abcam) was added. Reaction was stopped with 0.2 M H2SO4, and the absorbance was measured at 450 nm and 630 nm using a FLUOstar Omega microplate reader (BMG Labtech).
Luminex assay
Lung samples were acquired from a weighed portion of murine lung, previously homogenized in 1 mL of DMEM using a GentleMACS Dissociator (Miltenyi), and then clarified by collecting the supernatant after centrifugation at 450 rpm for 5 min. Before analysis, samples were inactivated by the addition of 0.5% Triton X100 and incubated for 30 min at 4 °C. Protease inhibitor cocktail (Complete, Roche) was added to prevent protein degradation during incubation. Samples were stored at −80 °C until the Luminex assay was conducted.
Cytokine/Chemokine array analysis was carried out using the Luminex Mouse Discovery Assay 11-plex kit (R&D Systems). Samples were centrifuged at 1000×g for 10 min and then diluted 1:2 in calibrator diluent, and the assay was conducted following the manufacturer’s instructions. Measurements were carried out using Luminex LABSCAN 100 and quantified by comparison to a standard curve. Data collection and analysis were performed utilizing LUMINEX 100 IS software.
Flow cytometry analysis of Tfh and GC B cells
Lymph node single cell suspensions were incubated with mouse FcR blocking reagent (Miltenyi, 130-092-575) for 15 min at 4 °C. Cells were washed and incubated with anti-mouse antibodies for surface staining for 15 min at 4 °C. Cells were washed and fixed with 4% paraformaldehyde for 30 min at room temperature and resuspended in PBS buffer. Acquisition was performed using a Gallios flow cytometer (Beckman Coulter).
Tfh cells were defined as CD45R-CD4+, CD44high, CXCR5+, PD-1+, and the following antibodies were used for surface staining: CD45R (B220)-APC-Vio770 (Miltenyi, 130-110-849), CD4-VioBright FITC (Miltenyi, 130-118-692), CD44-PerCP-Vio700 (Miltenyi, 130-128-625), CD185 (CXCR5)-APC (Miltenyi, 130-119-129), and CD279 (PD1)-PE (Miltenyi, 130-111-953). GC B cells were defined as CD19+GL7+CD95+, and the following antibodies were used for surface staining: CD19-APC (Miltenyi, 130-112-036), CD95 (FAS)-PE (Miltenyi, 130-112-036), and GL7-Alexa Fluor 488 eBioscience (Thermofisher, 53-5902-82).
Pseudotyped SARS‑CoV‑2 neutralization assay
The viral particles used in the neutralization assays were synthesized using an HIV-based second-generation lentiviral system, characterized by the utilization of three plasmids. The transfer plasmid (pwGFP) housed the viral particle’s genome (eGFP). The packaging plasmid (EMP0) was used to encode essential viral proteins, including Gag (structural proteins), Pol (polymerase), Rev (gene expression regulator), and Tat (transcriptional activator). The envelope plasmid encoded the full-length S protein of SARS-CoV-2. Two envelope plasmids were employed to pseudotype viral particles with the S protein variants of WT and XBB strain of SARS-CoV-2, respectively.
To produce the viral particles, 2.75 × 106 HEK293T cells were seeded in a T75 flask containing 13 mL of complete DMEM. The following day, the cells were transfected with 12.5 µg each of pwGFP and EMP0 plasmids, along with 2.5 µg of the different envelope plasmids, using FuGENE transfection reagent (Promega) according to the manufacturer’s instructions. After 48 h the viral supernatant was collected and filtered through an Acrodisc® Supor® 0.45 µm filter before being stored at −80 °C.
The capacity of each pseudovirus variant to infect ACE2-TMPRSS2 cells was determined empirically with an infectivity assay. The 2 × 105 cells/mL HEK293T-ACE2-TMPRSS2 cells were infected with an equal volume of serially diluted pseudovirus in DMEM. The cells were seeded in a black 96-well microplate (Thermo Scientific) and incubated at 37 °C and 5% CO2 for 48 h. Following incubation, cells were fixed with 4% paraformaldehyde in PBS for 1 h. The plates were washed twice with PBS and dried. The fluorescent spots were counted using a C.T.L. S6 Ultra-V analyzer with the FluoroSpot-X suite selected. The dilution of the pseudovirus chosen was the one that yielded 500–1000 infected cells per well (Supplementary Fig. 3).
