Abstract
SARS-CoV-2 infection of human cells is initiated by the binding of the viral Spike protein to its cell-surface receptor ACE2. We conducted a targeted CRISPRi screen to uncover druggable pathways controlling Spike protein binding to human cells. Here we show that the protein BRD2 is required for ACE2 transcription in human lung epithelial cells and cardiomyocytes, and BRD2 inhibitors currently evaluated in clinical trials potently block endogenous ACE2 expression and SARS-CoV-2 infection of human cells, including those of human nasal epithelia. Moreover, pharmacological BRD2 inhibition with the drug ABBV-744 inhibited SARS-CoV-2 replication in Syrian hamsters. We also found that BRD2 controls transcription of several other genes induced upon SARS-CoV-2 infection, including the interferon response, which in turn regulates the antiviral response. Together, our results pinpoint BRD2 as a potent and essential regulator of the host response to SARS-CoV-2 infection and highlight the potential of BRD2 as a therapeutic target for COVID-19.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Sequencing data are available from the NCBI Gene Expression Omnibus (GEO) with the following accession numbers: GSE165025 (RNA-sequencing data associated with Fig. 4), GSE182993 (CUT&RUN data associated with Fig. 5) and GSE182994 (RNA-sequencing data associated with Fig. 6f–h). Previously published BRD2 ChIP-seq data that were re-analysed here are available under accession codes GSE113714 and GSE104481. All other data supporting the findings of this study are available from the corresponding author on reasonable request. Source data are provided with this paper.
Code availability
Analysis of the CRISPRi screen results was carried out using custom code (MAGeCK-iNC) developed in the Kampmann laboratory. This has been described previously49 and is freely available at https://kampmannlab.ucsf.edu/mageck-inc.
References
Ziegler, C. G. K. et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell 181, 1016–1035 (2020).
Chu, H. et al. Comparative replication and immune activation profiles of SARS-CoV-2 and SARS-CoV in human lungs: an ex vivo study with implications for the pathogenesis of COVID-19. Clin. Infect. Dis. 71, 1400–1409 (2020).
Gutiérrez-Chamorro, L. et al. SARS-CoV-2 infection suppresses ACE2 function and antiviral immune response in the upper respiratory tract of infected patients. Preprint at bioRxiv https://doi.org/10.1101/2020.11.18.388850 (2020).
Blanco-Melo, D. et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 181, 1036–1045 (2020).
Bastard, P. et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science 370, eabd4585 (2020).
Hadjadj, J. et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 369, 718–724 (2020).
Zhang, Q. et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 370, eabd4570 (2020).
Samuel, R. M. et al. Androgen signaling regulates SARS-CoV-2 receptor levels and is associated with severe COVID-19 symptoms in men. Cell Stem Cell 27, 876–889 (2020).
Daniloski, Z. et al. Identification of required host factors for SARS-CoV-2 infection in human cells. Cell 184, 92–105 (2020).
Wang, R. et al. Genetic screens identify host factors for SARS-CoV-2 and common cold coronaviruses. Cell 184, 106–119.e14 (2020).
Schneider, W. M. et al. Genome-scale identification of SARS-CoV-2 and pan-coronavirus host factor networks. Cell 184, 120–132 (2021).
Wei, J. et al. Genome-wide CRISPR screens reveal host factors critical for SARS-CoV-2 infection. Cell 184, 76–91 (2020).
Shi, J. & Vakoc, C. R. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol. Cell 54, 728–736 (2014).
Fujisawa, T. & Filippakopoulos, P. Functions of bromodomain-containing proteins and their roles in homeostasis and cancer. Nat. Rev. Mol. Cell Biol. 18, 246–262 (2017).
Lui, I. et al. Trimeric SARS-CoV-2 Spike interacts with dimeric ACE2 with limited intra-Spike avidity. Preprint at bioRxiv https://doi.org/10.1101/2020.05.21.109157 (2020).
