Abstract
Lung-resident memory T (TRM) cells respond rapidly and effectively to respiratory pathogen invasion, suppressing pathogen proliferation. Previously, we identified a defined TLR3 agonist called Nexavant (NVT) and developed a vaccine platform that utilizes it to induce lung TRM. In this study, we aimed to determine whether the protective effect of TRM cells is observed in tuberculosis (TB), a chronic bacterial respiratory disease. We synthesized a peptide vaccine by elongating the CD4+ T cell epitopes from Mycobacterium tuberculosis antigens ESAT-6, CFP-10, and HspX, adjuvanted it with NVT and administered the vaccine intranasally or intramuscularly to mice. We demonstrated that intranasal administration of an NVT-formulated peptide vaccine induced the generation of CD4+ TRM cells in the lungs, and that our vaccine platform, containing a limited number of CD4 epitopes, provided protective efficacy comparable to that of the BCG vaccine, which contains multiple T cell epitopes. Furthermore, the peptides used in the vaccine were reactive in 23 out of 24 (95.8%) human PBMCs, indicating that they contain promiscuous epitopes. Our results suggest a straightforward approach to controlling pulmonary TB more effectively through the induction of lung CD4+ TRM cells, even when using the same target antigen. Additionally, this study supports a theoretical basis for developing an inhalable TB vaccine using synthetic peptides.
Similar content being viewed by others
Introduction
Tuberculosis (TB) is a chronic infectious disease caused by Mycobacterium tuberculosis (Mtb) and remains one of the deadliest infectious diseases in human history. Mtb infects and proliferates within the host’s macrophages by suppressing apoptosis, a process crucial for the host immune response, thereby enabling its intracellular survival1. The only existing TB vaccine, BCG (Bacille Calmette-Guerin), was developed nearly 100 years ago and its effectiveness in preventing TB ranges from 0 to 80%, with significant individual variation2. BCG is reported to be almost ineffective in preventing pulmonary TB in adults and has limited use in immunocompromised individuals, such as those living with HIV3,4,5. Given these limitations, the need for a more effective TB vaccine is increasingly emphasized, especially with the long duration of treatment and the rise of multidrug-resistant and extensively drug-resistant TB6.
In the development of a preventive vaccine, it is crucial to identify the immunological factors that contribute to disease protection7. Th1 cells, a subset of T helper cells, play a vital role in defense against TB8,9,10. They activate macrophages by secreting IFN-γ and generating reactive oxygen species or nitric oxide to eliminate Mtb. Many previous studies have reported that BCG also induces a protective immune response primarily through Th1-type CD4⁺ T cells11,12. As a result, extensive research has focused on identifying CD4 T cell epitopes that can trigger a Th1 response, contributing to TB protection, and many such epitopes have already been discovered.
A peptide-based vaccine platform that utilizes a minimal number of T cell epitopes represents a potentially safer and controllable approach due to its ability to provide precise control over the immune response and its relatively simple production through synthesis. However, this platform faces challenges, including weak immunogenicity and MHC restriction when used as a preventive vaccine13,14. While adding an adjuvant can address low immunogenicity, overcoming MHC restriction remains difficult, particularly when trying to identify promiscuous epitopes through screening15. Generally, MHC class II, which binds CD4 epitopes, is considered more promiscuous than MHC class I, which binds CD8 epitopes14,16. Additionally, CD4 epitopes from Mtb are extensively used not only for vaccines but also for diagnosing latent TB, leading to numerous studies in this area. As a result, we were able to identify several sequences of promiscuous CD4 epitopes located on target proteins relevant to vaccine development through a literature search17,18,19,20,21.
Poly(I:C) is a TLR3 agonist that induces a strong T cell immune response; however, its clinical application has been limited due to challenges such as structural heterogeneity leading to inconsistent responses, instability, toxicity, and difficulties in pharmacokinetic analysis22,23. To overcome these limitations, Nexavant (NVT), a dsRNA-based adjuvant, was developed for clinical applications. Structurally, NVT is a well-defined 424 bp dsRNA molecule with five-nucleotide 3′ overhangs, synthesized via PCR-coupled bidirectional in vitro transcription and purified using RNase T1. This defined structure enables consistent manufacturing, high purity, and precise pharmacokinetic tracking via RT-qPCR—features that are not achievable with the structurally heterogeneous poly(I:C)23. Functionally, both NVT and poly(I:C) activate TLR3. However, mechanistic analysis using HEK-293 dual reporter cells revealed that NVT activates TLR3 and RIG-I, but not MDA5, even when transfected with lipid carriers24. This selective engagement of TLR3 and RIG-I, but not MDA5, may indicate that NVT elicits a more targeted immunostimulatory response compared to poly(I:C), potentially contributing to its improved safety profile. Supporting this, our previous work demonstrated that NVT exhibits improved stability and safety compared to poly(I:C), while inducing humoral and cellular responses as potent as those elicited by poly(I:C)23. Furthermore, when incorporated into a commercial influenza vaccine and delivered to the lungs, NVT induced the formation of lung CD4+ TRM, providing cross-protection against heterologous viruses25. Based on these findings, we hypothesized that lung CD4+ TRM could serve as a defense mechanism against other respiratory pathogens, such as Mtb, and may provide a foundation for the development of a TB vaccine.
In this study, we formulated five well-characterized CD4⁺ T cell epitopes derived from three Mtb proteins (ESAT-6, CFP-10, and HspX) with the adjuvant NVT. Intranasal administration of this vaccine induced robust antigen-specific CD4⁺ T cell responses in both the lung and spleen, and promoted the formation of lung CD4+ TRM in mice. Notably, this vaccine conferred protective efficacy comparable to BCG against Mtb infection. Furthermore, the peptide antigens elicited cellular immune responses in human peripheral blood mononuclear cells (PBMCs), suggesting translational relevance. Collectively, these findings highlight the potential of our NVT-based peptide vaccine platform as a promising candidate for next-generation TB vaccines.
Results
Selection of MHC class II epitopes on ESAT-6, CFP-10, and HspX
To confirm the protective function of CD4+ TRM against Mtb in the lungs, it is crucial to select appropriate epitopes that can induce a CD4+ T cell response and formulate them with the adjuvant system. Many programs are available to predict epitopes suitable for human MHC, and we used the Immune Epitope Database (IEDB; www.iedb.org) to identify potential epitopes. We focused on ESAT-6, CFP-10, and HspX, which are TB antigens commonly used in vaccine research and have been extensively studied for their epitopes. The host species was set to human. For selection, we chose epitopes based on extensive data showing positive responses to IFN-γ in human PBMCs, with particular attention to promiscuously restricted epitopes that could cover multiple HLAs. The sequences were extended as much as possible to include as many epitopes as feasible, and when necessary, glutamic acid (E) was added at the end of the sequence to enhance solubility. As a result, two epitopes each from ESAT-6 and CFP-10, along with one from HspX, were selected (Table 1). Given that the three source proteins from which the epitopes were selected (ESAT-6, CFP-10, and HspX) are conserved26,27, we further evaluated the sequence identity of the five selected epitope peptides against the corresponding proteins in the Mtb H37Rv challenge strain using CLC Sequence Viewer. All five peptides were found to be 100% identical to the H37Rv sequences, confirming their complete conservation in the challenge strain.
