Abstract
Increasing access to water quality tests in low-income communities is a crucial strategy toward achieving global water equality. Recent studies in the Water Sanitation and Hygiene (WASH) sector underscore the importance of addressing practical concerns in water testing, such as robustness and results communication. In response, we present the WaterScope testing kit; an open-source, validated platform for drinking water quality assessment. It modernises the testing process with the inclusion of a unique cartridge/slider mechanism, machine-learning-enhanced classification and full digitalisation of results. WaterScope’s equivalency to conventional methods for quantifying E. coli is established through extensive validation experiments in both laboratory and field environments. This versatile platform provides potential to expand its applications to test other bacteria, perform colorimetric assays, and analyse clinical samples such as blood/urine samples. We anticipate that the system’s ease-of-use, portability, affordability, robustness, and digital nature will accelerate progress toward global water equality.
Similar content being viewed by others
Introduction
Approximately one in four people lack access to safely managed drinking water1. This problem is especially prevalent in low-income countries, where diarrhoeal diseases caused by faecal contamination of water are responsible for approximately 1 million preventable deaths per year2,3,4. While progress is being made, with over 687 million people having gained access to safely managed drinking water since 20155, this progress would need to be more than six times faster6 in the remainder of the decade to meet the United Nations’ target of safe water for all by 20307. Microbiological water monitoring has been identified as a key factor in improving humanitarian water, sanitation and hygiene (WASH) interventions8. By improving access to affordable, reliable, locally-available and easy-to-use water testing technologies, communities can be empowered to respond proactively to outbreaks and live with increased confidence in their drinking water supply.
Microbial water testing typically focuses on the enumeration of E. coli as an effective indicator of faecal contamination9, with the World Health Organisation (WHO) guidelines for drinking water stating that E. coli colony forming units (CFUs) ‘must not be detectable in any 100 ml sample’10. UNICEF’s target product profile outlines an ideal humanitarian water testing kit, describing how it would incorporate high sensitivity, rapid detection, low cost, portability, robust design, ease-of-use and low environmental footprint11. The most established testing methods involve cultivating samples until the E. coli grows sufficiently to produce a readily measurable signal; either through the evolution of gas in the multiple-tube fermentation technique12,13, or through a colour change brought about by the use of chromogenic nutrient media14,15. Novel innovations in E. coli testing have seen the development of biosensors16,17, E. coli specific bacteriophages18, DNA amplification methods19,20, and flow cytometry21 amongst others (see reviews by Nurliyana 201816 or Tambi 202322). However, existing implementations of these testing methods are not ideal for low-resource settings23 where the majority of institutions do not meet their water testing targets24.
Studies have repeatedly shown that human-centred design (HCD) is key to identifying and overcoming practical barriers within the WASH sector25,26,27. Through HCD, key issues can be identified such as: the high cost of logistics and labour in comparison to testing materials23,28, or the poor integration of systems for reporting, digitising and interpreting data29. One way of encouraging HCD and participatory development, is to make kit designs open-source30,31,32; this has the added benefit of making it easier to repair kits and reducing reliance on proprietary supplies of consumable components. To address these issues, this work introduces an open-source, digital, and portable testing kit for the quantification of E. coli and other parameters in water, referred to as the WaterScope (WS) test kit. We provide a comprehensive overview and validation of the WS kit, demonstrate its use for environmental monitoring in a year-long study of the River Cam (UK), and showcase its application for monitoring the quality of drinking water in East Africa.
Results and discussion
Overview of the WaterScope kit
To comprehensively understand the challenges in water testing, particularly in resource-constrained areas, we engaged in numerous discussions with various stakeholders in the water, sanitation, and hygiene (WASH) sector (from programme directors to scientists and field technicians, identified in collaboration with local academics, charities, citizen science groups and water utility companies) and reviewed existing literature on water quality monitoring in low-resource settings. In addition to these informal discussions, we conducted four workshops in diverse geographic locations including India, Tanzania, and South Sudan, where we engaged directly with potential end users. These workshops involved a total of 75 individuals and provided an opportunity to compare the WaterScope system with existing water testing methods through hands-on training. In addition to open discussions and structured interviews, we solicited written feedback from participants using prompts that focused on usability, areas for improvement, aspects they favoured, comparison to existing systems, and desired integration features.
For instance, during a workshop at Miranda House - University College for Women in India, participants primarily composed of students with an interest in WASH noted that the manual filtration process was too laborious. This feedback was instrumental in our decision to integrate an electric vacuum pump into the WaterScope system, significantly reducing the effort required for sample processing. Another workshop in Tanzania, attended by various WASH professionals from government and academia, highlighted the need for increased testing capacity. Participants indicated that the existing incubator could benefit from handling more samples simultaneously. In response, we upgraded the system to accommodate 14 samples per batch, doubling the previous capacity and enhancing the kit’s utility in handling larger sample numbers during field deployments.
These insights were crucial in adopting an iterative Human-Centered Design (HCD) approach for developing the WaterScope system. Each feature specifically addresses challenges identified in existing testing methods, correlating directly with distinct user needs as detailed in Table 1. This design strategy ensures that WaterScope is not only technologically innovative but also perfectly aligned with the practical needs and challenges of end-users. By integrating stakeholder feedback throughout the development process, we have significantly enhanced the effectiveness, usability and responsiveness of the WaterScope kit to meet the real-world demands of those it is designed to serve.