Lan, J. et al. Structure of the SARS-CoV-2 Spike receptor-binding domain bound to the ACE2 receptor. Nature 581, 215–220 (2020).
Chua, R. L. et al. COVID-19 severity correlates with airway epithelium–immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 38, 970–979 (2020).
Tseng, C.-T. K. et al. Apical entry and release of severe acute respiratory syndrome-associated coronavirus in polarized Calu-3 lung epithelial cells. J. Virol. 79, 9470–9479 (2005).
Kuchi, S., Gu, Q., Palmarini, M., Wilson, S. J. & Robertson, D. L. Meta-analysis of virus-induced host gene expression reveals unique signatures of immune dysregulation induced by SARS-CoV-2. Preprint at bioRxiv https://doi.org/10.1101/2020.12.29.424739 (2020).
Gilbert, L. A. et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451 (2013).
Gilbert, L. A. et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159, 647–661 (2014).
Horlbeck, M. A. et al. Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. eLife 5, e19760 (2016).
Deffieu, M. S. et al. Rab7-harboring vesicles are carriers of the transferrin receptor through the biosynthetic secretory pathway. Sci. Adv. 7, 1–17 (2021).
Doroshow, D. B., Eder, J. P. & LoRusso, P. M. BET inhibitors: a novel epigenetic approach. Ann. Oncol. 28, 1776–1787 (2017).
Xu, Y. & Vakoc, C. R. Targeting cancer cells with BET bromodomain inhibitors. Cold Spring Harb. Perspect. Med. 7, a026674 (2017).
Filippakopoulos, P. et al. Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010).
Faivre, E. J. et al. Selective inhibition of the BD2 bromodomain of BET proteins in prostate cancer. Nature 578, 306–310 (2020).
Winter, G. E. et al. BET bromodomain proteins function as master transcription elongation factors independent of CDK9 recruitment. Mol. Cell 67, 5–18 (2017).
Shi, C. et al. PROTAC induced-BET protein degradation exhibits potent anti-osteosarcoma activity by triggering apoptosis. Cell Death Dis. 10, 815 (2019).
Bermejo, J. A. P. et al. SARS-CoV-2 infection of human iPSC derived cardiac cells reflects cytopathic features in hearts of patients with COVID-19. Sci. Transl. Med. 13, 1–15 (2021).
Mulay, A. et al. SARS-CoV-2 infection of primary human lung epithelium for COVID-19 modeling and drug discovery. Cell Rep. 35, 109055 (2021).
Gordon, D. E. et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583, 459–468 (2020).
Skene, P. J. & Henikoff, S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 6, e21856 (2017).
Handoko, L. et al. JQ1 affects BRD2-dependent and independent transcription regulation without disrupting H4-hyperacetylated chromatin states. Epigenetics 13, 410–431 (2018).
Wang, S. et al. Target analysis by integration of transcriptome and ChIP-seq data with BETA. Nat. Protoc. 8, 2502–2515 (2013).
Au-Yeung, N. & Horvath, C. M. Histone H2A.Z suppression of interferon-stimulated transcription and antiviral immunity is modulated by GCN5 and BRD2. iScience 6, 68–82 (2018).
Robinot, R. et al. SARS-CoV-2 infection induces the dedifferentiation of multiciliated cells and impairs mucociliary clearance. Nat. Commun. 12, 4354 (2021).
Osterrieder, N. et al. Age-dependent progression of SARS-CoV-2 infection in Syrian hamsters. Viruses 12, 94301 (2020).
Imai, M. et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl Acad. Sci. USA 117, 16587–16595 (2020).
Sia, S. F. et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature 583, 834–838 (2020).
Rosenke, K. et al. Defining the Syrian hamster as a highly susceptible preclinical model for SARS-CoV-2 infection. Emerg. Microbes Infect. 9, 2673–2684 (2020).
Qiao, Y. et al. Targeting transcriptional regulation of SARS-CoV-2 entry factors ACE2 and TMPRSS2. Proc. Natl Acad. Sci. USA 118, e2021450118 (2020).
