Abstract
The pathophysiology of acute viral diseases is complex. It is characterized by strong inflammatory responses driven by immune cells, leading to tissue damage. Currently available in vitro models mainly recapitulate the viral life cycle but fail to model immune cell-mediated pathogenesis. Here we build macrophage-augmented organoids (MaugOs) by integrating macrophages into primary organoids that are cultured from human liver tissues. We test the infections of two RNA viruses, hepatitis E virus and SARS-CoV-2, and one DNA virus, monkeypox virus, which either primarily or secondarily affect the human liver. In all three models of acute viral diseases, MaugOs recapitulate infection and the resulting inflammatory response, although to different levels. We use this system to dissect the multifunctional role of human bile on hepatitis E virus replication and the inflammatory response through distinct mechanisms of action. We also show that MaugOs recapitulate features of inflammatory cell death triggered by hepatitis E virus infection when integrated with pro-inflammatory macrophages. Furthermore, we demonstrate a proof of concept in MaugOs for development of multitarget therapeutics that simultaneously target the virus, inflammatory response and the resultant inflammatory cell death.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The main data supporting the results in this study are available within the paper and its Supplementary Information. RNA sequencing data are publicly available on Data Archiving and Networked Services (DANS) at https://doi.org/10.17026/LS/W57FUZ (ref. 70). Other raw and analysed datasets generated during the study are available for research purposes from the corresponding authors on reasonable request. Source data are provided with this paper.
References
Chiang, J. Y. L. & Ferrell, J. M. Bile acid metabolism in liver pathobiology. Gene Expr. 18, 71–87 (2018).
Ma, Z., de Man, R. A., Kamar, N. & Pan, Q. Chronic hepatitis E: advancing research and patient care. J. Hepatol. 77, 1109–1123 (2022).
Zhao, Y. et al. Mechanistic insight of SARS-CoV-2 infection using human hepatobiliary organoids. Gut 72, 216–218 (2023).
Luxenburger, H. & Thimme, R. SARS-CoV-2 and the liver: clinical and immunological features in chronic liver disease. Gut 72, 1783–1794 (2023).
Wang, Y. et al. SARS-CoV-2 infection of the liver directly contributes to hepatic impairment in patients with COVID-19. J. Hepatol. 73, 807–816 (2020).
Li, X., Fan, C., Tang, J. & Zhang, N. Meta-analysis of liver injury in patients with COVID-19. Medicine 102, e34320 (2023).
Muller, G. et al. Monkeypox virus in liver and spleen of child in Gabon. Lancet 1, 769–770 (1988).
Uphoff, C. C., Pommerenke, C., Denkmann, S. A. & Drexler, H. G. Screening human cell lines for viral infections applying RNA-Seq data analysis. PLoS ONE 14, e0210404 (2019).
Han, Y., Yang, L., Lacko, L. A. & Chen, S. Human organoid models to study SARS-CoV-2 infection. Nat. Methods 19, 418–428 (2022).
Puschhof, J., Pleguezuelos-Manzano, C. & Clevers, H. Organoids and organs-on-chips: insights into human gut-microbe interactions. Cell Host Microbe 29, 867–878 (2021).
Casanova, J. L. & Abel, L. Mechanisms of viral inflammation and disease in humans. Science 374, 1080–1086 (2021).
Merad, M. & Martin, J. C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 20, 355–362 (2020).
Ginhoux, F. & Guilliams, M. Tissue-resident macrophage ontogeny and homeostasis. Immunity 44, 439–449 (2016).
Mosser, D. M. & Edwards, J. P. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 8, 958–969 (2008).
Li, Y. et al. Seasonal coronavirus infections trigger NLRP3 inflammasome activation in macrophages but is therapeutically targetable. Antivir. Res. 216, 105674 (2023).
Paerewijck, O. & Lamkanfi, M. The human inflammasomes. Mol. Asp. Med. 88, 101100 (2022).
Coll, R. C., Schroder, K. & Pelegrin, P. NLRP3 and pyroptosis blockers for treating inflammatory diseases. Trends Pharmacol. Sci. 43, 653–668 (2022).