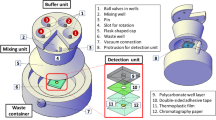
The WS kit (Fig. 1a) is based on the membrane filtration (MF) method15,33,34, due to it being the preferred method of organisations such as the US Environmental Protection Agency35, ISO15, UNICEF11, and Oxfam36. One of the main challenges with existing MF testing systems is around their complexity and multiple processing steps required to conduct a valid test without false-positives and false-negatives. To mitigate this, the system is designed around the use of a reusable sample cartridge37 into which a single-use membrane ‘slider’ can be inserted. The cartridge can be easily sterilised for multiple uses whereas the ‘slider’ comes pre-sterilised. The cartridge technology centralises the process of filtration, medium addition, incubation, and imaging in a single controlled environment, significantly simplifying the process and reducing the risk of user error. Additionally, preparing and adding nutrient medium to the cartridge is more straightforward compared to traditional systems, which typically necessitate prior lengthy preparation and pipetting with sterile, single-use plastics.
a Key components shown packed inside the system’s case, weighing 6 kg with a size of 29 cm × 23 cm × 26 cm. b Key steps required for a single E. coli test. Note that a single power bank provides sufficient energy for parallel processing of 14 samples with a 21-h incubation period. Results are uploaded to cloud storage via WiFi, via an embedded IoT module with cellular coverage or via an Android App.
Existing systems depend on the user’s ability to manually identify and count bacterial colonies, a task that can be prone to human error. Additionally, these systems necessitate the external management of test results, adding another layer of complexity to the process. The embedded microscope in the WS kit is an important part of modernising the whole testing process38 as it both enables automated classification of CFUs (making the device easier to use with lower training requirements), but also facilitates the immediate sharing of results into an online web dashboard (improving data transparency and notification speed for stakeholders). In addition, the microscope can be used for other purposes such as analysis of urine samples, as a photometer or to analyse colorimetric test strips when an appropriate modified cartridge/slider is used (see SI Section 9 for more information). Automatic CFU classification software routines have been described in several previous works39,40,41,42,43,44,45, but in our experience are unsuitable for use in the field for one of two reasons: a) the software is incapable of performing to the high standards required to meet WHO guidelines, or b) the software is too complicated to be used by users with minimal training. A discussion of automated testing software that we trialled can be found in Section 2 of the supplementary information. To our knowledge, the WaterScope kit is the first work that incorporates reliable automated classification into an E. coli testing procedure for use in low-resource settings. The kit achieves this through a combination of four things: (i) controlled lighting and imaging conditions; (ii) a classification model tailored specifically to our nutrient media and imaging geometry; (iii) a collection of 30,000 individual labels to train from, and (iv) integration of the software into an intuitive interface for users.
Validation 1: controlled lab experiments
To test the microbiological equivalency of the WS kit to other methods, a lab study was performed broadly following the protocol suggested by ISO 1799446. A 5-fold dilution series was prepared from a concentrated E. coli (EC) stock to create samples of approximately 10, 20, 40, 80 and 160 EC CFUs per 100 ml. Negative controls of autoclaved EC stock were run at the same time to check for false positives. Four methods were compared: the WS approach, the ISO 9308-1 method which uses Chromogenic Coliform Agar (CCA) media15, the ISO 9308-2 method which uses Colilert-18 with IDEXX’s Most Probable Number (MPN) approach13, and a final approach using Membrane Lauryl Sulphate Broth (MLSB) media which is utilised in field kits such as that of Delagua47. At each dilution, for each method, 14 repeats were measured. All negative controls for all methods resulted in no growth. The experimental results are shown in Fig. 2.
Test were performed on lab-strain E. coli across a five-fold dilution series with 14 samples at each dilution for WaterScope (WS - red), CCA (blue, ISO 9308-1), Colilert-18 (orange, ISO 9308-2) and MLSB (green) approaches. a Scatterplots of WS counts vs reference method counts, with linear trendline fitted. b Mean-difference plots comparing WS and reference methods, right-panel shows mean value (circle) and 95% confidence interval (error bars) with kernel density approximation (violin plot) for each reference. c Target CFU of the dilution series plotted against average counts from each method. Error bars represent one standard deviation. d Correlation matrix showing Spearman rank for different methods. Note that the Most Probable Number (MPN) approach was used with Colilert-18.
A strong linear relationship is identified between WS and each reference method (Fig. 2a), with coefficients of determination (R2) measured to be 0.92, 0.84 and 0.75 between WS and CCA, Colilert and MLSB methods, respectively. The WS method is found to be not significantly different at 95% confidence from any of the three reference methods (Fig. 2b). We do, however, replicate the well-studied observation that MLSB typically recovers fewer viable EC than other methods48. Spearman ranks also suggest that the WS method correlates strongly with all other methods (Fig. 2d). Notably the correlation between WS and CCA/Colilert (0.94/0.93), is very similar to the correlation between CCA/Colilert methods (0.95). Taken together, these results suggest the WS approach to recovery of E. coli performs similarly to the recommended ISO 9308 methodologies, something which is backed up by similar validations performed by an ISO 17025 accredited independent external laboratory (see SI Section 6).
Validation 2: controlled environmental validation
Environmental samples add a significant level of complexity to microbiological testing methods: interactions between different microbes, changes in physicochemical attributes of the water such as pH/chlorine/metal levels, and the presence of sediment can all drastically alter the growth characteristics of EC. To validate the WaterScope approach in this more challenging context, we conducted a year-long study in 2022 of water from the River Cam (Cambridge, UK). Samples were collected biweekly from Byron’s Pool (52°10′17“N, 0°5′55“E) and analysed using WS, CCA and Colilert methodologies, with 2–5 replicates performed for each method. Results are shown in Fig. 3.
