Abstract
Autophagy deficiency in breast cancer promotes metastasis through the accumulation of the autophagy cargo receptor NBR1. Here we show that autophagy normally suppresses breast cancer metastasis by enabling the clearance of NBR1–p62/SQSTM1 complexes that instruct p63-mediated pro-metastatic basal differentiation programmes. When autophagy is inhibited, the autophagy cargo receptors NBR1 and p62/SQSTM1 accumulate within biomolecular condensates in cells, which drives basal differentiation in both mouse and human breast cancer models. Mechanistically, these NBR1–p62/SQSTM1 complexes sequester ITCH, a ubiquitin ligase that degrades and negatively regulates p63 in breast cancer cells, thereby stabilizing and activating p63. Accordingly, mutant forms of NBR1 unable to sequester ITCH into NBR1–p62/SQSTM1 complexes do not promote basal differentiation and metastasis in vivo. Overall, our findings illuminate how proteostatic defects arising in the setting of therapeutic autophagy inhibition modulate epithelial lineage fidelity and metastatic progression.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The MS proteomics data associated with this study have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the identifier PXD047557. Published scRNA-seq data32,33 were obtained from Gene Expression Omnibus sources GSE161529 and GSE186344. Source data are provided with this paper. Other information related to the data reported in this paper and requests for reagents can be directed to the corresponding author.
References
Celia-Terrassa, T. & Kang, Y. Distinctive properties of metastasis-initiating cells. Genes Dev. 30, 892–908 (2016).
Liu, J. & Debnath, J. The evolving, multifaceted roles of autophagy in cancer. Adv. Cancer Res. 130, 1–53 (2016).
Assi, M. & Kimmelman, A. C. Impact of context-dependent autophagy states on tumor progression. Nat. Cancer 4, 596–607 (2023).
Amaravadi, R. K., Kimmelman, A. C. & Debnath, J. Targeting autophagy in cancer: recent advances and future directions. Cancer Discov. 9, 1167–1181 (2019).
White, E., Lattime, E. C. & Guo, J. Y. Autophagy regulates stress responses, metabolism, and anticancer immunity. Trends Cancer 7, 778–789 (2021).
Komarla, A., Dufresne, S. & Towers, C. G. Recent advances in the role of autophagy in endocrine-dependent tumors. Endocr. Rev. 44, 629–646 (2023).
Marsh, T. et al. Autophagic degradation of NBR1 restricts metastatic outgrowth during mammary tumor progression. Dev. Cell 52, 591–604 (2020).
Vera-Ramirez, L., Vodnala, S. K., Nini, R., Hunter, K. W. & Green, J. E. Autophagy promotes the survival of dormant breast cancer cells and metastatic tumour recurrence. Nat. Commun. 9, 1944 (2018).
Aqbi, H. F. et al. Autophagy-deficient breast cancer shows early tumor recurrence and escape from dormancy. Oncotarget 9, 22113–22122 (2018).
La Belle Flynn, A. et al. Autophagy inhibition elicits emergence from metastatic dormancy by inducing and stabilizing Pfkfb3 expression. Nat. Commun. 10, 3668 (2019).
Marsh, T., Tolani, B. & Debnath, J. The pleiotropic functions of autophagy in metastasis. J. Cell Sci. 134, jcs247056 (2021).
Badve, S. et al. Basal-like and triple-negative breast cancers: a critical review with an emphasis on the implications for pathologists and oncologists. Mod. Pathol. 24, 157–167 (2011).
Derakhshan, F. & Reis-Filho, J. S. Pathogenesis of triple-negative breast cancer. Annu. Rev. Pathol. 17, 181–204 (2022).
Cheung, K. J. et al. Polyclonal breast cancer metastases arise from collective dissemination of keratin 14-expressing tumor cell clusters. Proc. Natl Acad. Sci. USA 113, E854–E863 (2016).
Truong, A. B., Kretz, M., Ridky, T. W., Kimmel, R. & Khavari, P. A. p63 Regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev. 20, 3185–3197 (2006).
Kirkin, V. et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell 33, 505–516 (2009).
Rasmussen, N. L., Kournoutis, A., Lamark, T. & Johansen, T. NBR1: the archetypal selective autophagy receptor. J. Cell Biol. 221, e202208092 (2022).
Wei, H., Wang, C., Croce, C. M. & Guan, J. L. p62/SQSTM1 synergizes with autophagy for tumor growth in vivo. Genes Dev. 28, 1204–1216 (2014).