Verification of five peptide epitopes through a cellular immune response test on human PBMCs
Although we prioritized the selection of promiscuous epitope that can bind to multiple HLAs during the peptide selection process, we conducted an experiment to verify the peptide epitopes used in this study with human PBMC samples due to a sequence change compared to the references. Patients with active or latent TB typically have a memory response to the Mtb epitopes, resulting in interferon gamma production when PBMCs are stimulated with these epitopes. This principle underlies the IGRA test, which is widely used to diagnose latent TB. Protein-spanning mixtures of overlapping peptides from the ESAT-6 and CFP-10 proteins (TB Pepmix) were used as the control group. PBMCs from a total of 24 individuals were stimulated with either the TB Pepmix or the five peptides (Pep 5’) identified in this study. This group included 8 patients with active TB (AMC01, AMC04, AMC05, AMC06, AMC09, AMC11, AMC16, and AMC18), 11 individuals with latent TB infection (AMC02, AMC03, AMC08, AMC12, AMC13, AMC14, AMC15, AMC17, AMC19, AMC21, and AMC22), and 5 healthy controls (AMC07, AMC10, AMC20, AMC23, and AMC24). Immune reactivity was compared among these groups. Reactivity to Pep 5’ was observed in all eight patients with active TB and in 10 of the 11 individuals with latent TB infection. In the case of the AMC19 patient, where no reaction was detected, there was also a minimal response to the TB Pepmix control. Therefore, it is difficult to conclude that the peptide was ineffective in this case (Fig. 1a, b). As expected, healthy individuals without a memory response to TB showed little reactivity, although reactivity was observed in two healthy control subjects (AMC23 and AMC24). However, the value in AMC23 was so low that it is difficult to determine whether a reaction occurred. For AMC24, the result is likely a false negative of conventional diagnostic of latent TB, as a reaction was also observed with the TB Pepmix group. Interestingly, the overall responsiveness to Pep 5′ was higher than that observed when the samples were stimulated with TB Pepmix (Fig. 1c).
Human PBMCs were isolated from blood samples obtained from individuals with active TB (a), latent TB infection (b), or healthy donors (c), and were stimulated in vitro with TB PepMix (ESAT-6 and CFP-10, each at 5 μg/ml) or with Pep 5′ (a mixture of five peptides, each at 5 μg/ml) at 37 °C for 72 h. IFN-γ levels in the culture supernatants were measured using a human IFN-γ ELISA kit. d Antigen-specific T cell expansion was performed by stimulating healthy human PBMC with TB PepMix or Pep 5’ as described in “Materials and Methods”. On day 12, IFN-γ levels in the supernatants were quantified using a human IFN-γ ELISA kit. a–d Responses were considered positive when IFN-γ levels in the antigen-stimulated group were at least twice those in the unstimulated control. Values are presented as IFN-γ levels after subtraction of the unstimulated control for each individual.
Even in individuals without a memory response to TB, a T cell response specific to the peptide should be detectable if the correct HLA is present, provided the T cells are expanded with the peptide in vitro. To confirm this, an in vitro T cell expansion test was performed on five healthy individuals. As a result, peptide-specific T cell responses were observed in all five healthy PBMC samples (Fig. 1d). Notably, PBMCs from healthy individuals did not exhibit elevated IFN-γ production in the initial stimulation experiment (Fig. 1c) but did show more than a two-fold increase in IFN-γ secretion after 9 days of in vitro expansion, suggesting successful expansion of low-frequency, antigen-specific T cell populations. These findings support the notion that the five identified peptides are promiscuous T cell epitopes capable of eliciting responses from multiple HLAs, providing a solid foundation for their potential use in a human TB vaccine.
Two of the selected epitopes have an MHC sequence suitable for mice and demonstrate better immunogenicity compared to protein vaccine
Since the peptide epitopes were selected based on human MHC, it is essential to conduct a challenge experiment to confirm whether the epitope-specific T cell response contributes to protection against Mtb. To achieve this, it is necessary to verify whether the selected peptide epitopes can be presented on the MHC of mice and induce T cell responses.
To investigate whether these epitopes could stimulate T cell responses in mice, we immunized mice intramuscularly twice with NVT-adjuvanted recombinant TB proteins (ESAT-6, CFP10, and HspX). We then stimulated spleen cells from the immunized mice with each in individual epitope peptide (ESAT61, ESAT62, CFP101, CFP102, and Rv2031c) or a mixture of all five peptides (Pep 5′) to analyze T cell responses (Fig. 2a and Supplementary Fig. 2). We observed that ESAT61 and ESAT62 induced significantly higher levels of CD4+ T cell responses compared to unstimulated controls whereas the Rv2031c failed to induce T cell responses. CFP101 and CFP102 also elicited CD4⁺ T cell responses; however, the increase was not statistically significant compared to unstimulated controls. We also found that stimulation with Pep 5’ induced a higher CD4+ T cell response than stimulation with the single peptide alone (Fig. 2b, c). This response appears to result primarily from specific responses to ESAT61 and ESAT62, suggesting that the inclusion of multiple epitopes may be advantageous for inducing robust CD4+ T cell responses. Meanwhile, these five peptides did not induce CD8+ T cell responses in C57BL/6 mouse model (Fig. 2d, e).
a Experimental design for T cell response analysis. C57BL/6 mice (n = 4 per group) were immunized intramuscularly (i.m.) twice at 3-week intervals with either recombinant TB proteins or Pep 5′ (a peptide mixture consisting of ESAT61, ESAT62, CFP101, CFP102, and Rv2031c), both formulated with NVT. Spleen were harvested 4 weeks after the final immunization for T cell response analysis. b–e IFN-γ⁺CD4⁺ and IFN-γ⁺CD8⁺ T cell responses were analyzed by flow cytometry after in vitro stimulation with ESAT61, ESAT62, CFP101, CFP102, Rv2031c, or Pep 5′ of splenocytes from mice immunized with TB proteins. Representative dot plots (b) and percentages (c) of IFN-γ⁺CD4⁺ T cells. Representative dot plots (d) and percentages (e) of IFN-γ⁺CD8⁺ T cells. Statistical analyses were performed using one-way ANOVA with Tukey’s multiple comparisons test. f, g IFN-γ⁺CD4⁺ T cell responses were analyzed by flow cytometry after in vitro stimulation with Pep 5′ of splenocytes isolated from mice immunized with either recombinant TB proteins or Pep 5′. Representative dot plots (f) and percentages (g) of IFN-γ⁺CD8⁺ T cells. The two sided Mann–Whitney U test was used for statistical analysis of differences between two groups. c, e, g The box-and-whisker plots display the median (center line), 25th and 75th percentiles (box), and the minimum and maximum values (whiskers). Data are representative of two independent experiments. Unstim., unstimulated; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant.
Next, we examined the advantages of using peptides as vaccine antigens in terms of their ability to induce CD4+ T cell responses compared to protein antigens. To this end, we immunized mice intramuscularly twice with NVT-adjuvanted recombinant TB proteins or Pep 5′ and then stimulated spleen cells from the immunized mice with Pep 5′ to analyze T cell responses. We found that the target epitope-specific CD4+ T cell response was stronger when peptides containing the target epitopes were used, compared to when proteins containing the corresponding epitopes were used as a vaccine antigen. (Fig. 2f, g). This may be a natural consequence of the greater number of administered epitopes, but it can also be considered an advantage of peptides. In summary, we confirmed that two out of five peptides selected for human MHC were reactive in C57BL/6 mice, and based on these results, a TB challenge test was performed using this mouse strain.