Marsee, A. et al. Building consensus on definition and nomenclature of hepatic, pancreatic, and biliary organoids. Cell Stem Cell 28, 816–832 (2021).
Cao, X., van den Hil, F. E., Mummery, C. L. & Orlova, V. V. Generation and functional characterization of monocytes and macrophages derived from human induced pluripotent stem cells. Curr. Protoc. Stem Cell Biol. 52, e108 (2020).
Wang, W. et al. The RNA genome of hepatitis E virus robustly triggers an antiviral interferon response. Hepatology 67, 2096–2112 (2018).
Beer, A. et al. Chronic hepatitis E is associated with cholangitis. Liver Int. 39, 1876–1883 (2019).
Li, P. et al. Recapitulating hepatitis E virus-host interactions and facilitating antiviral drug discovery in human liver-derived organoids. Sci. Adv. 8, eabj5908 (2022).
Steiner, S. et al. SARS-CoV-2 biology and host interactions. Nat. Rev. Microbiol. 22, 206–225 (2024).
Guan, J., Fan, Y., Wang, S. & Zhou, F. Functions of MAP3Ks in antiviral immunity. Immunol. Res. 71, 814–832 (2023).
Broutier, L. et al. Culture and establishment of self-renewing human and mouse adult liver and pancreas 3D organoids and their genetic manipulation. Nat. Protoc. 11, 1724–1743 (2016).
Hamming, I. et al. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 203, 631–637 (2004).
Shiratori, H. et al. THP-1 and human peripheral blood mononuclear cell-derived macrophages differ in their capacity to polarize in vitro. Mol. Immunol. 88, 58–68 (2017).
Legrand, C. et al. Lactate dehydrogenase (LDH) activity of the cultured eukaryotic cells as marker of the number of dead cells in the medium [corrected]. J. Biotechnol. 25, 231–243 (1992).
Chiang, J. Y. L. & Ferrell, J. M. Bile acid receptors FXR and TGR5 signaling in fatty liver diseases and therapy. Am. J. Physiol. Gastrointest. Liver Physiol. 318, G554–G573 (2020).
Yin, X., Li, X. & Feng, Z. Role of envelopment in the HEV life cycle. Viruses 8, 229 (2016).
Hanafi, N. I., Mohamed, A. S., Sheikh Abdul Kadir, S. H. & Othman, M. H. D. Overview of bile acids signaling and perspective on the signal of ursodeoxycholic acid, the most hydrophilic bile acid, in the heart. Biomolecules 8, 159 (2018).
Li, Y. et al. Hepatitis E virus infection activates NOD-like receptor family pyrin domain-containing 3 inflammasome antagonizing interferon response but therapeutically targetable. Hepatology 75, 196–212 (2022).
Wang, Y. et al. Combating pan-coronavirus infection by indomethacin through simultaneously inhibiting viral replication and inflammatory response. iScience 26, 107631 (2023).
Group, R. C. et al. Dexamethasone in hospitalized patients with Covid-19. N. Engl. J. Med. 384, 693–704 (2021).
Li, P. et al. Clinical features, antiviral treatment, and patient outcomes: a systematic review and comparative analysis of the previous and the 2022 mpox outbreaks. J. Infect. Dis. 228, 391–401 (2023).
Mitja, O. et al. Mpox in people with advanced HIV infection: a global case series. Lancet 401, 939–949 (2023).
Bruhn, P. J., Osterballe, L., Hillingso, J., Svendsen, L. B. & Helgstrand, F. Posttraumatic levels of liver enzymes can reduce the need for CT in children: a retrospective cohort study. Scand. J. Trauma Resusc. Emerg. Med. 24, 104 (2016).
Duarte-Neto, A. N. et al. Main autopsy findings of visceral involvement by fatal mpox in patients with AIDS: necrotising nodular pneumonia, nodular ulcerative colitis, and diffuse vasculopathy. Lancet Infect. Dis. 23, 1218–1222 (2023).
Li, P. et al. Mpox virus infection and drug treatment modelled in human skin organoids. Nat. Microbiol. 8, 2067–2079 (2023).