Tests were performed biweekly throughout 2022 on the River Cam (52°10′17“N, 0°5′55“E) with 2 to 5 samples each for WaterScope (WS - red), CCA (blue, ISO 9308-1) and Colilert-18 (orange, ISO 9308-2) approaches. a Variation in mean EC counts throughout the year, with semi-transparent fills showing standard deviation. b Scatterplots of WS counts vs reference method counts, with linear trendlines fitted. c Correlation matrix showing Spearman rank for different methods. d Mean-difference plots comparing WS and reference methods, right-panel shows mean value (circle) and 95% confidence interval (error bars) with kernel density approximation (violin plot) for each reference. Note that the Most Probable Number (MPN) approach was used with Colilert-18.
EC counts from all methods were found to observe a typical seasonal variation, peaking in months with heavy rainfall and falling to a minimum during summer months (Fig. 3a), as has been observed in studies on other rivers49. However, it should be noted that EC levels are typically well above the 900 CFU/100 ml UK threshold for sufficient bathing water quality. The WaterScope method shows excellent agreement with CCA and Colilert methods; giving R2 values of 0.95 and 0.94 respectively (Fig. 3b), and Spearman ranks of 0.98 and 0.94 respectively (Fig. 3c). Mean-difference analysis again suggests no significant difference between WaterScope and the CCA/Colilert approaches to 95% confidence (Fig. 3d). All methods showed no growth on negative control samples (autoclaved EC stock). Interestingly, WS correlates with reference methods slightly more strongly in this study than in the above lab study; this is likely due to the fact that sample volumes were chosen such that individual test counts were close to/below 100 CFU, whereas the lab study included samples with up to 160 CFU, where colony overlap can start to make accurate counting more challenging. Taken together, these analyses suggest that the accuracy of the WaterScope testing kit performs just as strongly with environmental samples as it does with lab samples.
Validation 3: international field trials
Whilst the preceding validation studies demonstrate equivalence of the WaterScope system as compared with conventional lab-based water-testing techniques, the true test of the system is to see how it performs in the context of humanitarian water quality testing. In contrast to the controlled UK-based validation experiments in sections 1 and 2, here we evaluate WaterScope’s performance in three field trials based in East Africa where efficacy is contingent not only on technical performance, but also a mix of other factors. These factors range from the effectiveness of user-training to system robustness in different climates, to the ease-of-use of the system. Specifically, we carried out three field trials: first in Juba, South Sudan in May 2022; second in Kawangware, Kenya in September 2022; third in Addis Ababa, Ethiopia in June 2023. Between each field trial feedback was assessed and modifications/improvements were made. Overall, 301 WaterScope samples were taken, each with a Compact Dry™ measurement acquired for reference. A majority of samples came from ‘Improved’ water-sources (36% boreholes, 24% municipal water decanted to containers, 13% piped and 7% other), with the remainder coming from environmental surface water (16%) and bottled water (4%) to ensure a range of high and low counts (Fig. 4a).
Validation field trials were conducted using WaterScope systems in Juba (May 2022), Kawangware (September 2022), and Addis Ababa (June 2023), with slight modifications made between each trial. a Maps showing water sample types and locations for each trial. Scale bars are 10 km, 1 km and 5 km from left to right. b Compiled E. coli counts split into risk categories for WaterScope and Compact Dry testing methods. c ROC curves plotted for each field trial, generated by calculating binary classifications as the limit of detection is varied stepwise from 0 to 20, using Compact Dry results as reference. Legend values correspond to Area Under Curve (AUC) calculations.
Overall, a strong correlation was observed between WaterScope and Compact Dry™ measurements, however a notable exception to this was the presence of false positives (FPs) for our first field trial in Juba; we observed a Spearman rank of 0.75 including FPs and 0.91 excluding FPs, collated across all trials (Fig. 4b). We identified that the likely cause was residual E. coli contamination of WaterScope’s reusable funnels, and so took multiple steps to improve the sterilisation process. With the second trial in Kawangware we updated our training procedure, highlighting the need for careful sterilisation and in particular stressing that ethanol sterilisation is most effective at ca. 80% concentration (users had previously been spray sterilising using 100% ethanol50). Subsequently in the third trial in Addis Ababa we additionally introduced a UVC sterilisation step as an extra safeguard against residual contamination. We found the combination of these two changes to be extremely effective, achieving an FP rate of just 5% in the Addis Ababa field trial. Similarly, performing ROC Area Under Curve (AUC) calculations (a metric that gives a combined measure of specificity and sensitivity), we find substantial improvements between each field trial (Fig. 4c). Through this process of participatory research and iterative development, we demonstrate that the WaterScope system meets the equivalent standard of existing conventional procedures not only in controlled settings but also in dynamic and challenging field environments.
Validation 4: User satisfaction, ML performance, cost analysis and versatility
In addition to the scientific validation of the system, the WaterScope system underwent a rigorous evaluation for its practical applicability in the field. This evaluation included a direct comparison with conventional portable membrane filtration (MF) kits, a comparative analysis between manual and machine-assisted colony counting and the system’s overall flexibility in addressing varied analytical challenges.