Lung CD4+ TRM formed by an intranasal peptide vaccine provides robust protective efficacy against TB in the lungs
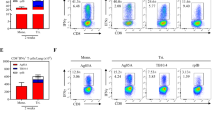
Previously, we showed that intranasal administration of vaccine antigens formulated with NVT confers a robust protective effect against infection, accompanied by pulmonary TRM formation25. To determine whether this could also be applied to peptide-based TB vaccines, we immunized mice intramuscularly or intranasally twice with NVT-adjuvanted Pep 5′ (Pep 5′ + NVT) and analyzed Pep 5′-specific T cell responses (Fig. 3a). We first monitored weight changes and clinical signs following immunization to evaluate the potential toxicity of our vaccine and the effect of the administration route. The results showed that, compared to the unvaccinated group, which exhibited no weight loss, both vaccine groups experienced a transient decrease in body weight following immunization, with the intranasal group showing a milder reduction than the intramuscular group. Notably, the weight loss observed after the second immunization was milder than that observed after the first dose. However, all animals returned to their baseline weight within a few days (Fig. 3b). Clinical scores, including activity level and respiratory condition, showed no significant differences between either vaccine group and the unvaccinated group. We observed that intranasal administration induced CD4+ T cell responses in both the lung and spleen to a similar extent as intramuscular injection (Fig. 3c-f). To evaluate the impact of vaccination route on lung TRM formation, we defined pulmonary CD4⁺ TRM as CD44⁺CD62L⁻CD69⁺CD4⁺ cells as shown in Supplementary Fig. 3. Despite comparable overall CD4⁺ T cell responses in the lungs between the two administration groups, that only intranasal administration induced the formation of lung CD4+ TRM (Fig. 3g-i). In contrast, CD8⁺ TRM were not detected in any group, consistent with the absence of detectable antigen-specific CD8⁺ T cell responses (Supplementary Fig. 4). To better delineate which CD4⁺ T cell subsets contribute to TRM formation and IFN-γ⁺ CD4⁺ T cell responses, we subdivided CD4⁺ T cells into naïve (TN; CD44⁻CD62L⁺), central memory (TCM; CD44⁺CD62L⁺), effector (TE; CD44⁻CD62L⁻), and effector memory (TEM; CD44⁺CD62L⁻) subsets for comparison. We found that the absolute number of TEM cells was significantly increased in the intranasally vaccinated group compared to the intramuscular group. These results suggest that the CD4⁺ T cell responses may have originated from multiple circulating T cell subsets. Moreover, given that TEM cells can act as precursors for TRM differentiation, their preferential expansion in the intranasal group suggests a contribution to the establishment of pulmonary tissue-resident memory and the enhancement of local immune protection (Fig. 3j).
a Experimental design. C57BL/6 mice (n = 14 per group) were immunized intramuscularly (i.m.) or intranasally (i.n.) twice at 3-week intervals with NVT-formulated Pep 5′. Four weeks after the final immunization, immune response analysis (n = 5 per group) or aerosol challenge with H37Rv (n = 9 per group) were performed. b Body weight was monitored daily for 5 days after both the first and second immunizations. c–f IFN-γ+CD4+ T cell responses to Pep 5’ in the lungs and spleen were analyzed by flow cytometry. Representative dot plots (c) and percentages (d) of IFN-γ⁺CD4⁺ T cells in the lung. Representative dot plots (e) and percentages (f) of IFN-γ⁺CD4⁺ T cells in the spleen. CD4+ TRM cells (CD4+CD44+CD62L-CD69+) in the lungs were analyzed by flow cytometry. Representative dot plots (g), percentages (h), and absolute numbers (i) of CD4+ RM cells in the lung. j Lung CD4⁺ T cell subsets based on CD44 and CD62L expression. CD4⁺ T cells were classified into naïve (TN; CD44⁻CD62L⁺), central memory (TCM; CD44⁺CD62L⁺), effector (TE; CD44⁻CD62L⁻), and effector memory (TEM; CD44⁺CD62L⁻) subsets. k–m The immunized mice (n = 9 per group) were infected with H37Rv via aerosol exposure. At 4 weeks post-infection, mice were sacrificed for bacterial counts (n = 5 per group) and lung histopathology (n = 4 per group). Bacterial loads in the lungs (k) and spleen (l). H&E staining (m) and inflamed area (n) in lung tissues. The box-and-whisker plots display the median (center line), 25th and 75th percentiles (box), and the minimum and maximum values (whiskers). Statistical analyses were performed using one-way ANOVA with Tukey’s multiple comparisons test. Data are representative of two independent experiments. Unstim., unstimulated; *P < 0.05; **P < 0.01; ****P < 0.0001; ns, not significant.
Next, to assess whether this difference affect protection against TB, we challenged immunized mice with Mtb strain H37Rv (ATCC 25618) and measured bacterial loads in the lungs and spleen at 4 weeks post-infection (Fig. 3a). In the lungs, no significant difference was observed between the intramuscular and intranasal groups; however, only the intranasal group showed a significant reduction in bacterial burden compared to the mock group (Fig. 3k). In the spleen, both the intramuscular and intranasal administration groups exhibited significant protective efficacy compared to the control group (Fig. 3l). Consistent with the bacterial load results, lung inflammation was significantly reduced only in the intranasal group compared to the mock group, although no significant difference was observed between the intranasal and intramuscular groups (Fig. 3m, n). These results suggest that intranasal administration of the same vaccine component may provide at least as much protection as intramuscular injection, and potentially even greater protection.
Intranasal peptide vaccine provides a protective effect comparable to that of BCG against the highly pathogenic Mtb strain HN878
After confirming the efficacy of the intranasal vaccine, we aimed to compare its protective efficacy to that of BCG, the only existing TB vaccine. Specifically, we sought to assess its effectiveness against the highly pathogenic strain HN878. This experiment was feasible because the target proteins we used, ESAT6 and CFP10, are highly conserved among mycobacteria28. To this end, we immunized mice intranasally twice with NVT, Pep 5′, or Pep 5′ + NVT at 3-week intervals on days 0 (week 0) and 21 (week 3). We used the BCG vaccine as a comparison group in the preventive model. Given the challenges associated with the approval of intranasal administration of BCG, a live vaccine, due to safety concerns29,30, and previous studies demonstrating that subcutaneous administration of BCG effectively protects against TB in mice31, we immunized mice subcutaneously with a single dose of BCG vaccine on day 0. On day 49 (week 7), all mice were infected with the highly pathogenic Mtb strain HN878 via aerosol exposure, and bacterial loads in the lungs and spleen, as well as lung inflammation, were assessed 4 weeks post-infection (Fig. 4a). We observed that the Pep 5′ + NVT group had significantly lower bacterial loads in the lungs than the NVT, Pep 5′, or unvaccinated group, and these levels were comparable to those observed in the BCG group (Fig. 4b). The Pep 5′ + NVT group also tended to have lower bacterial loads in the spleen than the unvaccinated group, while the BCG group had the lowest bacterial loads in the spleen (Fig. 4c). Both the Pep 5′ + NVT and BCG groups exhibited significantly reduced lung inflammation compared to the NVT or unvaccinated group. (Fig. 4d, e). This result suggests that if lung TRM is well-formed, two CD4 epitopes alone can provide efficacy similar to that of BCG vaccine, which consists of many T cell epitopes.
a Experimental design. C57BL/6 mice (n = 10 per group) were immunized subcutaneously (s.c.) once (at week 0) with BCG, or intranasally (i.n.) twice (at weeks 0 and 3) with NVT, Pep 5′, or NVT-formulated Pep 5′ (Pep 5′ + NVT). Four weeks after the final immunization, mice were infected with HN878 via aerosol exposure. At 4 weeks post-infection, mice were sacrificed for bacterial counts (n = 5 per group) and lung histopathology (n = 5 per group). Bacterial loads in the lungs (b) and spleen (c). H&E staining (d) and inflamed area (e) in lung tissues. The box-and-whisker plots display the median (center line), 25th and 75th percentiles (box), and the minimum and maximum values (whiskers). Statistical analyses were performed using one-way ANOVA with Tukey’s multiple comparisons test. This experiment was performed once. *P < 0.05; ***P < 0.001; ****P < 0.0001; ns not significant.