Li, P. et al. Mpox virus infects and injures human kidney organoids, but responding to antiviral treatment. Cell Discov. 9, 34 (2023).
Li, P. et al. Recapitulating infection, thermal sensitivity and antiviral treatment of seasonal coronaviruses in human airway organoids. eBioMedicine 81, 104132 (2022).
Lamers, M. M. & Haagmans, B. L. SARS-CoV-2 pathogenesis. Nat. Rev. Microbiol. 20, 270–284 (2022).
Vazquez-Armendariz, A. I. et al. Multilineage murine stem cells generate complex organoids to model distal lung development and disease. EMBO J. 39, e103476 (2020).
Vazquez-Armendariz, A. I. & Tata, P. R. Recent advances in lung organoid development and applications in disease modeling. J. Clin. Invest. 133, e170500 (2023).
Recaldin, T. et al. Human organoids with an autologous tissue-resident immune compartment. Nature 633, 165–173 (2024).
Huch, M. et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 160, 299–312 (2015).
Roos, F. J. M. et al. Human branching cholangiocyte organoids recapitulate functional bile duct formation. Cell Stem Cell 29, 776–794.e713 (2022).
Castejon-Ramirez, S., Pennington, J., Beene, H., Hysmith, N. & Ost, S. A case of neonatal monkeypox treated with oral tecovirimat. Pediatrics https://doi.org/10.1542/peds.2023-061198 (2023).
Yakubovsky, M. et al. Mpox presenting as proctitis in men who have sex with men. Clin. Infect. Dis. 76, 528–530 (2023).
Arfi, V. et al. Characterization of the early steps of infection of primary blood monocytes by human immunodeficiency virus type 1. J. Virol. 82, 6557–6565 (2008).
MacParland, S. A. et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat. Commun. 9, 4383 (2018).
Hakim, M. S. et al. The global burden of hepatitis E outbreaks: a systematic review. Liver Int. 37, 19–31 (2017).
Davis, H. E., McCorkell, L., Vogel, J. M. & Topol, E. J. Long COVID: major findings, mechanisms and recommendations. Nat. Rev. Microbiol. 21, 133–146 (2023).
Wong, A. C. et al. Serotonin reduction in post-acute sequelae of viral infection. Cell 186, 4851–4867.e4820 (2023).
Pan, Q. et al. Mobilization of hepatic mesenchymal stem cells from human liver grafts. Liver Transpl. 17, 596–609 (2011).
Ettayebi, K. et al. Replication of human noroviruses in stem cell-derived human enteroids. Science 353, 1387–1393 (2016).
Reese, V. C., Oropeza, C. E. & McLachlan, A. Independent activation of hepatitis B virus biosynthesis by retinoids, peroxisome proliferators, and bile acids. J. Virol. 87, 991–997 (2013).
Chhatwal, P. et al. Bile acids specifically increase hepatitis C virus RNA-replication. PLoS ONE 7, e36029 (2012).
Lloyd, G., Atkinson, T. & Sutton, P. M. Effect of bile salts and of fusidic acid on HIV-1 infection of cultured cells. Lancet 1, 1418–1421 (1988).
Winkler, E. S. et al. The intestinal microbiome restricts alphavirus infection and dissemination through a bile acid-type I IFN signaling axis. Cell 182, 901–918 e918 (2020).
Luo, L. et al. Chenodeoxycholic acid from bile inhibits influenza A virus replication via blocking nuclear export of viral ribonucleoprotein complexes. Molecules 23, 3315 (2018).
Kong, F., Saif, L. J. & Wang, Q. Roles of bile acids in enteric virus replication. Anim. Dis. 1, 2 (2021).
Haselow, K. et al. Bile acids PKA-dependently induce a switch of the IL-10/IL-12 ratio and reduce proinflammatory capability of human macrophages. J. Leukoc. Biol. 94, 1253–1264 (2013).
Arbel, R. et al. Nirmatrelvir use and severe Covid-19 outcomes during the Omicron surge. N. Engl. J. Med. 387, 790–798 (2022).
Desai, A. N. et al. Compassionate use of tecovirimat for the treatment of monkeypox infection. JAMA 328, 1348–1350 (2022).