User satisfaction was quantified during a workshop in South Sudan in June 2022 that included participants with diverse levels of WASH experience (recent, previous, or none) across 16 different metrics detailed in the Supplementary Information (SI) section 8. The participants engaged in a self-guided training session using the manuals provided for each system, which was then followed by a hands-on training session. According to the Post-Study System Usability Questionnaire (PSSUQ) results shown in Fig. 5a, the WaterScope system achieved favourable usability scores among those who had previous experience with water testing systems. The workshop revealed an interesting nuance among participants without previous WASH experience: they faced challenges with the WaterScope system during the self-training phase due to the manual’s lack of detail when compared to conventional MF methods. Despite this hurdle, the preference for WaterScope became pronounced following the hands-on training sessions. Participants favoured WaterScope for its better accuracy, shorter hands-on time, straightforward teaching process, and overall trustworthiness. The outcomes of the workshop underscore the efficacy of the HCD approach employed in the design process of the WaterScope system and highlights the importance of robust training programmes, which can significantly improve the usability of WaterScope, especially for those new to water testing.
a Comparative assessment of user satisfaction between WaterScope and generic membrane filtration (MF) across varied experience levels in water, sanitation, and hygiene (WASH) sectors. b Correlation between manual and automated colony forming unit (CFU) counts, validating the machine learning (ML) based identification. c Versatility demonstrated through the development of prototypes for early contamination detection, urinary tract infection (UTI) analysis, and chemical assessments using colorimetric assays.
For automated identification accuracy, we validated the machine learning (ML) algorithm used by WaterScope for colony counting against manual counting methods. As depicted in Fig. 5b, the CFU counts from both methods are strongly correlated, indicating the reliability of the automated system in providing consistent and accurate measurements. Additionally, the integrated microscope facilitates early colony detection as it can detect smaller colonies (normally not seen by eye) and subtle colour changes. The field trials in Juba and Kawangware with 130 environmental samples indicated that WaterScope could reliably detect bacterial contamination within 8 h for 67% of the cases, demonstrating its potential for prompt responses in emergency situations (see Supplementary Information Section 7).
In our cost analysis, we compared the WaterScope system with existing portable water testing kits such as the WagTech kits and the CompactDry adapted field method, as well as variations of these methods. The analysis encompassed different campaign durations to assess cost efficiency across various operational scales. WaterScope’s pricing, at $1499 for the kit and $2.95 per test, was taken from WaterScope’s online store, the costs for the other methods were estimated based on Aquaya’s internal experience with suppliers. Our findings reveal that WaterScope is significantly more cost-effective than the equivalent WagTech portable kit with Nutridisks for E. coli specificity, offering 26% cost savings over a two-year campaign. It is a similar cost to CompactDry (WaterScope is $0.25 more expensive per test when logistics, labour and equipment costs are accounted for). Notably, a locally produced, open-source version of WaterScope would further reduce costs, making it substantially cheaper than both the WagTech and CompactDry methods. For detailed cost calculations and assumptions, please refer to the Supplementary Information 10 provided in this paper.
The versatility of the cartridge technology in the WaterScope system is further demonstrated by its adaptability to different applications, such as colorimetric assays. Utilising the integrated microscope, the system can perform various assays, including the DPD (N,N-diethyl-p-phenylenediamine) method for chlorine measurement and phenol red for pH testing (see SI section 9 for more information). Theoretically, it is capable of analysing any colorimetric assay, including those that utilise test strips. Moreover, with modifications to the slider and media components, the system can be repurposed for clinical applications, as illustrated in Fig. 5c(iii) where a modified slider is employed for detecting urinary tract infections and assessing pathogen antibiotic sensitivity. Each segment of the slider is loaded with the urine sample, nutrient media and an antibiotic, purple segment means there is bacterial growth unaffected by the antibiotic and yellow means that the bacteria are susceptible to the antibiotic.
Discussion
In summary, we have developed an accurate, portable and digital testing kit for drinking water quality. Through a series of validation experiments in both lab and field settings, we have established that the WaterScope kit achieves equivalent performance in quantifying E. coli when compared to conventional reference methods. However, by employing HCD principles, the WaterScope system incorporates significant innovation when compared to conventional methods. This includes a unique cartridge/slider mechanism, ML-aided results classification, complete digitalisation of results, and a carefully designed sterilisation process. These elements enable the WaterScope kit to provide fully quantified water analysis whilst also being easy-to-use, portable, cost-effective, fully digital, and environmentally conscious.
By sharing the system design as open-source, we aim to foster innovation in water testing. It is our hope that the open-source community will help to further improve the WaterScope system; whether this is through simplifying the building process, reducing costs, improving training procedures, or even adding new functionalities. The flexibility of a system capable of portable incubation and imaging in low-resource locations presents numerous possibilities. For example, we are presently validating cartridges that allow users to conduct colorimetric assays (e.g. chlorine, arsenic, heavy metals), and plan to develop further slider/media combinations for clinical uses. We also anticipate integrating the WaterScope web dashboard with established disease and water monitoring efforts such as SORMAS51 and mWater to further enhance connectivity and the system’s impact. Ultimately, in this work, we present an innovative method for water testing that aims to open new pathways toward achieving global water equality.
Methods
WaterScope system design
Here we provide a brief description of the composition and design of the WaterScope kit. Further details are available on request, from the WaterScope website or on our GitHub repository52.
The WaterScope system fits into a case of dimensions 29 cm (L) × 23 cm (W) × 26 (H) cm (rated for IP67 and IK08 protection) and weighs 5.8 kg when fully equipped.
One half of the case contains a combined imaging/incubation system. The incubation system comprises two racks (each with space for seven cartridges) suspended in vacuum flasks with temperature sensors (DS18B20) and 12 × 15 cm resistive (5 Ohm) heating pads. The vacuum flasks were selected due to their excellent insulation from the environment which reduces power consumption and allows the system to operate in extreme ambient temperatures. The incubators are controlled by an ATmega 328P microcontroller which runs a PID control loop to ensure that the incubator temperature is reached quickly and remains stable. The imaging system comprises a cartridge compartment and a 5 MP OV5647 CMOS camera with a motorised lens that allows for the system to focus on the sample and capture an image of the colonies. A ring of white LED lights for controlled illumination) and a resistive heating element for removing condensation from the sample lid complete the imaging system. An LCD screen/button display allows users to check the incubation temperature/time or run a sample analysis. The incubators, display, buttons and illumination ring are controlled by a microcontroller and the camera is controlled by a Raspberry Pi (3B+ or 4) computer. The Raspberry Pi allows for the ML sample analysis to run directly on it without any Internet connection and it was selected for its low cost and general availability. The Raspberry Pi is switched off by the microcontroller when the analysis is not run to preserve power. The imaging/incubation system is contained within a custom 3D-printed housing which has been through extensive vibration/shock/drop robustness validation.