Gilham, D. et al. Bromodomain and extraterminal protein inhibitor, apabetalone (RVX-208), reduces ACE2 expression and attenuates SARS-CoV-2 infection in vitro. Biomedicines 9, 437 (2021).
Lee, H. K., Jung, O. & Hennighausen, L. JAK inhibitors dampen activation of interferon-stimulated transcription of ACE2 isoforms in human airway epithelial cells. Commun. Biol. 4, 654 (2021).
Mills, R. J. et al. BET inhibition blocks inflammation-induced cardiac dysfunction and SARS-CoV-2 infection. Cell 184, 2167–2182 (2021).
Ribero, M. S., Jouvenet, N., Dreux, M. & Nisole, S. Interplay between SARS-CoV-2 and the type I interferon response. PLoS Pathog. 16, e1008737 (2020).
Lei, X. et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 11, 3810 (2020).
Xia, H. et al. Evasion of type I interferon by SARS-CoV-2. Cell Rep. 33, 108234 (2020).
Tian, R. et al. CRISPR interference-based platform for multimodal genetic screens in human iPSC-derived neurons. Neuron 104, 239–255 (2019).
Stoner, R., Maures, T. & Conant, D. Methods and systems for guide RNA design and use US patent application 16/418,893 (2019).
Hsiau, T. et al. Inference of CRISPR edits from Sanger trace data. Preprint at bioRxiv https://doi.org/10.1101/251082 (2019).
Adamson, B. et al. A multiplexed single-cell CRISPR screening platform enables systematic dissection of the unfolded protein response. Cell 167, 1867–1882 (2016).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017).
Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA 95, 14863–14868 (1998).
Saldanha, A. J. Java Treeview—extensible visualization of microarray data. Bioinformatics 20, 3246–3248 (2004).
Skene, P. J., Henikoff, J. G. & Henikoff, S. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat. Protoc. 13, 1006–1019 (2018).
Meers, M. P., Tenenbaum, D. & Henikoff, S. Peak calling by sparse enrichment analysis for CUT&RUN chromatin profiling. Epigenet. Chromatin 12, 42 (2019).
Acknowledgements
We thank members of the Kampmann, Vignuzzi and Conklin laboratories, as well as V. Ramani, D. Ruggero, M. Ott and her laboratory and other members of the UCSF QBI Coronavirus Research Group (QCRG) for helpful discussions. We thank K. Leng for feedback on the manuscript. We acknowledge the Gladstone Stem Cell Core for help with cardiomyocyte production. A.J.S. is supported by NIH grant F32AG063487. G.N.R. is supported by the NSF Graduate Research Fellowship Program (GRFP) and UCSF Discovery Fellowship. S.A.L. was a Merck Fellow of the Helen Hay Whitney Foundation. I.L. was supported by an NSF GRFP award. M.K. is a Chan Zuckerberg Biohub Investigator.
Author information
Authors and Affiliations
Contributions
R.T., A.J.S. and M.K. conceptualized the overall project, analysed the results and prepared the manuscript, with input from all co-authors. V.V.R., A.M.K. and Q.D.T. performed and analysed live-virus experiments in Calu-3 cells, with guidance from M.V. R.R. performed and analysed human nasal epithelia experiments with guidance from L.A.C. L.C. performed and analysed the Syrian hamster experiments, with guidance from B.R.T. G.N.R. and S.J.R. performed and analysed the experiments with cardiomyocytes, with guidance from B.R.C. R.T., A.J.S., M.C. and X.G. performed and analysed all other experiments, with guidance from M.K. J.W. performed and analysed basal interferon signalling knockdown experiments in Calu-3 cells, with guidance from R.T. N.L. analysed the QuantSeq data, with guidance from R.T. S.A.L., I.L. and J.A.W. generated Spike-RBD. J.K.N. and J.S.W. generated the Calu-3 CRISPRi cell line. J.C.-S., J.O., T.M. and K.H. designed and provided sgRNAs to generate the ACE2 knockout cell line.