Evaluation of the synergistic effect of TB protection between lung CD4+ TRM induced by intranasal peptide vaccine and systemic immunity generated by BCG
The commercial BCG vaccine does not form TRM in the lungs when administered subcutaneously, which is the currently widely used route of administration32. Because the intranasal peptide vaccine developed in this study utilizes lung CD4+ TRM as the main factor for a protective immune response, which differs from existing vaccines, we sought to determine whether there is a synergistic effect between these different protective immune responses. To confirm this, we used BCG prime and vaccine boost model. We divided mice into BCG-primed and non-primed groups, and both groups were intranasally twice with NVT or Pep5′+NVT at 3-week intervals, starting 13 weeks after priming. Four weeks after the last vaccination, we measured T cell responses in the lungs and spleen of the vaccinated mice, or infected a separate group of mice with Mtb via aerosol exposure and assessed bacterial loads in the lung and spleen, along with lung inflammation, at 4 and 16 weeks post-infection (Fig. 5a). We found that BCG priming did not enhance Pep 5′-specific T cell responses in the lung. As expected, BCG alone or BCG-primed NVT boosting failed to elicit Pep 5′-specific T cell response because BCG vaccine does not contain the T cell epitope used in our vaccine (Fig. 5b, c). We also found that Pep 5′ + NVT significantly reduced lung bacterial burden to levels similar to BCG. However, BCG-primed Pep 5′ + NVT boosting did not show a synergistic effect in reducing lung bacterial loads compared to BCG or Pep 5′ + NVT alone at 4 weeks post-infection (Fig. 5d). Meanwhile, both Pep 5′ + NVT and BCG significantly reduced the bacterial burden in the spleen at 4 weeks post-infection. Unlike in the lung, BCG significantly reduced the bacterial burden in the spleen compared to Pep 5′ + NVT (Fig. 5e). At 16 weeks post-infection, all vaccinated groups exhibited significantly lower bacterial burdens in both the lungs and spleen compared to the unvaccinated group. Notably, the bacterial loads among the vaccinated groups were comparable (Fig. 5f, g). Finally, we examined the degree of inflammation in the lungs. Lung inflammation was effectively reduced in all vaccine groups at both 4 and 16 weeks post-infection (Fig. 5h–k). Taken together, no synergistic effect was observed between the protective immunity provided by lung TRM and that provided by the BCG vaccine.
a Experimental design. C57BL/6 mice (n = 23 per group) were divided into BCG-primed and non-primed groups, and mice in the BCG-primed group were immunized subcutaneously (s.c.) with BCG. At 13 weeks post-priming, all mice were immunized intranasally (i.n.) twice at 3-week intervals with NVT or Pep 5′ + NVT. Four weeks after the final immunization, immune response analysis (n = 5 per group) or aerosol challenge with HN878 (n = 20 per group) were performed. At 4 and 16 weeks post-infection (wpi), mice (n = 9 per group at each time point) were sacrificed for bacterial counts (n = 5 per group) and lung histopathology (n = 4 per group). b, c IFN-γ+CD4+ T cell responses to Pep 5′ in the lungs were analyzed by flow cytometry. Representative dot plots (b) and percentages (c) of IFN-γ⁺CD4⁺ T cells. Bacterial loads in the lungs (d) and spleen (e) at 4 wpi. f, g Bacterial loads in the lungs (f) and spleen (g) at 16 wpi. H&E staining (h) and inflamed area (i) in lung tissues at 4 wpi. j, k H&E staining (j) and inflamed area (k) in lung tissues at 16 wpi. The box-and-whisker plots display the median (center line), 25th and 75th percentiles (box), and the minimum and maximum values (whiskers). Statistical analyses were performed using one-way ANOVA with Tukey’s multiple comparisons test. This experiment was performed once. **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant.
Discussion
In this study, we demonstrated that formulating the CD4 epitope of Mtb with our vaccine adjuvant platform and delivering it to the lungs leads to the formation of antigen-specific lung CD4+ TRM, resulting in superior anti-TB efficacy. The adjuvant NVT utilized in this study is a defined TLR3 agonist that, even when administered intramuscularly, induces a robust T cell response comparable to poly(I:C)23, making it an effective adjuvant for intramuscular administration. In fact, a comparable level of CD4+ T cell response was observed in the group receiving this vaccine intramuscularly. However, CD4+ TRM population was not detected in the lungs, highlighting the additional benefit of lung CD4+ TRM. The vaccine used in this study, which consists of five peptides that induce the formation of lung CD4+ TRM, demonstrated efficacy comparable to BCG, despite BCG containing numerous epitopes. This efficacy was achieved with only two administrations and was confirmed against the highly pathogenic strain HN878. Given that most adjuvanted subunit TB vaccines currently in development require three doses, this approach is believed to improve in both efficacy and convenience.
The excellent protective efficacy conferred by lung TRM has already been studied in relation to BCG25,32. However, since BCG is a live, albeit attenuated, bacterium, there are concerns about potential side effects when it is administered directly to the lung mucosa, making it potentially difficult to use in humans. To address this issue, this study utilized synthetic peptide as an antigen, which is considered a safer option as it contains only the minimum epitope necessary for immune induction. Additionally, safety was a primary concern; therefore, adjuvants that have been thoroughly characterized for their role in the innate immune response, the formation mechanism of lung TRM and the chemokine and inflammatory cytokine profiles that induce immunopathology were used25.
In this study, we experimentally confirmed that two of the five peptides we selected (ESAT61 and ESAT62) also contain sequences that match the MHC of C57BL/6 mice. In fact, some of the sequences in ESAT61 and ESAT62 were identified in previous studies, which confirmed their reactivity in C57BL/633,34,35. On the other hand, we could not find any previous research showing that the sequences in CFP101 and CFP102 were reactive in C57BL/6 mice. Interestingly, for the sequences in Rv2031c, there were conflicting result: some studies indicated reactivity36, while others did not reactive37. However, when comparing to the literature, the sequence in the marginal region where processing could be an issue, was different, the adjuvant used was different, and since we could not confirm a response in mice in our experiment, we concluded that Rv2031c does not contain a sequence that matches the mouse MHC. Therefore, we can conclude that the CD4+ TRM measured in the mouse experiment in this study were specific only to ESAT61 and ESAT62, and the protective efficacy against TB observed in this study can be attributed to CD4+ T cell responses specific to these two epitopes.
Since many countries mandate BCG vaccination at birth, many next-generation TB vaccines are being studied with a BCG boosting strategy38. In this study, efficacy was evaluated in the BCG booster model, but no synergistic effect from boosting was observed. Although the vaccine used in this study targeted multiple antigens, only ESAT-6 elicited a response in the mouse model due to the MHC restriction characteristics of the peptide. On the other hand, since BCG does not contain ESAT-639, This result suggests that while the dosing schedule involved a boost, it did not constitute an immunological boost. Other vaccines in the literature that showed efficacy in the BCG booster model contained additional antigens present in BCG, alongside ESAT-6, which create a situation where BCG was immunologically boosted40,41,42. Therefore, to better understand the role of lung TRM in boosting BCG, further experiments with other epitopes present in BCG are needed.
In the present study, we investigated the additional preventive efficacy of TB-specific lung CD4+ TRM. This efficacy is attributed to the inherent characteristics of TRM, which enable them to respond quickly by pre-positioning themselves in local areas. For instance, no significant protective effect was observed in a treatment model where mice were pre-infected with TB bacteria, treated with antibiotics, and then administered the vaccine (Supplementary Fig. 5). Furthermore, in a model of latent TB reactivation, where the vaccine was administered after pre-infection with TB bacteria and antibiotic treatment, no significant difference was observed between the vaccinated and unvaccinated groups (Supplementary Fig. 6). All pathogens have factors that can activate the host’s innate immunity, as well as proteins that can act as antigens. Since the TB bacteria were airborne in advance during the creation of the latent TB model, it is thought that pulmonary TRM could be formed even in the unvaccinated group, and that this TRM likely showed a similar degree of relapse-suppression efficacy as the vaccinated group. In fact, we confirmed that relapse was significantly suppressed in all groups compared to pre-treatment levels. However, since some individuals who have previously had TB do relapse, this raises questions about whether the formation of TRM actually genuinely in preventing TB. To address this question, further research is needed to investigate the characteristics and quantity of TRM formed by natural infections compared to those induced by this vaccine.
The most ideal vaccine is one that closely mimics the target pathogen while ensuring safety. In conclusion, this study has developed a vaccine that induces protective immune factors by mimicking the natural infection route of TB, using a synthetic peptide that poses minimal safety concerns as an antigen. Our results support the potential of not only the peptides used in this study, but also new target antigens that can be incorporated into this platform to enhance its efficacy. We suggest this approach may represent a promising direction for enhancing the effectiveness of other TB vaccines currently in development.