For the neutralization assay, mouse sera were serially diluted in complete DMEM and added to black 96-well microplates, alongside the previously established volume of pseudovirus. The mixture was then incubated at 37 °C and 5% CO2 for 1 h to allow neutralization. After incubation, 2 × 104 HEK293T-ACE2-TMPRSS2 cells were added to each well, followed by 48 h of further incubation. The cells were then fixed as previously described, and the fluorescent spots were counted using a C.T.L. S6 Ultra-V analyzer with the FluoroSpot-X suite selected. The NT50 titer was defined as the reciprocal of the highest dilution at which a 50% or higher reduction in the number of spots compared to the untreated condition was achieved.
In vivo biodistribution
In vivo biodistribution assays were performed using Indium-111 radiolabeled LNPs. Lipid nanoparticle suspensions were incubated with 55 MBq Indium-111 oxine for 15 min at 37 °C. Labeled LNPs were afterwards purified using centrifugal concentrators (Vivaspin 500, 10.000 MWCO PES).
In vivo biodistribution assays were performed on BALB/c mice (n = 3 male, n = 3 female) on the U-SPECT6/E-class (MILabs) single photon emission computed tomography (SPECT) system. Radiolabeled LNPs were administered via intramuscular injection as described above. At 1 h, 3 h, 6 h, 24 h, 48 h, 72 h, 96 h, and 168 h post-administration, animals were anesthetized using isoflurane (2% in 100% O2), and SPECT images were acquired. Computer tomography (CT) scan was performed immediately after adjusting voltage and current to 55 kV and 0,33 mA, respectively. After image acquisition, these were reconstructed using indium-111 photopeaks with a window of 20% and a calibration factor to obtain the exact activity information (MBq/mL).
Images were analyzed using PMOD v3.2 software (PMOD Technologies, Switzerland). Signal values were corrected using the specific indium-111 radioactive decay correction factor and then transformed to standardized uptake value (SUV) units using the formula:
SUV = [tisular activity concentration (MBq/cm3)/dose (MBq)] × bodyweight (g).
Quantitative analysis was performed on organs/areas where signal could be correctly detected, setting a volume of interest (VOI) and quantifying over time, calculating the average signal in the VOI (Suvmean). Due to the difficulty in quantifying bone marrow signal, a visual analysis was performed to identify the timing of signal appearance in various bones.
At 168 h, the animals were euthanized, and the following organs/tissues were dissected: lungs, spleen, liver, kidneys, bone, muscle at the injection site, contralateral muscle, presacral lymph nodes, inguinal lymph nodes, and brain. Tissue activity was measured using a gamma counter (Hidex) to determine counts per minute (cpm) for each tissue. To quantitatively assess the amount of radioactivity in each sample, a ratio was calculated using the brain (background) as the reference tissue, and the values were normalized to tissue weight (g).
Serum biochemical analysis
Serum samples were collected as specified above. Biochemical analysis of AST and ALT levels was performed using the Cobas c-311 automatic analyzer (Roche Diagnostics).
SARS-CoV-2 challenge
Mice were anaesthetized with isoflurane (5% for induction and 2.5% for maintenance) and infected intranasally with the corresponding amount of virus to achieve the indicated doses, which was diluted in PBS in a total volume of 40 µL. After virus challenge, mice were periodically weighed and assessed using a clinical scoring system evaluating various parameters, including mouse appearance, level of consciousness, activity, response to stimuli, eye appearance, and frequency and quality of respiration61. Humane endpoints were established on a weight loss threshold of 25% or greater, alongside clinical scoring criteria.
Viral load determination by TCID50 assay
Lungs were harvested, weighed, and homogenized in 1 mL of DMEM using a GentleMACS Dissociator (Miltenyi). Homogenates were centrifuged at 450×g for 5 min, and the supernatant was taken for titration.
Virus titration was determined by TCID50 in Vero E6 cells. Vero E6 cells were seeded in 96-well plates at a density of 104 cells/well and cultured overnight at 37 °C and 5% CO2. Lung homogenates serial 1-log dilutions were prepared in complete DMEM with only 2% FBS and added to the cultured cells. 72 h after cell infection, plates were evaluated for cell death, and TCID50 was calculated using Ramakrishan's newly proposed method formula62 and normalized to the weight (g) of the lung and the amount of buffer (mL) used for lung homogenization.
Statistical analysis
In experimental studies, GraphPad Prism 10 software was used for representation and statistical analyses.
We used ordinary one-way or two-way ANOVA with Tukey’s multiple comparison post-test to compare experimental groups. Statistical significances are denoted in the Figures by asterisks, as follows: *p-value < 0.05, **p-value < 0.01, ***p-value < 0.001, or ****p-value < 0.0001. The absence of asterisks indicates non-statistical significance with a p-value > 0.05. Outliers were identified and removed using Grubbs’ test, applying a significance threshold of p < 0.05.