The other half of the case contains all of the remaining components. A diaphragm pump (max 150 kPa) with a vacuum gauge is housed inside custom 3D-printed casing to minimise size. The filtration set comprises a stackable collection of two 100 ml funnels, a 200 ml collection cup, the filtration base, and a filtration manifold with inlets to filter either one or two cartridges simultaneously. The UV steriliser contains a custom UVC LED circuit board combined with a cooling fan, contained within another custom 3D-printed housing. Additional components include a 74 Wh battery, spray bottle, cleaning brush, and foam padding. Every electronic component in the system is powered by a USB-C connector and operates at 5 V which simplifies the operation and the kits adaptability in field environments. Our GitHub repository contains more details on each hardware component and a comprehensive 20-page assembly guide on how to put the parts together.
The cartridge37 has two intended slider positions: one for pumping (with ports that connect to the filtration set), and another for incubation/imaging (with a media reservoir on one side and an imaging window on the other). The slider can be moved between positions by clamping/unclamping flaps on both sides of the cartridge. The cartridge is machined from aluminium, though injection moulded/3D-printed cartridges also work well (but have a shorter lifespan). The slider frame is either inject-moulded or 3D-printed and contains a 13 mm diameter absorbent pad topped with a 0.45 µm pore-size cellulose acetate filter.
Our preferred medium is the same composition as CCA53 without the agar. It contains: sodium chloride (5 g L−1), disodium hydrogen phosphate (2.7 g L−1), sodium dihydrogen phosphate (2.2 g L−1), yeast extract (2 g L−1), casein digest (1 g L−1), sodium pyruvate (1 g L−1), sorbitol (1 g L−1), tryptophan (1 g L−1), salmon GAL (0.2 g L−1), Tergitol® 15-S-7 surfactant (0.15 g L−1), IPTG (0.1 g L−1) and X-glucuronide (0.1 g L−1).
WaterScope system operation
To perform a single water test using pre-sterilised components, there are five key steps for a user. First, a slider is inserted into the filtration position of a cartridge and clamped in place. Second, the cartridge is mounted onto the filtration manifold and a funnel placed on top. The desired quantity of water is poured into the funnel and the pump is connected to the power bank and switched on until all water has passed through the filter. Third, the cartridge is removed from the filtration apparatus and the slider is moved into the incubation position. Additionally, the media cap on the back of the cartridge is filled half-full (ca. 300 µL) with pre-prepared media. Fourth, the cartridge is placed into the incubator and left for the desired amount of time (8 h for rapid presence determination or 21 h for fully quantified results). Fifth, the cartridge is removed and placed into the imaging compartment. Then either via the screen/buttons on the device, or via the WaterScope Android app, the user initiates the imaging procedure and will receive results within ca. 2–3 min. The user may view the results of all their tests on the WaterScope dashboard where they can additionally manually correct any sample counts or add further sample information.
WaterScope sterilisation processes
Sterilisation procedures for the WaterScope kit can be split into ‘pre-excursion’ methods (i.e. to be prepared before heading into the field), and ‘field’ methods (i.e. carried out for each individual water test).
Pre-excursion sterilisation methods are used to prepare cartridges and media. Cartridges are rinsed and steam-sterilised during a 1-h steam/dry cycle, or alternatively can be boiled/dried for the same amount of time. We are working on validating the embedded UVC steriliser for quicker sterilisation of the cartridges. The WaterScope media comes pre-sterilised, and the preparation takes less than 1 minute as it only involves mixing a powder with a liquid – significant improvement from the lengthy (>20 min) preparation step for existing portable kits. Both dry medium and sliders are normally sterilised in bulk using ethylene oxide, or gamma irradiation. Media can additionally be sterilised by mixing the powder/water together and bringing to the boil in a microwave three times in a row.
Field sterilisation methods are used to sterilise the operator’s hands and to prepare funnels/collection cups between samples. Operators should use 80% ethanol spray to sterilise their hands before each sample, and also if sample water contacts their hands. Care should be taken to avoid touching the slider membrane when clamping, the inside of the funnel during filtration, or the inside of the cartridge during media addition. Funnels/collection cups should be thoroughly sprayed/wiped with 80% ethanol, paying attention to the interior and funnel tip. The funnel/collection cup should then be placed into the UVC steriliser for 1 min prior to use. The UVC steriliser can be used as a sterile surface during the test procedure.
WaterScope automated image classification
Images go through five stages of processing: cropping, CFU identification, CFU classification, outlier detection, and manual review. First, circular cropping is achieved through the Hough gradient method to restrict the original image to only the outline of the filter membrane. Second, we use a neural network built with YOLOv854, trained on over 30,000 expert-labelled CFU instances to recognise the outline of CFU’s on membrane filters. The model is applied to the image in small overlapping patches (416×416 pixels) to achieve a balance between speed and resolution. Third, colonies are assigned a classification (e.g., EC, OC, premature) based on their HSV colour profile. Fourth, k-means clustering is used to identify the dominant colours in an image. We find that an automated analysis of dominant colours can routinely flag anomalous images which are typically either too numerous to count (TNTC) or covered with sediment. Fifth, the original image and its classification results are uploaded to the WaterScope dashboard where users can manually check and modify the results using a custom VIA project55. Additional information on the automated image classification is provide in the Supplementary Information Section 2.