Methods
Ethics statement
All animal studies were conducted in accordance with the guidelines of the Korean Food and Drug Administration. Animals were anesthetized with isoflurane administered via inhalation to ensure adequate sedation and minimize stress during the experimental procedure or euthanasia. For euthanasia, animals were deeply anesthetized with 5% isoflurane, and the depth of anesthesia was confirmed by the absence of all reflexes. The experimental protocols for animal studies were reviewed and approved by the Ethics Committee and the Institutional Animal Care and Use Committee (IACUC, Permit Number: 2023-0023) of the Laboratory Animal Research Center at Yonsei University College of Medicine (Seoul, Korea) and IACUC (Permit Number: NAVI-2022-0004) of NA Vaccine Institute (NAVI, Seoul, Korea). All human studies were approved by the Institutional Review Board (Permit Number: IRB 2022-0111) of Asan Medical Center (Seoul, Korea), and written informed consent was obtained from all donors prior to sample collection.
Reagents and mice
The FACS antibodies used in this study are listed in Supplementary Table 1. Nexavant (NVT) was produced by in vitro transcription as previously described23. All peptides were synthesized at >90% purity by Dandicure (Ochang, Korea) and stored in lyophilized form. PepMix™ M. tuberculosis (ESAT-6) and PepMix™ M. tuberculosis (CFP-10) were purchased from JPT Peptide Technologies GmbH (Berlin, Germany). DNase I was purchased from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany). Human IFN gamma Uncoated ELISA Kit, R848 (Resiquimod), and LPS were purchased from InvivoGen (San Diego, CA, USA). Human GM-CSF and IL-15 were purchased from PeproTech (Rocky Hill, NJ, USA). Human IL-1β, IL-2, IL-4, IL-7, and Flt3-L were purchased from R&D Systems (Minneapolis, MN, USA). Mouse Anti-Human CD28 and Mouse Anti-Human CD49d antibodies were purchased from BD Biosciences (San Jose, CA, USA). Specific pathogen-free C57BL/6 female mice at 6 weeks of age were purchased from Samtako Bio Korea (Kyounggi, Korea) or Orient Bio (Gyeonggi, Korea). Mice used for immune response analysis were bred in the NAVI animal facility, while mice for Mtb infection studies were bred under barrier conditions in the ABSL-3 facility at Yonsei University College of Medicine. All mice were fed a sterile commercial mouse diet and had ad libitum access to water.
Cloning, expression, and purification of recombinant proteins
To produce the recombinant proteins, the corresponding genes were amplified by PCR using genomic DNA from Mtb H37Rv ATCC 27294 as the template. For HspX, the following primers were used: forward, 5′-AAGCTTTACATCGAGTACATGGCCACCACCCTTCCC-3′, and reverse, 5′-GCTCGAGTGCGGCCGCGTTGGTGGACCGGATCTG-3′. The resulting PCR product was inserted into the pET-22b (+) vector (Novagen, Madison, WI, USA) after digesting both using HindIII and NotI restriction enzymes. For ESAT-6, the gene was amplified using the primers: forward, 5′-AAGCTTATGACAGAGCAGCAGTGGAAT-3′, and reverse, 5′-CTCGAGTGCGAACATCCCAGTGACGTT-3′. The PCR product was inserted into the pET-22b (+) vector after digestion with HindIII and XhoI restriction enzymes. For CFP-10, the gene was amplified using the primers: forward, 5′-GGCCGGGGATCCATGGCAGAGATGAAGACCG-3′, and reverse, 5′-GGCCGGGAATTCGAAGCCCATTTGCGAGGAC-3′. The PCR product was inserted into the pET-28a expression vector after digestion with BamHI and EcoRI restriction enzymes. The recombinant proteins were expressed in Escherichia coli BL21 (DE3) cells. Transformed E. coli cells were grown in LB broth containing appropriate antibiotics (100 μg/ml ampicillin for pET-22b constructs or 50 μg/ml kanamycin for pET-28a constructs) at 37 °C until the OD600 reached 0.6–0.8. Protein expression was induced by adding isopropyl β-D-1-thiogalactopyranoside (IPTG) to a final concentration of 1 mM, and the cultures were further incubated at 37 °C for 4 h. Cells were harvested by centrifugation at 6000 × g for 20 min at 4 °C. For protein purification, the cell pellets were resuspended in lysis buffer (20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 5 mM imidazole) and disrupted by sonication. After centrifugation at 15,000 × g for 30 min at 4 °C, the supernatants containing soluble proteins were applied to a Ni-NTA agarose column (Qiagen, Hilden, Germany) pre-equilibrated with lysis buffer. The columns were washed with washing buffer (20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 20 mM imidazole), and the His-tagged recombinant proteins were eluted with elution buffer (20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 250 mM imidazole). The purified proteins were dialyzed against phosphate-buffered saline (PBS, pH 7.4) and concentrated using Amicon Ultra centrifugal filters (Millipore, Bedford, MA, USA). Protein concentrations were determined using the Bradford assay (Bio-Rad, Hercules, CA, USA), and protein purity was assessed by SDS-PAGE. The amount of residual LPS in the protein preparations was evaluated using the Limulus amoebocyte lysate (LAL) test kit (Lonza, Basel, Switzerland) according to the manufacturer’s instructions.
Bacterial culture
Mtb HN878 was obtained from the strain collections of the International Tuberculosis Research Center (Changwon, Korea). Mycobacterium bovis BCG (Pasteur strain 1173P2) was kindly provided by the Pasteur Institute (Paris, France). Mtb was grown in Middlebrook 7H9 broth (Difco, Detroit, MI, USA) supplemented with 0.05% Tween-80 and ADC enrichment at 37 °C.
Antigen characteristics and vaccine formulation
The vaccine used in this study consisted of a mixture of antigens and the adjuvant NVT. In the protein subunit vaccine, recombinant proteins derived from Mtb H37Rv (ATCC 27294)—specifically ESAT-6, CFP-10, and HspX—were used as antigens. The three recombinant proteins, each at an equal dose (5 µg per mouse), were mixed with NVT (10 µg per mouse) in PBS to prepare the final aqueous suspension for administration. For the peptide vaccine, five peptide sequences were selected based on known CD4⁺ T cell epitopes derived from Mtb antigens ESAT-6, CFP-10, and HspX. Candidate peptides were synthesized as linear, unmodified sequences via solid-phase peptide synthesis at a length of 36–38 amino acids to include overlapping epitope regions (Table 1). The five candidate peptides were mixed at equal doses (5 µg per mouse for each) with NVT (10 µg per mouse) in PBS to prepare the final aqueous suspension for administration.
Isolation of human PBMCs
Human blood samples were collected at Asan Medical Center (Seoul, Korea) from 24 individuals (5 healthy donors, 8 with active TB, and 11 with latent TB). PBMCs were isolated from human blood samples using density gradient centrifugation with Ficoll-Paque PLUS (Cytiva, Marlborough, UK). Briefly, the whole blood was diluted with an equal volume of PBS with 1% heat-inactivated human serum and carefully layered onto Ficoll-Paque in a 15 mL conical tube. The samples were centrifuged at 400 × g for 30 min at room temperature (RT) without brake. The PBMC layer was carefully collected, washed twice with PBS, and then frozen and stored using cryopreservation solution (containing 90% heat-inactivated human serum and 10% DMSO) for future experiments.
In vitro stimulation of human PBMCs
Human PBMCs were thawed and suspended in X-VIVO 15 medium containing DNase I (1 μg/ml). After centrifugation at 400 × g for 10 min at RT, the cells were incubated in 20% RPMI medium (RPMI-1640 supplemented with 20% heat-inactivated human serum) at 37 °C with 5% CO2 for 7 h without stimulation. After resting, the cells were resuspended in R10 medium (RPMI-1640 supplemented with 10% heat-inactivated human serum, 0.1 mg/ml gentamicin, 1% GlutaMAX, and 10 mM HEPES) and seeded in 96-well round-bottom plates at a density of 5 × 10⁵ cells per well. The cells were then stimulated with PepMix (ESAT-6 and CFP-10, each at 5 μg/ml) or with Pep 5′ (a mixture of five peptides, each at 5 μg/ml) at 37 °C with 5% CO2 for 72 h. After stimulation, culture supernatants were collected and stored at –80 °C until analysis. Human IFN-γ levels in the supernatants were quantified using ELISA kit according to the manufacturer’s instructions. The cut-off value used to distinguish positive responses from background was set at twice the IFN-γ level measured in the unstimulated control. For each individual, the response value was calculated by subtracting the IFN-γ level of the unstimulated control from that of the antigen-stimulated group.