Data availability
The data supporting the conclusions of this research are accessible within the article and its supplementary files. The source data is included in this paper. The design of the SARS-CoV-2 Spike sequence, including all modifications relative to the publicly available Comirnaty sequence, is described in detail in the "DNA template design" of the Methods section. Additional information is available from the corresponding author upon request.
Change history
08 October 2025
In the Acknowledgements section, the funding source and grant number ‘Ministerio de Ciencia e Innovación’(Spain) through project STABVAC4COV’ should have read ‘CDTI and financed by the European Union – NextGenerationEU through project STABVAC4COV – MIP-20211002’. The original article has been corrected.
References
Tregoning, J. S., Flight, K. E., Higham, S. L., Wang, Z. & Pierce, B. F. Progress of the COVID-19 vaccine effort: viruses, vaccines and variants versus efficacy, effectiveness and escape. Nat. Rev. Immunol. 21, 626–636 (2021).
Verbeke, R., Lentacker, I., De Smedt, S. C. & Dewitte, H. The dawn of mRNA vaccines: the COVID-19 case. J. Control. Release J. Control. Release Soc. 333, 511–520 (2021).
Pardi, N., Hogan, M. J., Porter, F. W. & Weissman, D. mRNA vaccines—a new era in vaccinology. Nat. Rev. Drug Discov. 17, 261–279 (2018).
Polack, F. P. et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 383, 2603–2615 (2020).
Baden, L. R. et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 384, 403–416 (2021).
Pawlowski, C. et al. FDA-authorized mRNA COVID-19 vaccines are effective per real-world evidence synthesized across a multi-state health system. Med 2, 979–992.e8 (2021).
Scheaffer, S. M. et al. Bivalent SARS-CoV-2 mRNA vaccines increase breadth of neutralization and protect against the BA.5 Omicron variant in mice. Nat. Med. 29, 247–257 (2023).
Ye, Q. et al. Rational development of a combined mRNA vaccine against COVID-19 and influenza. NPJ Vaccines 7, 84 (2022).
Cheng, L., Chan, W. K., Zhu, L., Chao, M. H. & Wang, Y. Confronting inequalities and bridging the divide: a retrospective study assessment of country-level COVID-19 vaccine equality with a Cox regression model. Vaccines 12, 552 (2024).
Acharya, K. P., Ghimire, T. R. & Subramanya, S. H. Access to and equitable distribution of COVID-19 vaccine in low-income countries. npj Vaccines 6, 54 (2021).
Duroseau, B., Kipshidze, N. & Limaye, R. J. The impact of delayed access to COVID-19 vaccines in low- and lower-middle-income countries. Front. Public Heal. 10, 1087138 (2023).
Chi, W.-Y. et al. COVID-19 vaccine update: vaccine effectiveness, SARS-CoV-2 variants, boosters, adverse effects, and immune correlates of protection. J. Biomed. Sci. 29, 82 (2022).
Echaide, M. et al. mRNA vaccines against SARS-CoV-2: advantages and caveats. Int. J. Mol. Sci. 24, 5944 (2023).
Kim, S. C. et al. Modifications of mRNA vaccine structural elements for improving mRNA stability and translation efficiency. Mol. Cell. Toxicol. 18, 1–8 (2022).
Luna-Cerralbo, D. et al. A statistical-physics approach for codon usage optimisation. Comput. Struct. Biotechnol. J. 23, 3050–3064 (2024).
Broset Blasco, E. & Martínez Oliván, J. E. Artificial polynucleotides for expressing proteins, WO2024110381A1. (2022).
Ramachandran, S., Satapathy, S. R. & Dutta, T. Delivery strategies for mRNA vaccines. Pharm. Med. 36, 11–20 (2022).
Hou, X., Zaks, T., Langer, R. & Dong, Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 6, 1078–1094 (2021).
Hald Albertsen, C. et al. The role of lipid components in lipid nanoparticles for vaccines and gene therapy. Adv. Drug Deliv. Rev. 188, 114416 (2022).
Hassett, K. J. et al. Optimization of lipid nanoparticles for intramuscular administration of mRNA vaccines. Mol. Ther. Nucleic Acids 15, 1–11 (2019).
Zhang, L. et al. Effect of mRNA-LNP components of two globally-marketed COVID-19 vaccines on efficacy and stability. NPJ Vaccines 8, 156 (2023).
Gimenez-Warren, J. Ionizable lipids and lipid nanoparticles containing thereof, WO2024110381A1. (2022).
Peña, Á et al. Multicomponent thiolactone-based ionizable lipid screening platform for efficient and tunable mRNA delivery to the lungs. Commun. Chem. 8, 116 (2025).