Validation study designs
In each study, counts of EC CFU were predicted by WaterScope’s classification software and manually verified by two independent experts; the average count from each expert was taken to produce a final count thus isolating ML algorithm performance from the actual microbiological recovery. For non-WaterScope methods (CCA, Colilert, MLSB, Compact Dry) we followed manufacturer-specified protocols and again counts were averaged from two independent expert reviewers.
In validation study 1, all samples used a 100 ml processing volume. For validation studies 2 and 3, sample volume was determined by an initial turbidity check; 100 ml was processed for turgidities ranging 0 NTU to 0.5 NTU, 50 ml up to 1.5 NTU, 20 ml up to 5 NTU, 5 ml up to 50 NTU, and 1 ml for over 50 NTU. Note this was relevant to WS, CCA and Compact Dry methods, whilst for Colilert 100 ml was used in every case. Calculation of CFU volume concentration was correspondingly adjusted according to sample volume (WS samples processed through the Android phone application allow sample volumes to be stored at the time of processing).
Validation study 1 comprised a total of 70 samples (14 repeats at five concentrations) for each of the four methods tested. Validation study 2 comprised 100 samples each for WS and CCA methods, and 71 samples for Colilert; a minimum of two repeats were performed on each sampling date for all three methods, but in some cases WS/CCA samples were acquired at multiple volumes to ensure countable results. Validation study 3 comprised 162 samples in Juba, 57 in Kawangware, and 82 in Addis Ababa for both WS and Compact Dry methods.
Samples in validation study 1 were prepared by diluting a concentrated stock of E. coli with all sample preparation carried out in sterile laboratory conditions. Samples for validation study 2 were collected using sterile sample dippers from the river bank, taking care to avoid disturbing sediment. Samples for validation study 3 were collected using either thio-bags for potentially chlorinated sources or the WaterScope collection cup for unchlorinated sources; the cup was sterilised between samples following the previously described protocol.
Statistical analyses
Linear regression was performed to assess the linearity between WS and reference methods in Figs. 2a and 3b. In Figs. 2b and 3d, we carry out a statistical equivalency test based on mean-difference plots, broadly following the protocol set out in ISO 1799446; the 95% confidence interval pertains to the standard deviation of the difference in log-10 mean counts multiplied by 1.96. Each validation study additionally uses the two-sided Spearman rank as a nonparametric measure of monotonicity between WS and reference methods. Figure 4c additionally shows an ROC plot for validation study 3; this was generated by calculating specificity and sensitivity from true positive, true negative, false positive, and false positive binary classifications at limits of detection varying stepwise from 1 to 20 (taking Compact Dry measurements as our reference for ‘True’ counts). Each of these calculations necessarily involves the pairing of individual WS and reference results; to avoid any bias, this pairing was performed randomly in cases where repeat measurements were taken.
Data availability
All results and associated plotting scripts generated during this study are openly available on Figshare at https://doi.org/10.6084/m9.figshare.25808815.v1. Additionally, all design files and software supporting this research are accessible on GitHub at https://github.com/WaterScope-Org.
Code availability
GitHub repository: https://github.com/WaterScope-Org.
References
WHO. Drinking Water Fact Sheet. https://www.who.int/news-room/fact-sheets/detail/drinking-water (2022).
Ritchie, H. & Roser, M. Clean Water and Sanitation. https://ourworldindata.org/water-access (2021).
Stanaway, J. D. et al. Global, regional, and national comparative risk assessment of 84 behavioural, environmental and occupational, and metabolic risks or clusters of risks for 195 countries and territories, 1990-2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 392, 1923–1994 (2018).
Wolf, J. et al. Burden of disease attributable to unsafe drinking water, sanitation, and hygiene in domestic settings: a global analysis for selected adverse health outcomes. Lancet 401, 2060–2071 (2023).
WHO & UNICEF. Joint Monitoring Programme for Water Supply, Sanitation and Hygiene. Preprint at https://washdata.org/ (2023).
UNICEF, JMP & WHO. Progress on Household Drinking Water, Sanitation and Hygiene 2000-2022. https://washdata.org/reports/jmp-2023-wash-households (2023).
UN. Sustainable development goal 6.1. United Nations Sustainable Development Preprint at https://sdgs.un.org/goals/goal6.
Ramesh, A., Blanchet, K., Ensink, J. H. J. & Roberts, B. Evidence on the effectiveness of water, sanitation, and hygiene (wash) interventions on health outcomes in humanitarian crises: a systematic review. PLoS One 10, e0124688 (2015).
Price, R. G. & Wildeboer, D. Escherichia coli - Recent Advances on Physiology, Pathogenesis and Biotechnological Applications (IntechOpen, 2017).
WHO. Guidelines for Drinking-Water Quality (World Health Organization, 2004).
Target Product Profile: Rapid water quality detection method or portable kit UNICEF Supply Division. Preprint at https://www.unicef.org/supply/documents/target-product-profile-rapid-water-quality-detection-method-or-portable-kit.
McCrady, M. H. The numerical interpretation of fermentation-tube results. J. Infect. Dis. 17, 183–212 (1915).
ISO 9308-2:2012 Water Quality — Enumeration of Escherichia Coli and Coliform Bacteria — Part 2: Most Probable Number Method. https://www.iso.org/standard/52246.html (2012).
Edberg, S. C. et al. National field evaluation of a defined substrate method for the simultaneous enumeration of total coliforms and Escherichia coli from drinking water: comparison with the standard multiple tube fermentation method. Appl. Environ. Microbiol. 54, 1595 (1988).