Kirkin, V., Lamark, T., Johansen, T. & Dikic, I. NBR1 cooperates with p62 in selective autophagy of ubiquitinated targets. Autophagy 5, 732–733 (2009).
Turco, E. et al. Reconstitution defines the roles of p62, NBR1 and TAX1BP1 in ubiquitin condensate formation and autophagy initiation. Nat. Commun. 12, 5212 (2021).
Zaffagnini, G. et al. p62 Filaments capture and present ubiquitinated cargos for autophagy. EMBO J. 37, e98308 (2018).
Sun, D., Wu, R., Zheng, J., Li, P. & Yu, L. Polyubiquitin chain-induced p62 phase separation drives autophagic cargo segregation. Cell Res. 28, 405–415 (2018).
Sanchez-Martin, P. et al. NBR1-mediated p62–liquid droplets enhance the Keap1–Nrf2 system. EMBO Rep. 21, e48902 (2020).
Petherick, K. J. et al. Pharmacological inhibition of ULK1 kinase blocks mammalian target of rapamycin (mTOR)-dependent autophagy. J. Biol. Chem. 290, 11376–11383 (2015).
Rebecca, V. W. et al. PPT1 promotes tumor growth and is the molecular target of chloroquine derivatives in cancer. Cancer Discov. 9, 220–229 (2019).
Kaizuka, T. et al. An autophagic flux probe that releases an internal control. Mol. Cell 64, 835–849 (2016).
Rosenbluth, J. M. et al. Organoid cultures from normal and cancer-prone human breast tissues preserve complex epithelial lineages. Nat. Commun. 11, 1711 (2020).
Gray, G. K. et al. A human breast atlas integrating single-cell proteomics and transcriptomics. Dev. Cell 57, 1400–1420 (2022).
Danieli, A. & Martens, S. p62-Mediated phase separation at the intersection of the ubiquitin–proteasome system and autophagy. J. Cell Sci. 131, jcs214304 (2018).
Sanchez-Martin, P. & Komatsu, M. p62/SQSTM1—steering the cell through health and disease. J. Cell Sci. 131, jcs222836 (2018).
Deosaran, E. et al. NBR1 acts as an autophagy receptor for peroxisomes. J. Cell Sci. 126, 939–952 (2013).
Pal, B. et al. A single-cell RNA expression atlas of normal, preneoplastic and tumorigenic states in the human breast. EMBO J. 40, e107333 (2021).
Gonzalez, H. et al. Cellular architecture of human brain metastases. Cell 185, 729–745 (2022).
Gu, Z., Eils, R. & Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016).
Barton, C. E. et al. Novel p63 target genes involved in paracrine signaling and keratinocyte differentiation. Cell Death Dis. 1, e74 (2010).
Komatsu, M. et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 12, 213–223 (2010).
Qiang, L. et al. Regulation of cell proliferation and migration by p62 through stabilization of Twist1. Proc. Natl Acad. Sci. USA 111, 9241–9246 (2014).
Kageyama, S. et al. p62/SQSTM1-droplet serves as a platform for autophagosome formation and anti-oxidative stress response. Nat. Commun. 12, 16 (2021).
Rossi, M. et al. The E3 ubiquitin ligase Itch controls the protein stability of p63. Proc. Natl Acad. Sci. USA 103, 12753–12758 (2006).
Kim, G. et al. Easy and accurate protein structure prediction using ColabFold. Nat. Protoc. 20, 620–642 (2025).
Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682 (2022).
Tan, C. T. et al. MOAP-1-mediated dissociation of p62/SQSTM1 bodies releases Keap1 and suppresses Nrf2 signaling. EMBO Rep. 22, e50854 (2021).
Lignitto, L. et al. Nrf2 activation promotes lung cancer metastasis by inhibiting the degradation of Bach1. Cell 178, 316–329 (2019).
Gatti, V. et al. p63 At the crossroads between stemness and metastasis in breast cancer. Int. J. Mol. Sci. 20, 2683 (2019).
Granit, R. Z. et al. Regulation of cellular heterogeneity and rates of symmetric and asymmetric divisions in triple-negative breast cancer. Cell Rep. 24, 3237–3250 (2018).
Towers, C. G. et al. Cancer cells upregulate NRF2 signaling to adapt to autophagy inhibition. Dev. Cell 50, 690–703 (2019).