Crommelin, D. J. A., Anchordoquy, T. J., Volkin, D. B., Jiskoot, W. & Mastrobattista, E. Addressing the cold reality of mRNA vaccine stability. J. Pharm. Sci. 110, 997–1001 (2021).
Schoenmaker, L. et al. mRNA-lipid nanoparticle COVID-19 vaccines: structure and stability. Int. J. Pharm. 601, 120586 (2021).
Oude Blenke, E. et al. The storage and in-use stability of mRNA vaccines and therapeutics: not a cold case. J. Pharm. Sci. 112, 386–403 (2023).
Albert, M.-C. et al. Identification of FasL as a crucial host factor driving COVID-19 pathology and lethality. Cell Death Differ. https://doi.org/10.1038/s41418-024-01278-6 (2024).
Lederer, K. et al. SARS-CoV-2 mRNA vaccines foster potent antigen-specific germinal center responses associated with neutralizing antibody generation. Immunity 53, 1281–1295.e5 (2020).
Verbeke, R., Hogan, M. J., Loré, K. & Pardi, N. Innate immune mechanisms of mRNA vaccines. Immunity 55, 1993–2005 (2022).
Sharma, P., Hoorn, D., Aitha, A., Breier, D. & Peer, D. The immunostimulatory nature of mRNA lipid nanoparticles. Adv. Drug Deliv. Rev. 205, 115175 (2024).
Dubuc, I. et al. Cytokines and lipid mediators of inflammation in lungs of SARS-CoV-2 infected mice. Front. Immunol. 13, 893792 (2022).
Winkler, E. S. et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 21, 1327–1335 (2020).
Tamura, T. et al. Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants. Nat. Commun. 14, 2800 (2023).
Tamura, T. et al. Virological characteristics of the SARS-CoV-2 Omicron XBB.1.5 variant. Nat. Commun. 15, 1176 (2024).
Yue, C. et al. ACE2 binding and antibody evasion in enhanced transmissibility of XBB.1.5. Lancet Infect. Dis. 23, 278–280 (2023).
Lewnard, J. A. et al. Increased vaccine sensitivity of an emerging SARS-CoV-2 variant. Nat. Commun. 14, 3854 (2023).
Azevedo, P. O. et al. Differential requirement of neutralizing antibodies and T cells on protective immunity to SARS-CoV-2 variants of concern. npj Vaccines 8, 15 (2023).
Powers, J. M. et al. Divergent pathogenetic outcomes in BALB/c mice following Omicron subvariant infection. Virus Res. 341, 199319 (2024).
Halfmann, P. J. et al. Characterization of Omicron BA.4.6, XBB, and BQ.1.1 subvariants in hamsters. Commun. Biol. 7, 331 (2024).
Uddin, M. N. & Roni, M. A. Challenges of storage and stability of mRNA-based COVID-19 vaccines. Vaccines 9, 1033 (2021).
Kis, Z. Stability modelling of mRNA vaccine quality based on temperature monitoring throughout the distribution chain. Pharmaceutics 14, 430 (2022).
Cheng, F. et al. Research advances on the stability of mRNA vaccines. Viruses 15, 668 (2023).
Alejo, T. et al. Comprehensive optimization of a freeze-drying process achieving enhanced long-term stability and in vivo performance of lyophilized mRNA-LNPs. Int. J. Mol. Sci. 25, 10603 (2024).
Kafetzis, K. N. et al. The effect of cryoprotectants and storage conditions on the transfection efficiency, stability, and safety of lipid-based nanoparticles for mRNA and DNA delivery. Adv. Healthc. Mater. 12, e2203022 (2023).
Muramatsu, H. et al. Lyophilization provides long-term stability for a lipid nanoparticle-formulated, nucleoside-modified mRNA vaccine. Mol. Ther. 30, 1941–1951 (2022).
Hogan, M. J. & Pardi, N. mRNA vaccines in the COVID-19 pandemic and beyond. Annu. Rev. Med. 73, 17–39 (2022).
Akinc, A. et al. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat. Nanotechnol. 14, 1084–1087 (2019).
Sabnis, S. et al. A Novel amino lipid series for mRNA delivery: improved endosomal escape and sustained pharmacology and safety in non-human primates. Mol. Ther. 26, 1509–1519 (2018).
Ansell, S. M. US 10, 166, 298 B2 (2019).
Ai, L. et al. Lyophilized mRNA-lipid nanoparticle vaccines with long-term stability and high antigenicity against SARS-CoV-2. Cell Discov. 9, 9 (2023).