Antigen-specific T cell expansion of human PBMCs
On day 0, human PBMCs were rested in 20% RPMI medium for 7 h, then suspended in R10 medium and seeded into 96-well round-bottom plates at a density of 1 × 105 cells/100 μl per well. Subsequently, 100 μl of 2x cytokine solution containing human GM-CSF (2000 IU/ml), human IL-4 (1000 IU/ml), and human Flt3-L (100 ng/ml) was added to each well, and the cells were incubated at 37 °C with 5% CO2. On day 1, 100 µl of the supernatant was removed, and 100 µl of a 2x adjuvant and stimulus solution containing R848 (20 µM), LPS (200 ng/ml), human IL-1β (20 ng/ml), and either PepMix (ESAT-6 and CFP-10, each 5 µg/ml) or Pep 5′ (50 µg/ml, with each peptide at 10 µg/ml) was added. On Days 2, 4, and 7, 100 µl of the supernatant was removed, and 100 µl of a 2x feeding solution containing human IL-2 (20 IU/ml), human IL-7 (20 ng/ml), and human IL-15 (20 ng/ml) was added to refresh the media. On Day 9, the cells were resuspended in R10 media and seeded in 96-well round-bottom plates at a density of 2 × 10⁵ cells per well. The cells were then stimulated with anti-CD28 (1 μg/ml) and anti-CD49d (1 μg/ml), either alone (unstimulated control), with PepMix (ESAT-6 and CFP-10, each at 5 μg/ml), or with Pep 5′ (each peptide at 5 μg/ml). On Day 12, the supernatants were collected and stored at –80 °C until analysis. Human IFN-γ levels in the supernatants were quantified using ELISA kit according to the manufacturer’s instructions. The cut-off value used to distinguish positive responses from background was set at twice the IFN-γ level measured in the unstimulated control. For each individual, the response value was calculated by subtracting the IFN-γ level of the unstimulated control from that of the antigen-stimulated group.
Immunization and challenge
In this study, aerosol infection of mice with H37Rv or HN878 was conducted using a Glas-Col aerosol apparatus (Terre Haute, IN, USA) adjusted to achieve an initial infectious dose of 200 CFUs. To assess the immunogenicity of the peptide candidates, mice were immunized intramuscularly twice at 3-week intervals with 10 μg of NVT-adjuvanted 15 μg of recombinant TB proteins (ESAT-6, CFP-10, and HspX, 5 μg of each, totaling 15 μg). T cell responses in the lungs and spleens were measured by flow cytometry 4 weeks after the final immunization.
To compare the protective efficacy between intramuscular and intranasal administration of the vaccine, mice were immunized either intramuscularly or intranasally twice at 3-week intervals with 10 μg of NVT-adjuvanted 25 μg of Pep 5′ (each peptide at 5 μg). Four weeks after the final immunization, lung TRM and T cell responses in the lungs and spleens of some immunized mice were measured by flow cytometry. Other immunized mice were challenged with H37Rv via aerosol exposure, and survival was monitored.
For the preventive model, mice were immunized subcutaneously once (on day 0) with 2 × 105 CFU of BCG, or intranasally twice (on days 0 and 21) with 10 μg of NVT, or with 10 μg of NVT-adjuvanted 25 μg of Pep 5′ (each peptide at 5 μg). Four weeks after the final immunization, they were infected with HN878 via aerosol exposure. At 4 weeks post-infection, bacterial counts were measured in the lungs and spleen of infected mice, and lung tissue was stained with hematoxylin and eosin (H&E).
For BCG prime and vaccine booster models, mice were divided into BCG-primed and non-primed groups, and mice in the BCG-primed group were immunized subcutaneously with 2 × 105 CFU of BCG. At 13 weeks post-priming, all mice were immunized intranasally twice at 3-week intervals with 10 μg of NVT, or with 10 μg of NVT-adjuvanted 25 μg of Pep 5′ (each peptide at 5 μg). Four weeks after the final immunization, the immunized mice were either sacrificed for immune response analysis or infected with HN878 via aerosol exposure. At 4 and 16 weeks post-infection, bacterial counts were measured in the lungs and spleen of infected mice, and lung tissue was stained with H&E.
For reactivation experiments, mice were infected with HN878 via aerosol exposure. The infected mice were orally treated with Isoniazid (INH) (10 mg/kg) and Pyrazinamide (PZA) (150 mg/kg) via gavage once a day and 5 times per week for 17 weeks, from 4 to 21 weeks post-infection, during which time some mice were immunized intranasally with NVT (10 μg) or NVT (10 μg)-adjuvanted Pep 5′ (25 μg, each peptide at 5 μg) at 17, 19, and 21 weeks post-infection. At 4, 21, and 33 weeks post-infection, bacterial counts in the lungs were measured.
To determine the efficacy of the vaccine during ethambutol (EMB) treatment, mice were infected with HN878 via aerosol exposure. The mice were divided into two groups and were either treated or not treated with EMB at 100 mg/kg orally once daily, 5 times a week, for 6 weeks between weeks 5 and 11 post-infection. Some mice were immunized intranasally with NVT (10 μg)-adjuvanted Pep 5′ (25 μg, each peptide at 5 μg) at weeks 7, 9, and 11 post-infection in combination with EMB treatment. Bacterial counts in the lungs were measured at weeks 5 and 14 post-infection.
Single cell preparation
Spleens and lungs were harvested from immunized mice and dissociated into single cells. Spleen tissues were mechanically dissociated by gently pressing them through a 70 μm cell strainer using the plunger of a syringe. The resulting cell suspension was centrifuged at 300 × g for 5 min at 4 °C, and red blood cells (RBCs) were lysed using 1× RBC lysis buffer (BioLegend, San Diego, CA, USA) for 4–5 min on ice. The cells were then washed with PBS and resuspended in complete RPMI-1640 medium (RPMI-1640 supplemented with 10% FBS, 1% penicillin-streptomycin, and 2 mM L-glutamine). Lung tissues were finely minced into ~1 mm³ fragments using sterile scissors and enzymatically digested in RPMI-1640 medium containing 1 mg/mL Collagenase Type IV (Worthington Biochemical Corporation, Lakewood, NJ, USA) at 37 °C for 30 min. After digestion, the tissue suspension was filtered through a 70 μm cell strainer to obtain single-cell suspensions. RBCs were removed using 1× RBC lysis buffer as described above. Cells were then washed with PBS and filtered through a 40 μm cell strainer to remove debris. Final single-cell suspensions were resuspended in complete RPMI-1640 medium.
Intracellular cytokine staining and flow cytometry analysis
Single-cell suspensions were stimulated overnight at 37 °C with 5 μg/ml of ESAT61, ESAT62, CFP101, CFP102, or Rv2031c; with Pep 5′ (a mixture of five peptides, each at 5 μg/ml); or with recombinant TB proteins (ESAT-6, CFP-10, and HspX), each at 5 μg/ml each. Then, 1 μg/ml of a protein transport inhibitor GolgiPlug (BD Bioscience, Franklin Lakes, NJ, USA) was added and further cultured for 4–6 h. After washing with PBS, the cells were stained with a viability dye, anti-CD4, anti-CD8, and anti-CD44 antibodies for 20 min at 4 °C. After washing with FACS buffer (PBS containing 1% FBS and 0.1% NaN3), the cells were fixed and permeabilized using the BD Cytofix/Cytoperm solution (BD Biosciences), and then stained with anti-IFN-γ antibody for 20 min at 4 °C. The cells were washed and resuspended with FACS buffer and analyzed by flow cytometry. To examine lung TRM cell subsets, lung cells were stained with surface antibodies specific to CD4, CD8a, CD44, CD62L, CD69, and CD103, and analyzed by flow cytometry.