Su, H. et al. Cancer cells escape autophagy inhibition via NRF2-induced macropinocytosis. Cancer Cell 39, 678–693 (2021).
Debnath, J., Gammoh, N. & Ryan, K. M. Autophagy and autophagy-related pathways in cancer. Nat. Rev. Mol. Cell Biol. 24, 560–575 (2023).
Martin, M. D. et al. Effect of ablation or inhibition of stromal matrix metalloproteinase-9 on lung metastasis in a breast cancer model is dependent on genetic background. Cancer Res. 68, 6251–6259 (2008).
Debnath, J., Muthuswamy, S. K. & Brugge, J. S. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 30, 256–268 (2003).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Meng, E. C. et al. UCSF ChimeraX: tools for structure building and analysis. Protein Sci. 32, e4792 (2023).
Acknowledgements
We thank L. Esserman (UCSF), J. Brugge (Harvard Medical School), J. Garber (Dana–Farber Cancer Institute) and D. Dillon (Brigham and Women’s Hospital) for human organoid resources employed in this study, and X. Wang (UCSF) for technical assistance. This study was supported by the UCSF Parnassus Flow Cytometry and Biological Imaging Development Colabs, the Center for Advanced Technology and Helen Diller Family Comprehensive Cancer Center Pathology Cores. We also thank T. Wittman (UCSF) and S. Y. Kim (UCSF) for assistance with the live-cell microscopy. UCSF ChimeraX was developed by the Resource for Biocomputing, Visualization, and Informatics at UCSF with support from NIH (grant number GM129325) and the Office of Cyber Infrastructure and Computational Biology, NIAID. Grant support includes the NIH (grant numbers CA126792, CA188404 and CA201849 to J.D.), Samuel Waxman Cancer Research Foundation (to J.D.), Mark Foundation for Cancer Research Endeavor Award (to J.D. and J.P.R.), and California Breast Cancer Research Program (grant number B26IB1494 to J.P.R. and Idea Award to M.J.M.B.). Bioinformatics analysis by H.G. was supported by Financiamiento Basal para Centros Científicos y Tecnológicos de Excelencia (Centro Ciencia & Vida; grant number FB210008), Fondo Nacional de Desarrollo Científico y Tecnológico (FONDECYT grant number 1230021) and Concern Foundation (Conquer Cancer Now 2024–2026). Support to J.M.R. for breast organoid generation includes the Doris Duke Charitable Foundation, NIH (grant number CA281361) and the Dana–Farber Cancer Institute/Harvard Cancer Center Breast SPORE (grant number 1P50CA168504).
Author information
Authors and Affiliations
Contributions
G.M. and J.D. conceived the study and designed the experiments. G.M., H.G., A.V., J.M.R. and J.D. developed key methodologies for the study. G.M., T.M., A.N., A.V., P.P. and S.B.E. performed the experiments. G.M., H.G., A.M.L., A.N., A.V., A.P.W., J.P.R. and J.D. analysed and interpreted the data. T.M., A.M.L., A.V., M.B., M.J.M.M., L.A.H., A.P.W., J.P.R. and J.M.R. provided key materials, reagents and technical support for the study. G.M., H.G., A.V. and J.D. wrote the paper with input from all authors. J.D. supervised the overall study.
Corresponding author
Ethics declarations
Competing interests
J.D. is a former member of the SAB of Vescor Therapeutics (concluded January 2022). The other authors declare no competing interests.
Peer review
Peer review information
Nature Cell Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
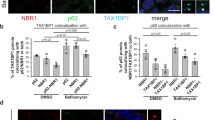
Extended Data Fig. 1 p62/SQSTM1 promotes basal differentiation upon autophagy inhibition in vivo.