ISO 9308-1:2014 Water Quality — Enumeration of Escherichia Coli and Coliform Bacteria — Part 1: Membrane Filtration Method for Waters with Low Bacterial Background Flora. https://www.iso.org/obp/ui/#iso:std:iso:9308:-1:ed-3:v1:en (2014).
Nurliyana, M. R. et al. The detection method of Escherichia coli in water resources: a review. J. Phys. Conf. Ser. 995, 012065 (2018).
Bedell, E., Harmon, O., Fankhauser, K., Shivers, Z. & Thomas, E. A continuous, in-situ, near-time fluorescence sensor coupled with a machine learning model for detection of fecal contamination risk in drinking water: design, characterization and field validation. Water Res. 220, 118644 (2022).
Alonzo, L. F. et al. Rapid, sensitive, and low-cost detection of Escherichia coli bacteria in contaminated water samples using a phage-based assay. Sci. Rep. 12, 7741 (2022).
Mendes Silva, D. & Domingues, L. On the track for an efficient detection of Escherichia coli in water: a review on PCR-based methods. Ecotoxicol. Environ. Saf. 113, 400–411 (2015).
Manzanas, C. et al. Molecular testing devices for on-site detection of E. coli in water samples. Sci. Rep. 13, 4245 (2023).
Cheswick, R. et al. Comparing flow cytometry with culture-based methods for microbial monitoring and as a diagnostic tool for assessing drinking water treatment processes. Environ. Int. 130, 104893 (2019).
Tambi, A., Brighu, U. & Gupta, A. B. Methods for detection and enumeration of coliforms in drinking water: a review. Water Supply 0, 1 (2023).
Bain, R. et al. A summary catalogue of microbial drinking water tests for low and medium resource settings. Int. J. Environ. Res. Public Health 9, 1609–1625 (2012).
Peletz, R. et al. Why do water quality monitoring programs succeed or fail? A qualitative comparative analysis of regulated testing systems in sub-Saharan Africa. Int J. Hyg. Environ. Health 221, 907–920 (2018).
Bourne, S. User-Centred Design and Humanitarian Adaptiveness. (ALNAP Case Study, 2019).
Burton, J., Patel, D., Landry, G., Anderson, S. M. & Rary, E. Failure of the “Gold Standard”: the role of a mixed methods research toolkit and human-centered design in transformative WASH. Environ. Health Insights 15, 11786302211018392 (2021).
Lubis, P. Y., Shahri, B. & Ramirez, M. Human-Centered Design For Water, Sanitation, And Hygiene: Insights From Jakarta. https://doi.org/10.5380/8SDS2021.ART57 (2021).
Delaire, C. et al. How much will it cost to monitor microbial drinking water quality in Sub-Saharan Africa? Environ. Sci. Technol. 51, 5869–5878 (2017).
Kumpel, E. et al. From data to decisions: understanding information flows within regulatory water quality monitoring programs. NPJ Clean. Water 3, 1–11 (2020).
Droujko, J. & Molnar, P. Open-source self-made sensors show high potential in river research. Nat. Water 1, 758–759 (2023).
Langhorst, T. et al. Increased scale and accessibility of sediment transport research in rivers through practical, open-source turbidity and depth sensors. Nat. Water 1, 760–768 (2023).
Collins, J. T. et al. Robotic microscopy for everyone: the OpenFlexure microscope. Biomed. Opt. Express 11, 2447–2460 (2020).
Dufour, A. P., Strickland, E. R. & Cabelli, V. J. Membrane filter method for enumerating Escherichia coli. Appl Environ. Microbiol 41, 1152–1158 (1981).
Goetz, A., Tsuneishi, N., Kabler, P. W., Streicher, L. & Neumann, H. G. Application of molecular filter membranes to the bacteriological analysis of water [with discussion]. J Am Water Works Assoc 43, 943–984 (1951).
EPA. Method 1603: Escherichia Coli (E. Coli) in Water by Membrane Filtration Using Modified Membrane-Thermotolerant Escherichia Coli Agar (Modified MTEC). https://www.epa.gov/sites/default/files/2015-08/documents/method_1603_2009.pdf.
Water Quality Analysis in Emergency Situations. Oxfam Policy & Practice Preprint at https://policy-practice.oxfam.org/resources/water-quality-analysis-in-emergency-situations-126714/.
Patto, A. et al. Preprint at https://patents.google.com/patent/US20220073855A1/en?q=(waterscope)&inventor=patto&oq=waterscope+patto (2022).
Egli, A., Schrenzel, J. & Greub, G. Digital microbiology. Clin. Microbiol. Infect. 26, 1324 (2020).
Berg, S. et al. ilastik: interactive machine learning for (bio)image analysis. Nat. Methods 16, 1226–1232 (2019).
Khan, A. U. M., Torelli, A., Wolf, I. & Gretz, N. AutoCellSeg: robust automatic colony forming unit (CFU)/cell analysis using adaptive image segmentation and easy-to-use post-editing techniques. Sci. Rep. 8, 1–10 (2018).
Geissmann, Q. OpenCFU, a new free and open-source software to count cell colonies and other circular objects. PLoS ONE 8, e54072 (2013).
Moucka, M. et al. Performance of four bacterial cell counting apps for smartphones. J. Microbiol. Methods 199, 106508 (2022).
Shamash, M. & Maurice, C. F. OnePetri: accelerating common bacteriophage petri dish assays with computer vision. Phage 2, 224 (2021).
Stolze, N. et al. Automated image analysis with ImageJ of yeast colony forming units from cannabis flowers. J. Microbiol. Methods 164, 105681 (2019).