Bacterial enumeration and lung histopathology
Lungs and spleens from Mtb-infected mice were harvested and homogenized in PBS containing 0.05% Tween 80. The tissue homogenates were serially diluted and plated on Middlebrook 7H10 agar (Difco, Detroit, MI, USA) supplemented with 10% OADC (BD, Franklin Lakes, NJ, USA) to count the CFU of Mtb. For histopathological analysis, the middle-cross section from the superior lobe of right lung was stained with H&E. The percentage of inflamed sections of the lungs were determined by using the ImageJ (National Institutes of Health, MD, USA) program.
Statistical analysis
All data were analyzed using GraphPad Prism version 10.4.0 for Windows (GraphPad Software, La Jolla, CA, USA; www.graphpad.com). Comparisons between two groups were statistically evaluated using the nonparametric Mann–Whitney U test. Significant differences among multiple groups were determined using one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparisons test. Significances were considered as follows: *p < 0.05, ** p < 0.01, ***p < 0.001, ****p < 0.0001.
Data availability
The protein sequences used in this study have been deposited in the UniProt database. The accession numbers are as follows: ESAT-6 (B5TV89), CFP-10 (P9WNK5), and HspX (P9WMK1). These sequences can be accessed via https://www.uniprot.org.
References
Gan, H. et al. Mycobacterium tuberculosis blocks crosslinking of annexin-1 and apoptotic envelope formation on infected macrophages to maintain virulence. Nat. Immunol. 9, 1189–1197 (2008).
Andersen, P. & Doherty, T. M. The success and failure of BCG—implications for a novel tuberculosis vaccine. Nat. Rev. Microbiol 3, 656–662 (2005).
Fine, P. E. Variation in protection by BCG: implications of and for heterologous immunity. Lancet 346, 1339–1345 (1995).
Colditz, G. A. et al. Efficacy of BCG vaccine in the prevention of tuberculosis. Meta-analysis of the published literature. JAMA 271, 698–702 (1994).
Nuttall, J. J. & Eley, B. S. BCG vaccination in HIV-infected children. Tuberc. Res Treat. 2011, 712736 (2011).
Fonseca, J. D., Knight, G. M. & McHugh, T. D. The complex evolution of antibiotic resistance in Mycobacterium tuberculosis. Int J. Infect. Dis. 32, 94–100 (2015).
Pollard, A. J. & Bijker, E. M. A guide to vaccinology: from basic principles to new developments. Nat. Rev. Immunol. 21, 83–100 (2021).
Lyadova, I. V. & Panteleev, A. V. Th1 and Th17 Cells in Tuberculosis: Protection, Pathology, and Biomarkers. Mediators Inflamm. 2015, 854507 (2015).
Nikitina, I. Y. et al. Th1, Th17, and Th1Th17 Lymphocytes during Tuberculosis: Th1 Lymphocytes Predominate and Appear as Low-Differentiated CXCR3(+)CCR6(+) Cells in the Blood and Highly Differentiated CXCR3(+/-)CCR6(-) Cells in the Lungs. J. Immunol. 200, 2090–2103 (2018).
Flynn, J. L., Chan, J. & Lin, P. L. Macrophages and control of granulomatous inflammation in tuberculosis. Mucosal Immunol. 4, 271–278 (2011).
Cowley, S. C. & Elkins, K. L. CD4+ T cells mediate IFN-gamma-independent control of Mycobacterium tuberculosis infection both in vitro and in vivo. J. Immunol. 171, 4689–4699 (2003).
Andersen, P. & Smedegaard, B. CD4(+) T-cell subsets that mediate immunological memory to Mycobacterium tuberculosis infection in mice. Infect. Immun. 68, 621–629 (2000).
Hamley, I. W. Peptides for vaccine development. ACS Appl Bio Mater. 5, 905–944 (2022).
Purcell, A. W., McCluskey, J. & Rossjohn, J. More than one reason to rethink the use of peptides in vaccine design. Nat. Rev. Drug Discov. 6, 404–414 (2007).
Rosa, D. S., Ribeiro, S. P. & Cunha-Neto, E. CD4+ T cell epitope discovery and rational vaccine design. Arch. Immunol. Ther. Exp. (Warsz.) 58, 121–130 (2010).
Marty Pyke, R. et al. Evolutionary pressure against MHC class II binding cancer mutations. Cell 175, 416–428.e413 (2018).
Lindestam Arlehamn, C. S. et al. A quantitative analysis of complexity of human pathogen-specific CD4 T cell responses in healthy M. tuberculosis infected South Africans. PLoS Pathog. 12, e1005760 (2016).
Caccamo, N. et al. Cytokine profile, HLA restriction and TCR sequence analysis of human CD4+ T clones specific for an immunodominant epitope of Mycobacterium tuberculosis 16-kDa protein. Clin. Exp. Immunol. 133, 260–266 (2003).
Yang, F. F. et al. Monitoring of peptide-specific and gamma interferon-productive T cells in patients with active and convalescent tuberculosis using an enzyme-linked immunosorbent spot assay. Clin. Vaccin. Immunol. 19, 401–410 (2012).
Arlehamn, C. S. et al. Dissecting mechanisms of immunodominance to the common tuberculosis antigens ESAT-6, CFP10, Rv2031c (hspX), Rv2654c (TB7.7), and Rv1038c (EsxJ). J. Immunol. 188, 5020–5031 (2012).
Vincenti, D. et al. Identification of early secretory antigen target-6 epitopes for the immunodiagnosis of active tuberculosis. Mol. Med. 9, 105–111 (2003).
Komal, A., Noreen, M. & El-Kott, A. F. TLR3 agonists: RGC100, ARNAX, and poly-IC: a comparative review. Immunol. Res 69, 312–322 (2021).
Ko, K. H. et al. A novel defined TLR3 agonist as an effective vaccine adjuvant. Front Immunol. 14, 1075291 (2023).
Lee, S. H. et al. The defined TLR3 agonist, nexavant, exhibits anti-cancer efficacy and potentiates anti-PD-1 antibody therapy by enhancing immune cell infiltration. Cancers (Basel) 15, 5752 (2023).
Ko, K. H. et al. A vaccine platform targeting lung-resident memory CD4(+) T-cells provides protection against heterosubtypic influenza infections in mice and ferrets. Nat. Commun. 15, 10368 (2024).
Lightbody, K. L. et al. Molecular features governing the stability and specificity of functional complex formation by Mycobacterium tuberculosis CFP-10/ESAT-6 family proteins. J. Biol. Chem. 283, 17681–17690 (2008).
Geluk, A. et al. T-cell recognition of the HspX protein of Mycobacterium tuberculosis correlates with latent M. tuberculosis infection but not with M. bovis BCG vaccination. Infect. Immun. 75, 2914–2921 (2007).
Gey Van Pittius, N. C. et al. The ESAT-6 gene cluster of Mycobacterium tuberculosis and other high G+C Gram-positive bacteria. Genome Biol. 2, RESEARCH0044 (2001).
Satti, I. et al. Safety of a controlled human infection model of tuberculosis with aerosolised, live-attenuated Mycobacterium bovis BCG versus intradermal BCG in BCG-naive adults in the UK: a dose-escalation, randomised, controlled, phase 1 trial. Lancet Infect. Dis. 24, 909–921 (2024).
Tree, J. A. et al. Intranasal bacille Calmette-Guerin (BCG) vaccine dosage needs balancing between protection and lung pathology. Clin. Exp. Immunol. 138, 405–409 (2004).
Kang, A. et al. Subcutaneous BCG vaccination protects against streptococcal pneumonia via regulating innate immune responses in the lung. EMBO Mol. Med 15, e17084 (2023).