a, Primary or metastatic tumour tissues generated from MMTV-PyMT; CAG-CreER;Atg12F/F or MMTV-PyMT; CAG-CreER;Atg5F/F tumours in Marsh et al.7 and stained for p63 (green), p62/SQSTM1 (red) and nuclei (DAPI, blue). Representative individual cells in boxed areas enlarged in Extended Data Fig. 1a–i (p62 high, p63 negative), 1a-ii (p62 high, p63 positive) and 1a-iii (p62 low, p63 positive) and enumerated in Extended Data Fig. 1b,c. Vehicle=autophagy-competent primary tumours or metastases, OHT=autophagy-deficient primary tumours and TAM= autophagy-deficient metastases. b, Quantification of tumour cells positive for p62. 600 or more tumour cells enumerated from each of four individual animals. Vehicle=autophagy-competent primary tumours or metastases, OHT=autophagy-deficient primary tumours and TAM= autophagy-deficient metastases. c, Quantification of p62 levels (high versus low) and p63 positivity in tumour cells from autophagy-deficient tumours. 600 or more tumour cells were enumerated from each of four individual animals. d, Number of metastases per section of lung in shCTRL (n = 10 mice), shATG7 (n = 22), shATG7 + shp62, #1 (n = 13), shATG7 + shp62, #2 (n = 13). e,f, Representative images of lung metastases stained for CK14 (green, e), p63 (green, f) and nuclei (DAPI, blue). Certain images in Extended Data Fig. 1f (for example, shATG7 and shATG7 + shp62, #1) also shown in Fig. 1d. g, Quantification of p63 (nuclear) positive and CK14 positive cells in the lung metastases for the indicated conditions. 250 or more cells from 4–8 metastases counted per cohort. 5 individual mice from three independent experiments evaluated. h, Immunoblot for the indicated proteins in MCF10A cells expressing the indicated shRNAs. Samples run on separate gels with equal loading of total proteins. i, PyMT R221A cells immunostained for p63 (red), p62 (green) and nuclei (DAPI, blue) for the indicated conditions. Statistics: Statistical significance was determined by either a 2-tailed Student’s t-test for (b) and (c) or by one-way ANOVA followed by Dunnett’s multiple comparisons test for (d) and (g). Data are represented by mean ± S.E.M; each data point represents one mouse in all graphs. Data and unprocessed blots are provided.
Extended Data Fig. 2 Autophagy inhibition promotes basal differentiation in mouse mammary cancer and human breast cancer cells.
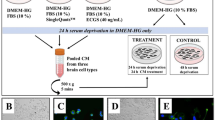
a, CK14 (red), CK5 (green) and nuclei (DAPI, blue) immunostaining of mouse PyMT R221A cells expressing the indicated shRNAs. b, Representative flow cytometry profile of p62/SQSTM1 positive (red) and p62, p63 double-positive (yellow) MDA-MB-231 cells. c, Dual reporter autophagic flux probe described in Kaizuka et al.26. d, Representative immunofluorescent staining of MDA-MB-231 cells for p63 (light blue) for the indicated cohorts. Expression of GFP (green) and RFP (red) in individual cells expressing dual reporter autophagic flux probe. Arrows mark representative p63 positive cells. Right offset: merged image with magnification corresponding to boxed area. e, Quantification of p63 positive cells in Extended Data Fig. 2d. Each data point indicates the percent of positive cells calculated from one individual experiment (n = 3 independent experiments). 200 or more cells were enumerated per experiment per condition. f, Summary of human prophylactic mastectomy (PM) and breast cancer (BC) organoids used in this study. g, Representative immunofluorescent images of primary cells generated from human prophylactic mastectomy (PM) organoids (Org 164) and treated with solvent, Ulk-i or HCQ for 18 h. Cells immunostained for CK14 (red) and nuclei (DAPI, blue) following treatment. h, Quantification of human BC organoid cells positive for p63 and CK5 in Extended Data Fig. 2h. 250 or more cells were counted from each of three individual organoids (N = 3). i, Representative immunofluorescent images of primary cells generated from human breast cancer (BC) organoids (Torg 40) and treated with solvent, Ulk-i or HCQ. Following treatment, cells immunostained as indicated for the following markers: CK5 (red, top row), p63 (green, top row), p62 (green, bottom row), and nuclei (DAPI, blue). Scale bar equals 20 µm. Right: magnification of p62 immunostaining in representative control or Ulk-i treated cells. Statistical significance determined by a 2-tailed Student’s t-test in (e) and (h). Data are represented as mean ± S.E.M. Each dot represents percentages calculated from an individual experiment (e) or a human patient organoid (h). Data and unprocessed blots are provided.