Corresponding authors
Ethics declarations
Competing interests
J.C.-S., J.O., T.M. and K.H. are employees and shareholders of Synthego Corporation. All other authors declare no competing interests.
Peer review information
Nature Cell Biology thanks Andrew Bowie, Ke Lan and the other, anonymous, reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
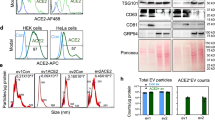
Extended Data Fig. 1 Calu-3 cells bind Spike-RBD specifically and were engineered to express CRISPRi machinery enabling CRISPRi screening.
a, Spike-RBD binding in different cell types at 20 nM and 200 nM Spike-RBD was quantified by flow cytometry. b, Expression of CRISPRi machinery (dCas9-BFP-KRAB) in the CRISPRi Calu-3 line indicated by the expression of BFP by flow cytometry. C, Enrichment of sgRNAs targeting specific genes (coloured dots) or non-targeting control sgRNAs plotted against the negative log of the P-value with a FDR of 0.1 shown (dashed lines).
Extended Data Fig. 2 Individual sgRNA re-test of screening hits.
a-i, Spike-RBD signal measured by flow-cytometry as a function of Spike-RBD concentration. Blue lines represent cells expressing the sgRNA targeting the gene of interest, black lines represent un-transduced control cells in the same well. Average of two technical replicates are shown.
Extended Data Fig. 3 BRD2 is effectively knocked down by CRISPRi.
Western blot for BRD2 and the loading control GAPDH in CRISPRi Calu-3 cells expressing no sgRNA or sgRNAs targeting ACE2 or BRD2. Three lanes represent samples from three independent wells.
Extended Data Fig. 4 Non-toxic concentration range of BRD2 inhibitors.
a, Calu-3 cells were treated with vehicle or the indicated concentrations of JQ1 or ABBV-744 for 5 days. Cell viability was then assayed with CellTiter-Glo 2.0 to calculate viability. Error bars represent the standard deviation of four biological replicates. Treatments are relative to untreated cells. b, Human iPSC-derived cardiomyocytes were treated for 72 hours with vehicle or the indicated concentrations of JQ1 or ABBV-744, and the percentage of dead cells was quantified as the ratio of propidium iodide-positive cells (dead cells) over Hoechst-positive cells (all cells). Error bars represent the standard deviation of three biological replicates (six biological replicates for the vehicle condition). c, Primary human bronchial epithelial (NHBE) cells were treated with ABBV-744 at the indicated concentrations for 72 hours and toxicity was assessed using CellTiter-Glo 2.0. Error bars represent the standard deviation of four biological replicates. P-values determined using Mann-Whitney two-tailed test. Treatments are relative to vehicle cells.
Extended Data Fig. 5 Validation of knockdown of interferon regulators by CRISPRi.
a-c, Calu-3 cells expressing sgRNAs knocking down genes essential for interferon signal transduction assayed for transcript levels of sgRNA targets relative to ACTB by qPCR. mRNA levels are fraction of control sgRNA. Error is the standard deviation of three biological replicates.
Extended Data Fig. 6 Viral replication in apical supernatants of reconstructed human nasal epithelia cultures and bodyweight of hamsters throughout the course of SARS-CoV-2 infection.
a-c, Calu-3 cells expressing sgRNAs knocking down genes essential for interferon signal transduction assayed for transcript levels of sgRNA targets relative to ACTB by qPCR. mRNA levels are fraction of control sgRNA. Error is the standard deviation of three biological replicates. A, Apical supernatants of either infected or mock-infected nasal epithelia treated with ABBV-744 at the indicated concentrations or not treated (NT) were isolated and assayed for SARS-CoV-2 N RNA content. Average of four biological quadruplicates are shown with error bars representing the standard deviation. b, Hamsters were weighed over the course of SARS-CoV-2 infection and weights were plotted as a percent of bodyweight on the day of infection. Inset, zoom in on body weight percent between 85 and 110 percent.