Perdomo, C. et al. Mucosal BCG vaccination induces protective lung-resident memory T cell populations against tuberculosis. mBio 7, e01686–16 (2016).
Woodworth, J. S. et al. Protective CD4 T cells targeting cryptic epitopes of Mycobacterium tuberculosis resist infection-driven terminal differentiation. J. Immunol. 192, 3247–3258 (2014).
Ashhurst, A. S. et al. Mucosal vaccination with a self-adjuvanted lipopeptide is immunogenic and protective against mycobacterium tuberculosis. J. Med Chem. 62, 8080–8089 (2019).
Platteel, A. C. M. et al. Strategies to enhance immunogenicity of cDNA vaccine encoded antigens by modulation of antigen processing. Vaccine 34, 5132–5140 (2016).
Kumar Das, D. et al. Targeting dendritic cells with TLR-2 ligand-coated nanoparticles loaded with Mycobacterium tuberculosis epitope induce antituberculosis immunity. J. Biol. Chem. 298, 102596 (2022).
Roupie, V. et al. Immunogenicity of eight dormancy regulon-encoded proteins of Mycobacterium tuberculosis in DNA-vaccinated and tuberculosis-infected mice. Infect. Immun. 75, 941–949 (2007).
Qu, M., Zhou, X. & Li, H. BCG vaccination strategies against tuberculosis: updates and perspectives. Hum. Vaccin Immunother. 17, 5284–5295 (2021).
Harboe, M., Oettinger, T., Wiker, H. G., Rosenkrands, I. & Andersen, P. Evidence for occurrence of the ESAT-6 protein in Mycobacterium tuberculosis and virulent Mycobacterium bovis and for its absence in Mycobacterium bovis BCG. Infect. Immun. 64, 16–22 (1996).
Dietrich, J. et al. Mucosal administration of Ag85B-ESAT-6 protects against infection with Mycobacterium tuberculosis and boosts prior bacillus Calmette-Guerin immunity. J. Immunol. 177, 6353–6360 (2006).
Lin, P. L. et al. The multistage vaccine H56 boosts the effects of BCG to protect cynomolgus macaques against active tuberculosis and reactivation of latent Mycobacterium tuberculosis infection. J. Clin. Invest 122, 303–314 (2012).
Kwon, K. W. et al. BCG-booster vaccination with HSP90-ESAT-6-HspX-RipA multivalent subunit vaccine confers durable protection against hypervirulent Mtb in mice. NPJ Vaccines 9, 55 (2024).
Arend, S. M. et al. Antigenic equivalence of human T-cell responses to Mycobacterium tuberculosis-specific RD1-encoded protein antigens ESAT-6 and culture filtrate protein 10 and to mixtures of synthetic peptides. Infect. Immun. 68, 3314–3321 (2000).
Mustafa, A. S. et al. Multiple epitopes from the Mycobacterium tuberculosis ESAT-6 antigen are recognized by antigen-specific human T cell lines. Clin. Infect. Dis. 30, S201–S205 (2000). Suppl 3.
Mustafa, A. S. et al. Human Th1 cell lines recognize the Mycobacterium tuberculosis ESAT-6 antigen and its peptides in association with frequently expressed HLA class II molecules. Scand. J. Immunol. 57, 125–134 (2003).
Tully, G. et al. Highly focused T cell responses in latent human pulmonary Mycobacterium tuberculosis infection. J. Immunol. 174, 2174–2184 (2005).
Kumar, M. et al. Immune response to Mycobacterium tuberculosis specific antigen ESAT-6 among south Indians. Tuberculosis (Edinb.) 90, 60–69 (2010).
Panda, S. et al. Identification of differentially recognized T cell epitopes in the spectrum of tuberculosis infection. Nat. Commun. 15, 765 (2024).
Nagai, H. et al. Immunological responses and epitope mapping by tuberculosis-associated antigens within the RD1 region in Japanese patients. J. Immunol. Res 2014, 764028 (2014).
Li Pira, G. et al. Evaluation of antigen-specific T-cell responses with a miniaturized and automated method. Clin. Vaccin. Immunol. 15, 1811–1818 (2008).
Oswald-Richter, K. et al. Mycobacterial ESAT-6 and katG are recognized by sarcoidosis CD4+ T cells when presented by the American sarcoidosis susceptibility allele, DRB1*1101. J. Clin. Immunol. 30, 157–166 (2010).
Shams, H. et al. Characterization of a Mycobacterium tuberculosis peptide that is recognized by human CD4+ and CD8+ T cells in the context of multiple HLA alleles. J. Immunol. 173, 1966–1977 (2004).
Mothe, B. R. et al. The TB-specific CD4(+) T cell immune repertoire in both cynomolgus and rhesus macaques largely overlap with humans. Tuberculosis (Edinb.) 95, 722–735 (2015).
Lindestam Arlehamn, C. S. et al. Memory T cells in latent Mycobacterium tuberculosis infection are directed against three antigenic islands and largely contained in a CXCR3+CCR6+ Th1 subset. PLoS Pathog. 9, e1003130 (2013).
Paul, S. et al. A population response analysis approach to assign class II HLA-epitope restrictions. J. Immunol. 194, 6164–6176 (2015).
Caccamo, N. et al. Th0 to Th1 switch of CD4 T cell clones specific from the 16-kDa antigen of Mycobacterium tuberculosis after successful therapy: lack of involvement of epitope repertoire and HLA-DR. Immunol. Lett. 98, 253–258 (2005).
Wilkinson, R. J. et al. Human T- and B-cell reactivity to the 16kDa alpha-crystallin protein of Mycobacterium tuberculosis. Scand. J. Immunol. 48, 403–409 (1998).
Agrewala, J. N. & Wilkinson, R. J. Differential regulation of Th1 and Th2 cells by p91-110 and p21-40 peptides of the 16-kD alpha-crystallin antigen of Mycobacterium tuberculosis. Clin. Exp. Immunol. 114, 392–397 (1998).
Agrewala, J. N. & Wilkinson, R. J. Influence of HLA-DR on the phenotype of CD4+ T lymphocytes specific for an epitope of the 16-kDa alpha-crystallin antigen of Mycobacterium tuberculosis. Eur. J. Immunol. 29, 1753–1761 (1999).
Wilkinson, K. A., Hudecz, F., Vordermeier, H. M., Ivanyi, J. & Wilkinson, R. J. Enhancement of the T cell response to a mycobacterial peptide by conjugation to synthetic branched polypeptide. Eur. J. Immunol. 29, 2788–2796 (1999).
Acknowledgements
This research was supported by a grant from the Korea Health Technology R & D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare (RS-2022-KH128236 and RS-2022-KH128243, Republic of Korea), and supported by Korea Drug Development Fund funded by Ministry of Science and ICT, Ministry of Trade, Industry, and Energy, and Ministry of Health & Welfare (RS-2024-00337146, Republic of Korea).
Author information
Authors and Affiliations
Contributions
S.B.C. and D.-H.K. conceived the work. K.H.K., S.B.C. and D.-H.K. designed the experiments. K.H.K., S.-H.B., H.S.B., Y.M.K., S.H.G., Y.Y.J., E.H.K. and T.S.S. conducted the experiments and performed data analysis. H.-G.C. and H.-J.K. provided critical reagents. D.-H.K. offered essential comments. K.H.K. and S.B.C. wrote and reviewed the manuscript. All authors have read and agreed to the submitted version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
D.-H.K. is the Chief Executive Officer of the NA Vaccine Institute. K.H.K., H.S.B. and S.B.C. are employees of the NA Vaccine Institute. T.S.S. is an employee of Asan Medical Center. The remaining authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ko, K.H., Baek, SH., Bae, H.S. et al. A vaccine targeting lung resident-memory CD4+ T cell phenotype protects against Mycobacterium tuberculosis in mice. npj Vaccines 10, 161 (2025). https://doi.org/10.1038/s41541-025-01225-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41541-025-01225-7