Wang, B. et al. Lyophilized monkeypox mRNA lipid nanoparticle vaccines with long-term stability and robust immune responses in mice. Hum. Vaccin. Immunother. 21, 2477384 (2025).
Ruppl, A. et al. Formulation screening of lyophilized mRNA-lipid nanoparticles. Int. J. Pharm. 671, 125272 (2025).
Meulewaeter, S., Nuytten, G., Cheng, M. H. Y. & Smedt, S. C. De. Continuous freeze-drying of messenger RNA lipid nanoparticles enables storage at higher temperatures. J. Control. Release 357, 149–160 (2023).
Kim, B. et al. Optimization of storage conditions for lipid nanoparticle-formulated self-replicating RNA vaccines. J. Control. Release 353, 241–253 (2023).
Li, M. et al. Lyophilization process optimization and molecular dynamics simulation of mRNA-LNPs for SARS-CoV-2 vaccine. NPJ Vaccines 8, 153 (2023).
Suzuki, Y. et al. Design and lyophilization of lipid nanoparticles for mRNA vaccine and its robust immune response in mice and nonhuman primates. Mol. Ther. Nucleic Acids 30, 226–240 (2022).
Hermosilla, J. et al. Analysing the in-use stability of mRNA-LNP COVID-19 vaccines ComirnatyTM (Pfizer) and SpikevaxTM (Moderna): a comparative study of the particulate. Vaccines 11, 1635 (2023).
WHO. Messenger RNA Encoding the Full-Length SARS-CoV-2 Spike Glycoprotein (WHO, 2021) https://berthub.eu/articles/11889.doc
GISAID. https://gisaid.org/wiv04/.
Martínez, J., Lampaya, V., Larraga, A., Magallón, H. & Casabona, D. Purification of linearized template plasmid DNA decreases double-stranded RNA formation during IVT reaction. Front. Mol. Biosci. 10, 1248511 (2023).
Shrum, B. et al. A robust scoring system to evaluate sepsis severity in an animal model. BMC Res. Notes 7, 233 (2014).
Ramakrishnan, M. A. Determination of 50% endpoint titer using a simple formula. World J. Virol. 5, 85–86 (2016).
Acknowledgements
We would like to extend our sincere gratitude to all the members of Certest Biotec, especially the Certest Pharma group, whose valuable insights and expertise significantly contributed to the success of this research. Authors would like to acknowledge the use of “Servicios Científico Técnicos” at CIBA (IACS-University of Zaragoza), especially the collaboration of the animal facilities and flow cytometry services, as well as the use of “Servicio de análisis Bioquímicos” at CIMA (University of Navarra). This study was supported by CDTI and financed by the European Union – NextGenerationEU through project STABVAC4COV – MIP-20211002 (Desarrollo de vacunas termoestables basadas en mRNA frente a la variante delta del SARS-CoV-2). Work in J.P. lab is funded by PID2020-113963RBI00 from AEI (Agencia Estatal de Investigación), Aragon Government (B29-20R), Postdoctoral Juan de la Cierva Contract (M.A.); the National Institute Carlos III (COV20-00308 and CIBERINFEC, CB21/13/ 00087).
Author information
Authors and Affiliations
Contributions
Conceptualization: E.M., E.B., E.P., and J.M. mRNA sequence design: E.B. mRNA synthesis: V.L. and D.C. Lipid molecular design, synthesis, and characterization: J.H., J.G.-W. LNP formulation and characterization: T.A., D.d.M. Biological and immune assays: C.M., A.S., S.A., A.L., B.A., C.G., A.C. Biodistribution study: I.P. and G.Q. Establishment of COVID-19 Infection models and in vivo vaccine-induced protection experiments: J.P., I.U., N.P., M.A., C.M., A.S., S.A., B.M., and J.B. Supervision: E.M., E.P., and J.M. Writing original draft and figure preparation: E.M., C.M., A.S., and A.L. Manuscript review: all authors.
Corresponding authors
Ethics declarations
Competing interests
E.M., E.B., C.M., A.S., S.A., T.A., A.L., B.A., D.d.M., J.H., J.G.-W., V.L., D.C., J.M., and E.P. are employees at the Certest Pharma Department, Certest Biotec S.L. E.B., J.H., D.d.M., J.M., and J.G.-W. are inventors on patents related to this publication. All other authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Mata, E., Broset, E., Matute, C. et al. Preserved efficacy of lyophilized SARS-CoV-2 mRNA vaccine incorporating novel ionizable lipids after one year at 25 °C. npj Vaccines 10, 135 (2025). https://doi.org/10.1038/s41541-025-01201-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41541-025-01201-1