Extended Data Fig. 3 p62/SQSTM1 is required for the effects of NBR1 overexpression on tumour metastasis and pro-metastatic basal differentiation.
a, Immunofluorescent staining for p62 (green) and CK5 (red) in MDA-MB-231 cells expressing indicated shRNAs and, when indicated, treated with Ulk-i. Offset: magnification of boxed area in Ulk-i+shCTRL cells. b, Immunoblot analysis of MDA-MB-231 and R221A cells from indicated cohorts. Loading control (actin) corresponds to same blot. c, Immunofluorescent staining of indicated MDA-MB-231 cell cohorts for FLAG (NBR1, green), p62 (red) and nuclei (DAPI, blue). d, Quantification of total number of metastases per section of lung for shCTRL (n = 7), WT-NBR1 (n = 6), WT-NBR1 + shp62, #1 (n = 8), wt-NBR1 + shp62, #2 (n = 9). e, Representative H&E staining of lung metastases from indicated groups. f,g, Lungs bearing metastases from indicated groups stained for p63 (green, f) or CK14 (green, g) and nuclei (DAPI, blue). h, PyMT R221A cells expressing the indicated constructs stained for p63 (green, left panel) and CK14 (green, right panel) and nuclei (DAPI, blue). i, Quantification of p63 (nuclear staining) and CK14 positive R221A cells. 400 or more cells from each of three individual experiments were counted per condition. j, Quantification of total number of metastases per section of lung in shCTRL (n = 12), WT-NBR1 (n = 11), D50R-NBR1 (n = 10). shCTRL and WT-NBR1 cohorts were also included as controls in d and Extended Data Fig. 7a. k, Lungs bearing metastases from the indicated groups stained for p63 (green, top), CK14 (green, bottom) and DAPI (blue). l, Quantification of p63 (nuclei) and CK14 positive cells within lung metastases for the indicated conditions. Minimum of 250 cells from multiple (4–8) metastases counted per cohort. A total of five (n = 5) individual mice from three independent experiments evaluated. Statistical significance determined by either one-way (d,j,l) or two-way (i) ANOVA, followed by Dunnett’s multiple comparisons test. Data are represented by mean ± S.E.M; each dot represents one animal (d,j,l) or an individual experiment (i). Data and unprocessed blots are provided.
Extended Data Fig. 4 Autophagy inhibition promotes transcription of p63 target genes and p63 protein stability.
a, qRT-PCR analysis of p63 and p63 target mRNA expression in mouse 4T1 cells for the indicated groups. b,c, qRT-PCR analysis of p63 mRNA expression in human triple-negative MDA-MB-231 cells (b) and mouse PyMT R221A (c) cells for the indicated groups. d, Quantification of p63 protein levels in MDA-MB-231 cells following cycloheximide treatment. Following ATG7 knockdown or 18 h treatment with indicated agents, cells were treated with cycloheximide for indicated times, lysed, subject to immunoblot analysis and integrated band intensity was calculated via densitometry. For each condition and timepoint, integrated band intensity of p63 protein was normalized to both its respective actin control and the corresponding t = 0 timepoint. e, Representative immunoblot from cycloheximide chase assays of MDA-MB-231 cells used for quantification in d. MG132 employed as a control to inhibit proteosomal degradation of p63. Samples were run on separate gels with equal loading of total proteins. f, Representative immunofluorescent staining of CK14 (red) and nuclei (DAPI, blue) in lung metastasis from indicated groups. g, Phase-contrast micrograph of human MDA-MB-231 and mouse PyMT R221A cells 24 h following treatment with CTRL or p63 shRNAs. h, Working model: autophagy inhibition in breast cancer cells results in accumulation of the ACRs NBR1 and p62/SQSTM1, which in turn, increases protein levels of the p63 transcription factor and enhanced expression of p63-target genes. Data (a–c) are represented by mean ± S.E.M. Each dot represents a technical replicate within one bio-replicate; one representative of three bio-replicates are shown. Each data point in d represents mean band intensity ± S.E.M from three (n = 3) independent experiments. Data and unprocessed blots are provided.
Extended Data Fig. 5 NBR1–p62/SQSTM1 complexes sequester the ubiquitin ligase ITCH in autophagy-deficient cells.