Arias, C. A., Miller, W. R., Olsen, R., Gollihar, J. & Armstrong, A. The response of mpox-associated inflammatory syndrome to steroid therapy. Lancet Infect. Dis. 23, e323–e324 (2023).
de Almeida, L. et al. Identification of immunomodulatory drugs that inhibit multiple inflammasomes and impair SARS-CoV-2 infection. Sci. Adv. 8, eabo5400 (2022).
Malireddi, R. K. S. et al. Inflammatory cell death, PANoptosis, screen identifies host factors in coronavirus innate immune response as therapeutic targets. Commun. Biol. 6, 1071 (2023).
Gautam, A. et al. Necroptosis blockade prevents lung injury in severe influenza. Nature 628, 835–843 (2024).
Pan, Q. Host response to hepatitis E virus. Data Archiving and Networked Services https://doi.org/10.17026/LS/W57FUZ (2024).
Acknowledgements
Part of this work is supported by the COVID-19 Programme (number 50-56300-98-2201 to Q.P.) and a VIDI grant (number 91719300 to Q.P.) from the Netherlands Organisation for Health Research and Development (ZonMw). We thank the Allen Cell Collection for providing the H2B-GFP hiPSC line.
Author information
Authors and Affiliations
Contributions
Conceptualization: Q.P., L.J.W.v.d.L. and K.L. Methodology, visualization and data curation: K.L., Yining Wang, J.L., J.Z., A.M.G.d.S., Z.D., R.S., K.O.-V., P.P.C.B., D.M.O., F.v.d.H., T.T. and M.E.v.R. Formal analysis and investigation: M.M.A.V., Yijin Wang, H.L.A.J., I.A., C.S., M.P.P., C.M.O., P.L., O.M., A.N.D.-N., V.V.O., L.J.W.v.d.L. and Q.P. Project administration: K.L. and Q.P. Supervision: Q.P. and L.J.W.v.d.L. Writing: Q.P., K.L. and A.N.D.-N. Funding acquisition: Q.P.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Biomedical Engineering thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Construction and characterization of macrophage-augmented organoids.
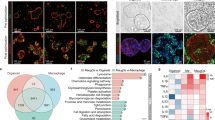
a, Immunofluorescence staining images of THP-1 monocytes and differentiated macrophages. CD68, a macrophage marker; DAPI, nuclei. b and c, Schematic illustration of isolation and representative images of primary macrophages during the differentiation process. The schematic was created in BioRender. van der Laan, L. (2025) https://BioRender.com/l3e28mw. d, Characterization of primary macrophages by the monocyte/macrophage markers. Expression of CD11b and CD14 was determined on living single CD45+ cells. e, Schematic overview of macrophage differentiation from GFP-labeled hiPSCs. The schematic was created in BioRender. van der Laan, L. (2025) https://BioRender.com/b94wwvt. f, Brightfield images of hiPSC-derived monocytes and macrophages. g, Characterization of hiPSC-derived macrophages by the monocyte/macrophage marker. Expression of CD14 was determined on living single CD45+ cells. h and i, Representative immunofluorescence images and quantification of hepatic macrophages (CD68) from different donors (Donor 1, n = 4; Donor 2, n = 4; Donor 3, n = 2, biological replicates). j and k, Immunofluorescence staining and quantification of individual organoids (from the same line) incorporated with THP-1 macrophages (M; labeled with CFSE; green), PBMC-derived primary macrophages (PriM; stained with CD68; green), and iPSC-derived macrophages (iPSM; tagged with GFP; green). Organoids were stained with EpCAM (red). Statistical analysis comparing pre- and post-optimized protocols was performed by Mann–Whitney U test. n = 12; ***P < 0.001. l, Gene expression analysis of MaugOs compared to their corresponding macrophage types: HM MaugO compared to hepatic macrophages (three donors), MaugO compared to THP-1 macrophages (n = 3, biological replicates), and iPS MaugO compared to iPSC-derived macrophages (n = 4, biological replicates). All gene expression thresholds were set to 1.