Supplementary information
Supplementary Information
Flow cytometry gating strategy, associated with Fig. 1.
Supplementary Table 1
Results from CRISPRi screens for Spike-RBD and anti-TFRC binding were analysed by the MAGeCK-iNC pipeline (see Methods for details) and are listed for all genes targeted by the H1 sgRNA library. Columns are: targeted gene, targeted transcription start site, knockdown phenotype (epsilon), P value, and gene score.
Supplementary Table 2
The first six tabs show the results of differential gene expression analyses for ACE2 knockdown, ABBV-744 treatment, BRD2 knockdown, JQ1 treatment, SARS-CoV-2 protein E overexpression and COMP knockdown, respectively, using edgeR (see Methods for details). Columns are: gene symbol, log2-fold change, log2 counts per million, F value, P value and FDR by the Benjamini–Hochberg method. The “TPM” tab shows the raw transcripts per million (TPM) values for all samples. Columns: treatment conditions with two replicates each. Rows: all genes in the human transcriptome reference. The last tab provides the numerical values underlying the heatmap in Fig. 4a. Columns: treatment conditions. Rows: genes that are among top 50 differentially expressed genes in any of the conditions.
Supplementary Table 3
Results of differential gene expression analyses using edgeR for Syrian hamster lungs. First tab, SARS-CoV-2 infected compared to uninfected Syrian hamster lungs; second tab, 100-nm ABBV-744 compared to vehicle-treated Syrian hamster lungs after SARS-CoV-2 infection. Columns are: gene symbol, log2-fold change, log2 counts per million, F value, P value and FDR by the Benjamini–Hochberg method.
Supplementary Table 4
BRD2 direct targets that are up- or downregulated in the BRD2 knockdown condition identified by the BETA analyses are listed. Columns are up-regulated targets and downregulated targets.
Supplementary Table 5
Protospacer sequences of individual sgRNAs used in Fig. 1g are listed.
Source data
Source Data Fig. 1
Statistical source data Fig. 1.
Source Data Fig. 2
Statistical source data Fig. 2.
Source Data Fig. 2
Raw unprocessed blots.
Source Data Fig. 3
Statistical source data Fig. 3.
Source Data Fig. 4
Statistical source data Fig. 4.
Source Data Fig. 6
Statistical source data Fig. 6.
Source Data Extended Data Fig. 1
Statistical source data Extended Data Fig. 1.
Source Data Extended Data Fig. 2
Statistical source data Extended Data Fig. 2.
Source Data Extended Data Fig. 3
Raw unprocessed blots.
Source Data Extended Data Fig. 4
Statistical source data Extended Data Fig. 4.
Source Data Extended Data Fig. 5
Statistical source data Extended Data Fig. 5.
Source Data Extended Data Fig. 6
Statistical source data Extended Data Fig. 5.
Rights and permissions
About this article
Cite this article
Samelson, A.J., Tran, Q.D., Robinot, R. et al. BRD2 inhibition blocks SARS-CoV-2 infection by reducing transcription of the host cell receptor ACE2. Nat Cell Biol 24, 24–34 (2022). https://doi.org/10.1038/s41556-021-00821-8
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41556-021-00821-8
This article is cited by
-
Large-scale CRISPRi screens link metabolic stress to glioblastoma chemoresistance
Journal of Translational Medicine (2025)
-
Identification of genetic modifiers enhancing B7-H3-targeting CAR T cell therapy against glioblastoma through large-scale CRISPRi screening
Journal of Experimental & Clinical Cancer Research (2024)
-
Altered ACE2 and interferon landscape in the COVID-19 microenvironment correlate with the anti-PD-1 response in solid tumors
Cellular and Molecular Life Sciences (2024)
-
SARS-CoV-2 E protein interacts with BRD2 and BRD4 SEED domains and alters transcription in a different way than BET inhibition
Cellular and Molecular Life Sciences (2024)
-
Machine learning-aided search for ligands of P2Y6 and other P2Y receptors
Purinergic Signalling (2024)