a, Coomassie staining of total cell lysates from indicated cells utilized for TMT-MS in Fig. 4c and b. b, Volcano plot of proteins identified by TMT-MS from α-FLAG immunoprecipitates from HEK293T cells expressing WT-NBR1 vs. D50R (both 3×FLAG-tagged). Proteins plotted according to −log10 P values as determined by two-tailed Student’s t-test and log2-fold enrichment (n = 3 biological replicates). Red: proteins significantly enriched in WT-NBR1, identified with P > 0.05 and log2-fold change > |0.5 | . Blue: significantly enriched in D50R. Grey: not enriched in either group. c, Immunoblot of α-FLAG immune complexes (lanes 1–3) and total cell lysates (lanes 4–6) from 293T cells expressing indicated constructs. Samples run on separate gels with equal loading; loading control (actin) corresponds to the NEDD4 blot. d,e, Immunofluorescent staining of 293T cells for FLAG (NBR1, green), nuclei (DAPI, blue) and TAX1BP1 (red; d) or ITCH (red; e) for the indicated conditions. f, Immunoblot analysis of total cell lysates of MDA-MB-231 cells used for the immunoprecipitation studies in Fig. 4d. g, Immunoblot analysis of total cell lysate of MDA-MB-231 cells for the immunoprecipitation studies in Fig. 4e. Actin loading control in f and g corresponds to same blot. h, Quantification of percent co-localization (relative to total number of puncta) of FLAG (NBR1) and ITCH puncta in MDA-MB-231 and MCF10A cells. For each condition, a minimum of 500 puncta from three individual high-power fields (n = 3) representing at least 30 individual cells were evaluated. i, Representative immunofluorescent staining for ITCH (red), FLAG (NBR1, green) and nuclei (DAPI, blue) of MDA-MB-231 cells expressing the indicated constructs and shRNAs. Each data point (h) represents percent of enumerated puncta co-localized per independent fields. Data represented by mean ± S.E.M. Statistical significance was determined by two-way ANOVA, followed by Dunnett’s multiple comparisons test. Data and unprocessed blots are provided.
Extended Data Fig. 6 Dynamic properties of p62 and ITCH in biomolecular condensates.
a, Immunoblotting of MDA-MB-231 cells expressing indicated FLAG-tagged NBR1 constructs. α-FLAG immune complexes (upper) and total cell lysates (lower) blotted for the indicated proteins. Samples run on separate gels with equal loading of immune complex (upper) or total proteins (lower). For α-FLAG immune complexes, NBR1 loading corresponds to p62 gel; for total lysate, loading control (actin) corresponds to the p63 gel. b, MDA-MB-231 cells expressing indicated ITCH constructs (GFP-tagged) immunostained for ITCH (green), p62/SQSTM1 (red) or DAPI (blue). c, Representative immunofluorescent staining of MDA-MB-231 cells for p62/SQSTM1 (green) and ITCH (red) for the indicated treatment groups. d, Representative selected frames of GFP–ITCH (green) and mCherry–p62 (red) puncta in MDA-MB-231 cells for the indicated time points. Frames demonstrate course of fission event of multiple (#1- #3) green (ITCH) and p62 (red) puncta; point of fission is indicated by arrow. Selected frames from this sequence of images are also shown in Fig. 7e. Also see Supplementary Video 3. Certain images in d are also shown in Fig. 7e. e, Representative selected frames of GFP–ITCH (green) and mCherry–p62 (red) puncta in MDA-MB-231 cells for the indicated time points. Frames demonstrate course of fusion event of ITCH (green, #1) and p62 (red, #2) puncta; point of fusion indicated by arrow. Selected frames of this from this sequence of images are also shown in Fig. 7f. Also see Supplementary Video 4. Certain images in e also shown in Fig. 7f. Unprocessed blots are provided.
Extended Data Fig. 7 Depletion of ITCH triggers p63-mediated basal differentiation and metastatic outgrowth of mammary tumours.
a, Quantification of total number of metastases per section of lung for CTRL (n = 12 mice), WT-NBR1 (n = 11) and del-ZnF-NBR1 (n = 10). shCTRL and WT-NBR1 cohorts were also used as controls in Extended Data Fig. 3d. b, Quantification of total number of metastases per section of lung for shCTRL (n = 13 mice), shATG7 (n = 6), shITCH, #1 (n = 25), shITCH, #2 (n = 24), shKEAP1, #1 (n = 12) and shKEAP1, #2 (n = 12). Mice in shCTRL and shATG7 cohorts are also used in Extended Data Fig. 1d. Related to Fig. 8g–l. c,d, Control (shCTRL) and ITCH-depleted MDA-MB-231 cells immunostained for p63 (green) and CK14 (red; c) or for p62 (green) and CK5 (red; d). Nuclei counterstained with DAPI (blue). e, Immunoblot analysis of MDA-MB-231 cells for the indicated proteins (left), 48 h following treatment with CTRL, NEDD4 or ITCH shRNAs. Samples were run on separate gels with equal loading of total proteins; loading control (actin) corresponds to ITCH blot. f, Model for breast cancer cells with normal autophagy (top) versus cells with autophagy inhibition (bottom). Normal autophagic degradation of ACRs reduces steady state levels of p63 due to ITCH-mediated UPS turnover. Inhibiting autophagy results in accumulation of NBR1–p62/SQSTM1 complexes, which sequester ITCH away from p63. As a result, stabilized p63 promotes the increased expression of basal cytokeratins and increased metastasis. Each dot (a,b) represents one animal. Statistical significance (a,b) was determined by one-way ANOVA, followed by Dunnett’s multiple comparisons test. Data are represented by mean ± S.E.M. Data and unprocessed blots are provided.