Extended Data Fig. 2 HEV infection in patient liver and responses to HEV infection in macrophages, organoids, and MaugOs.
a, Histology and pathology analysis in the liver tissue of a patient with HEV infection. Red circles indicate potentially affected hepatocytes and infiltrated macrophages, and green circle indicates possible involvement of cholangiocytes alone with infiltrated macrophages (left). Immunohistochemical staining of HEV ORF2 capsid protein (right). The schematic was created in BioRender. van der Laan, L. (2025) https://BioRender.com/preew29. b, Immunofluorescence staining to detect possible HEV infection in macrophages in liver tissues from three hepatitis E patients. Liver tissue from a patient without HEV infection served as a control. Macrophage marker CD68 (red), HEV ORF2 capsid protein (green), nuclear staining (blue). c, Quantification of HEV ORF2 and CD68 double-positive cells within the CD68 population in patient liver tissues. d, Confocal imaging of viral double-stranded RNA (dsRNA) in HEV infected organoids integrated with THP-1 macrophages for 48 hours. Images from the same view but different layer of Fig. 3b. e, Correlation matrix of transcriptomic profiles of HEV infected organoids, macrophages and MaugOs (HEVO, HEVM, and HEVOM) (n = 3, biological replicates). A value of 1 represents complete correlation, and a value of 0 represents no significant correlation. f-h, KEGG pathway analysis showing top 14 significantly regulated pathways for HEV infected macrophages (M), organoids (ICO), and MaugOs in comparison with their respective uninfected control groups (n = 3, biological replicates). Commentary to Fig. 3e.
Extended Data Fig. 3 Responses to HEV infection in macrophages, organoids, and MaugOs.
a and b, Quantification of inflammatory gene expression in THP-1 macrophages (n = 4) or organoids (n = 4) upon HEV infection for indicated time points. Commentary to Fig. 3e. The exact p value * = 0.028. c, Schematic illustration of experimental design. Cholangiocyte organoids or hepatic differentiated organoids were first integrated with THP-1 macrophages for 24 hours to form MaugOs or Hep MaugOs, and then inoculated with HEV particles. Creaded in BioRender. van der Laan, L. (2025) https://BioRender.com/ulh914d. d, HEV infection kinetics in organoids, macrophages, or MaugOs (n = 6). e, Quantification of IL-1β and TNF-α cytokines in supernatant of organoids (ICO), macrophages (M), or MaugOs (n = 4). f, Brightfield and immunofluoresent imaging of hepatic differentiated organoids. Albumin (red): hepatocyte marker; DAPI (blue): nuclear staining. g, qRT-PCR quantification of the expression of stem cell marker (LGR5) and hepatic markers (albumin/ALB and glucose-6-phosphatase catalytic subunit/G6PC) in undifferentiated (ICO) and hepatic differentiated ICOs (HepO) (n = 4). The exact p value * = 0.028. h, Relative levels of HEV RNA (12 h normalized as 1) in hepatic differentiated organoids upon infection (n = 4). i, Quantification of HEV RNA level in MaugOs and Hep MaugOs 24 hours after HEV infection for 24 hours (n = 4). j, Production of IL-1β and TNF-α cytokines in supernatant of hepatic differentiated organoids (HepO), and Hep MaugOs upon HEV infection for 24 hours (n = 4). k and l, Relative levels of HEV RNA (12 h normalized as 1) and the expression of inflammatory genes quantified by qRT-PCR in HEV infected hiPSC-derived macrophages (n = 4). The exact p value * = 0.028. m, Quantification of HEV viral RNA in hepatic macrophages derived from perfusates of two donors inoculated with HEV for indicated time points (n = 4). Hepatic macrophages inoculated with HEV for 12 hours set as control (normalized as 1; each donor normalized to the corresponding control). n, Quantification of inflammatory gene expression in hepatic macrophages inoculated with HEV for 12 hours (three donors) and 24 hours (four donors). Uninfected groups served as control (normalized as 1). o, Quantification of inflammatory gene expression in HEV infected organoids integrated with hepatic macrophages (from two donors) for 12 or 24 hours. Uninfected MaugOs served as control (normalized as 1). (a, b, e, g, h, i, j k and l) Data are mean ± SD. *p < 0.05; **p < 0.01; Two-tailed Mann-Whitney U test. Exact p values are labelled, ‘n’ refers biological replicates.