Stirling, D. R. et al. CellProfiler 4: improvements in speed, utility and usability. BMC Bioinform. 22, 1–11 (2021).
14:00-17:00. ISO 17994:2014 Water Quality — Requirements for the Comparison of the Relative Recovery of Microorganisms by Two Quantitative Methods. https://www.iso.org/standard/56617.html (2014).
DelAgua Water Testing Kits - DelAgua. Preprint at https://www.delagua.org/delagua-testing-kits/ (2021).
Alonso, J. L., Soriano, A., Carbajo, O., Amoros, I. & Garelick, H. Comparison and recovery of escherichia coli and thermotolerant coliforms in water with a chromogenic medium incubated at 41 and 44.5°C. Appl. Environ. Microbiol. 65, 3746 (1999).
Abia, A. L. K., Ubomba-Jaswa, E. & Momba, M. N. B. Impact of seasonal variation on Escherichia coli concentrations in the riverbed sediments in the Apies River, South Africa. Sci. Total Environ. 537, 462–469 (2015).
Sauerbrei, A. Bactericidal and virucidal activity of ethanol and povidone-iodine. Microbiologyopen 9, e1097 (2020).
Tom-Aba, D. et al. The Surveillance Outbreak Response Management and Analysis System (SORMAS): digital health global goods maturity assessment. JMIR Public Health Surveill. 6((2)), e15860 (2020).
WaterScope. WaterScope (UK) Preprint at https://www.waterscope.org/.
ISO 9308-1:2014 Water quality — Enumeration of Escherichia coli and coliform bacteria — Part 1: Membrane filtration method for waters with low bacterial background flora. Preprint at https://www.iso.org/obp/ui/#iso:std:iso:9308:-1:ed-3:v1:en (2014).
Jocher, G., Chaurasia, A., & Qiu, J. (2023). Ultralytics YOLO (Version 8.0.0) [Computer software] at https://github.com/ultralytics/ultralytics (2023).
Dutta, A. & Zisserman, A. The VIA Annotation Software for Images, Audio and Video. in Proceedings of the 27th ACM International Conference on Multimedia 2276–2279 (Association for Computing Machinery, 2019).
Metcalfe, C., Guppy, L. & Qadir, M. Global Barriers to Improving Water Quality: A Critical Review. (United Nations University Institute for Water, Environment and Health, 2017).
Pichel, N. et al. Field-testing solutions for drinking water quality monitoring in low-and middle-income regions and case studies from Latin American, African and Asian countries. J. Environ. Chem. Eng. 11, 111180 (2023).
Acknowledgements
The authors wish to express their sincere appreciation and extend their gratitude to the following organisations and individuals for their indispensable support and contributions to this research: Humanitarian Innovation Fund/Elrha: their substantial financial assistance and expert guidance have been crucial in the conceptualisation and execution of this study. Innovate UK: the funding and innovative insights provided by Innovate UK have been fundamental to the success and advancement of our project. Engineering and Physical Sciences Research Council (Grant Numbers: EP/L027151/1 and EP/P029426/1): we are indebted to the EPSRC for their significant financial support and the provision of essential resources that were pivotal in the early stages of our work. University of Cambridge: the authors are grateful for the academic support, extensive resources, and stimulating research environment provided by the University of Cambridge, which have collectively facilitated the successful completion of this research. Aquaya Institute: we would like to express our gratitude to the Aquaya Institute especially Caroline Delaire for conducting the field validations/usability workshops of our system in South Sudan and Kenya, and for helping with the data analysis. Academic collaborators, charities, makerspaces, citizen science groups, WASH professionals: for all their feedback, help and support, especially Action Against Hunger, Centre for Global Equality, UPES, Miranda House, Ifakara Health Institute, Triple-P, VigyanShaala, Oxfam, TearFund, SticLab, Life Science Factory. Residents of the Local Community: our heartfelt thanks are extended to the local residents, whose participation and invaluable insights have immeasurably enriched the scope and relevance of this study.
Author information
Authors and Affiliations
Contributions
Concept development: R.B., T.Z., N.P., A.P. and Sa.M.; Funding acquisition: A.D., So.M., A.P., M.A.H., J.J.B., N.P., R.B., T.Z. and Sa.M.; Study design: A.D., G.R.L., M.A., S.J.S., K.S., Li.A., La.A., A.P., M.A.H. and Sa.M.; Experimental work: A.D., A.P., G.R.L., A.E., M.A., S.J.S., E.L., K.S., Li.A. and Sa.M.; Software development: G.R.L., C.J., T.Z. and Sa.M.; Hardware development: C.H., A.P., A.E., J.S., R.B. and Sa.M.; Data analysis: A.D., G.R.L., K.S., Li.A., A.P., M.A.H. and Sa.M.; Manuscript writing: A.D., G.R.L. and Sa.M. in collaboration with all authors. A.D. and G.R.L. are joint first authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare the following Competing Financial Interests: Alexander Patto, Nalin Patel, Tianheng Zhao, Richard Bowman, and Sammy Mahdi are co-inventors on the WaterScope Cartridge patent (Patent Number: WO2020128480A2). There are no Competing Non-Financial Interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Dabrowska, A., Lewis, G.R., Atlabachew, M. et al. Expanding access to water quality monitoring with the open-source WaterScope testing platform. npj Clean Water 7, 68 (2024). https://doi.org/10.1038/s41545-024-00357-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41545-024-00357-y
This article is cited by
-
The relationship between concerns of local issues and water conservation behaviors: insights from Albuquerque, New Mexico, USA
Sustainable Water Resources Management (2025)