Supplementary information
Supplementary Video 1
FRAP of mCherry–p62 puncta. Fluorescence recovery of mCherry–p62 (red) puncta in MDA-MB-231 cells after photobleaching. Images were acquired every 10 s. Video plays at 2 frames s−1. Adjacent puncta are left unbleached as internal controls for overall fluorescence loss during imaging. Images are cropped and magnified at an arbitrary scale.
Supplementary Video 2
FRAP of GFP–ITCH puncta. Fluorescence recovery of GFP–ITCH (green) puncta in MDA-MB-231 cells after photobleaching. Images were acquired every 10 s. The video plays at 2 frames s−1. Adjacent puncta are left unbleached as internal controls for overall fluorescence loss during imaging. Images are cropped and magnified at an arbitrary scale.
Supplementary Video 3
Fission events involving GFP–ITCH and mCherry–p62 puncta. Inter- and intramolecular fission events between GFP–ITCH and mCherry–p62 puncta in control shRNA-treated MDA-MB-231 cells. Images were acquired every 0.5 s. Video plays at 2 frames s−1. Images are cropped and magnified at an arbitrary scale.
Supplementary Video 4
Fusion events involving GFP–ITCH and mCherry–p62 puncta. Inter- and intramolecular fusion events between GFP–ITCH and mCherry–p62 puncta in control shRNA-treated MDA-MB-231 cells. Images were acquired every 0.5 s. Video plays at 2 frames s−1. Images are cropped and magnified at an arbitrary scale.
Supplementary Tables 1 and 2
Supplementary Table 1. Quantitative TMT-MS data from immunoprecipitated NBR1 complexes. Proteins identified from anti-FLAG immune complexes generated from HEK293T cells overexpressing 3×FLAG-tagged empty vector, WT-NBR1 or D50R-NBR1. Data are organized into sheets as follows: (1) Complete MS data comparing HEK293T cells expressing WT-NBR1 versus empty vector from three independent biological replicates. (2) Complete MS data comparing HEK293T cells expressing D50R-NBR1 versus empty vector from three independent biological replicates. (3) Complete MS data comparing HEK293T cells expressing WT-NBR1 versus D50R-NBR1 from three independent biological replicates. Supplementary Table 2. Reagents and resources. Detailed list of primary and secondary antibodies, oligonucleotides/primers, plasmids and constructs, shRNA, software, web-based resources and algorithms, chemicals, kits and assays, cell lines, bacterial strains and mouse strains.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 1 and Extended Data Fig. 1
Unprocessed gels and western blots.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 2
Unprocessed gels and western blots.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 3
Unprocessed gels and western blots.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 4
Unprocessed gels and western blots.
Source Data Fig. 5
Unprocessed gels and western blots.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 6
Unprocessed gels and western blots.
Source Data Fig. 7
Statistical source data.
Source Data Fig. 8
Statistical source data.
Source Data Fig. 8
Unprocessed gels and western blots.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 3
Unprocessed gels and western blots.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 4
Unprocessed gels and western blots.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 5
Unprocessed gels and western blots.
Source Data Extended Data Fig. 6
Unprocessed gels and western blots.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 7
Unprocessed gels and western blots.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mondal, G., Gonzalez, H., Marsh, T. et al. Autophagy-targeted NBR1–p62/SQSTM1 complexes promote breast cancer metastasis by sequestering ITCH. Nat Cell Biol 27, 1098–1113 (2025). https://doi.org/10.1038/s41556-025-01689-8
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41556-025-01689-8
This article is cited by
-
LAMTOR5 promotes hepatoma growth in mice by disrupting LC3-p62-mediated autophagy and preventing p62 proteasome degradation
Acta Pharmacologica Sinica (2026)