Extended Data Fig. 4 SARS-CoV-2 infection and inflammatory response in macrophage-augmented organoids, related to Fig. 2.
a and b, Confocal imaging of SARS-CoV-2 infected organoids(SARS-CoV-2 ICO) and SARS-CoV-2 infected organoids integrated with THP-1 macrophages (SARS-CoV-2 MaugO). Yellow arrows point to SARS-CoV-2 transmission from infected organoids to macrophages based on positive for viral dsRNA or N-protein. Macrophages were pre-labeled by CFSE (green) in a. EpCAM marks epithelial (organoid) cells. c, Quantification of SARS-CoV-2 RNA level in infected organoids integrated with THP-1 macrophages for 48 hours. The MaugOs without infection served as control (CTR). d and e, Quantification of inflammatory gene expression (n = 6, biological replicates) and IL-β secretion (n = 6, biological replicates) into supernatant in SARS-CoV2 infected organoids integrated with THP-1 macrophages for 48 hours. Uninfected MaugOs served as control (CTR), and normalized as 1 in panel d. Data are mean ± SD. *p < 0.05; **p < 0.01; Two-tailed Mann-Whitney U test. Exact p values are labelled.
Extended Data Fig. 5 Characterizing macrophages and the response to treatment.
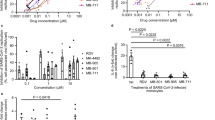
a, Gating strategies for flow cytometry analysis of macrophage markers. b, Expression of M1 markers (HLA DR, CD80) and M2 markers (DC-SIGN, CD163) in undifferentiated THP-1 monocytes, and M0, M1, M2 macrophages derived from THP-1 monocytes. c, Representative images of macrophage subtypes. d, The effects of the NF-κB inhibitor (BAY11-7085, Bay11), caspae-1 inhibitor (VX-765;VX) and NLRP3 inhibitor (MCC950; MCC) on IL-6, TNF-α, CCL2 and IL-10 RNA levels quantified by qRT-PCR in HEV infected MaugOs (n = 4), complementary to Fig. 6c. e, IL-10 cytokine production quantified by ELISA (n = 4),), complementary to Fig. 6i. f and g, Quantification of IL-1β and TNF-α gene expression in HEV infected MaugOs treated with ribavirin (RBV), dexamethasone (DEX) or the combination (n = 4), complementary to Figs. 6k and 6l. h, Quantification of IL-10 RNA and protein levels (n = 4). i, IL-10 cytokine production in HEV infected MaugOs (M1) treated with ribavirin, dexamethasone (DEX) or the combination (n = 4), complementary to Fig. 6o. (d-i) Data are mean ± SD. *p < 0.05; **p < 0.01; Two-tailed Mann-Whitney U test. Exact p values are labelled, ‘n’ refers biological replicates.
Extended Data Fig. 6 Human bile on HEV infection and inflammatory response.
a, Schematic illustration of HEV subgenomic replication model and HEV full-length infectious model with bile treatment. Creaded in BioRender. van der Laan, L. (2025) https://BioRender.com/ixrevi5. b, The effects of bile acids (100 μM CDCA or 100 μM CA) treatment for two days on organoids (ICO) harboring the HEV subgenomic replicon (n = 6), complementary to Fig. 5d. c, Brightfield images of 1% bile treatment for two days on organoids. d, Cell viability of organoids treated with indicated concentrations of human bile for two days (n = 6). e, Schematic illustration of bile treatment on HEV infected THP-1 macrophages. Creaded in BioRender. van der Laan, L. (2025) https://BioRender.com/p4z7u7z. f, Cell viability of THP-1 macrophages treated with indicated concentrations of bile for one day (n = 6). g and h, Quantification of HEV viral RNA (n = 4) and inflammatory gene expression in HEV infected macrophages treated with 1% bile for one day (IL6, n = 4; other genes, n = 6, biological replicates). i, The schemaitic of experiment design was creaded in BioRender. van der Laan, L. (2025) https://BioRender.com/ic5d3mq. j, Quantification of gene expression of IL-β, IL-6, TNF-α and IL-10 in 1 μg/mL LPS incubated MaugOs treated with bile (n = 8) or co-treated with ZGG for one day (n = 4), complementary to Figs. 5k and 5l. Data are mean ± SD. *p < 0.05; **p < 0.01; ***p < 0.01; Two-tailed Mann-Whitney U test. Exact p values are labelled, ‘n’ refers biological replicates.
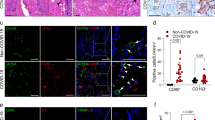
Extended Data Fig. 7 Characterizing MPXV infection in the liver of a patient and in organoids based models.
a, Image of the liver from a patient died from disseminated mpox. Here we specifically focused on liver involvement in one case who developed MPXV-hepatitis. In the liver, we observed steatosis, diffuse congestion, focal subcapsular hemorrhages and scattered lobular nodules. b, Histology (H&E) of the liver section from a patient without MPXV infection (as a negative control). c, Immunohistochemical staining against the MPXV antigens showed negative signal in this negative control liver tissue, indicating the specificity of the antibody. d, Biliary duct exhibiting cholangiocytes with pykontic nuclei and hypereosinophiliccytoplasm (H&E). e, Focal MPXV-hepatitis (arrow), and the adjacent hepatocytes showing reduced expression of E-cadherin by immunohistochemical staining. f, Immunohistochemical staining of bacterial lipopolysaccharide (LPS) detected bacterial fragments in the cytoplasm of Kupffer cells (black arrows) or entire bacilli in the sinusoids (red arrows). g, Confocal imaging of MPXV infection in ICOs complementary to Fig. 7e. EpCAM (yellow): epithelial marker; dsRNA (red): MPXV double-stranded RNA; DAPI (blue): nuclear staining. h, Quantification of MPXV genomic DNA in the supernatant harvested from infected adult ICOs (O), hepatic differentiated ICOs (HepO), and fetal ICOs (FetalO) 4 days post-inoculation (n = 4, biological replicates), complementary to Fig. 7j. i, Confocal imaging shows MPXV infection in macrophages of MaugOs constructed from hepatic differentiated organoids, complementary to Fig. 8b. CD68 (red): macrophage marker; MPXV (green): staining against the virions; DAPI (blue): nuclear staining. j, Confocal imaging of uninfected macrophages (M) and ICOs infected with MPXV for 2days, and then integrated with macrophages for 24 hours (MPXV MaugO). CK7 (red): marker of biliary epithelium; MPXV (green): staining against the virions; DAPI (blue): nuclear staining. k, Quantification of MPXV viral RNA in MaugOs constructed by different approaches (n = 4, biological replicates). ICOs were infected with MPXV for 2 or 5 days, and then integrated with macrophages for 24 or 48 hours. Relating to Fig. 8h, ICOs were infected with MPXV for 5 days, and then integrated with macrophages for 24 or 48 hours.; Two tailed Mann-Whitney U test. Exact p values are labelled. l, The expression of inflammatory genes in ICOs infected with MPXV for 2 days, and then integrated with macrophages for 24 hours (n = 4). (h, k, i) Data are mean ± SD. *p < 0.05; **p < 0.01.
Supplementary information
Supplementary Information
Supplementary Tables 1–3 and Videos 1–3.
Supplementary Video 1
Supplementary Video 1.
Supplementary Video 2
Supplementary Video 2.
Supplementary Video 3
Supplementary Video 3.
Source data
Source Data Figs. 4 and 6
Unprocessed western blots.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, K., Wang, Y., Li, J. et al. Macrophage-augmented organoids recapitulate the complex pathophysiology of viral diseases and enable development of multitarget therapeutics. Nat. Biomed. Eng 9, 1848–1868 (2025). https://doi.org/10.1038/s41551-025-01417-5
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41551-025-01417-5
This article is cited by
-
Status and outlook of mRNA therapeutics for viral diseases
EMBO Molecular Medicine (2026)
-
AI-driven discovery of antiretroviral drug bictegravir and etravirine as inhibitors against monkeypox and related poxviruses
Communications Biology (2025)