Abstract
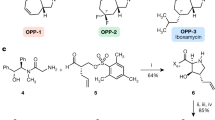
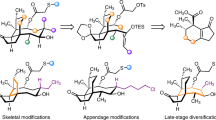
The emergence of bacterial antimicrobial resistance threatens to undermine the utility of antibiotic therapy in medicine. This threat can be addressed, in part, by reinventing existing antibiotic classes using chemical synthesis. Here we present the discovery of BT-33, a fluorinated macrobicyclic oxepanoprolinamide antibiotic with broad-spectrum activity against multidrug-resistant bacterial pathogens. Structure–activity relationships within the macrobicyclic substructure reveal structural features that are essential to the enhanced potency of BT-33 as well as its increased metabolic stability relative to its predecessors clindamycin, iboxamycin and cresomycin. Using X-ray crystallography, we determine the structure of BT-33 in complex with the bacterial ribosome revealing that its fluorine atom makes an additional van der Waals contact with nucleobase G2505. Through variable-temperature 1H NMR experiments, density functional theory calculations and vibrational circular dichroism spectroscopy, we compare macrobicyclic homologues of BT-33 and a C7 desmethyl analogue and find that the C7 methyl group of BT-33 rigidifies the macrocyclic ring in a conformation that is highly preorganized for ribosomal binding.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data supporting this study are made available in the main text or the Supplementary Information. Crystallographic data for the structure of intermediate 8 were deposited in the Cambridge Crystallographic Data Centre (deposition number 2376105) and are accessible free of charge. Coordinates and structure factors were deposited in the RCSB PDB for the wild-type T. thermophilus 70S ribosome in complex with mRNA, deacylated A-site tRNAPhe, aminoacylated P-site fMet-tRNAiMet, deacylated E-site tRNAPhe and BT-33 with accession code 9D0J. Reference structures used for analysis can be found in the RCSB PDB with accession codes 4V7V and 6XHW. Source data are provided with this paper.
References
Mason, D., Dietz, A. & DeBoer, C. Lincomycin, a new antibiotic. I. Discovery and biological properties. Antimicrob. Agents Chemother. 554–559 (1962).
Birkenmeyer, R. D. & Kagan, F. Lincomycin. XI. Synthesis and structure of clindamycin, a potent antibacterial agent. J. Med. Chem. 13, 616–619 (1970).
Leclercq, R. & Courvalin, P. Bacterial resistance to macrolide, lincosamide, and streptogramin antibiotics by target modification. Antimicrob. Agents Chemother. 35, 1267–1272 (1991).
Toh, S.-M. et al. Acquisition of a natural resistance gene renders a clinical strain of methicillin-resistant Staphylococcus aureus resistant to the synthetic antibiotic linezolid. Mol. Microbiol. 64, 1506–1514 (2007).
Murina, V., Kasari, M., Hauryliuk, V. & Atkinson, G. C. Antibiotic resistance ABCF proteins reset the peptidyl transferase centre of the ribosome to counter translational arrest. Nucleic Acids Res. 46, 3753–3763 (2018).
Crowe-McAuliffe, C. et al. Structural basis of ABCF-mediated resistance to pleuromutilin, lincosamide, and streptogramin A antibiotics in Gram-positive pathogens. Nat. Commun. 12, 3577 (2021).
Obana, N. et al. Genome-encoded ABCF factors implicated in intrinsic antibiotic resistance in Gram-positive bacteria: VmlR2 Ard1 and CplR. Nucleic Acids Res. 51, 4536–4554 (2023).
Brisson-Noël, A. & Courvalin, P. Nucleotide sequence of gene linA encoding resistance to lincosamides in Staphylococcus haemolyticus. Gene 43, 247–253 (1986).
Bozdogan, B. et al. A new resistance gene, linB, conferring resistance to lincosamides by nucleotidylation in Enterococcus faecium HM1025. Antimicrob. Agents Chemother. 43, 925–929 (1999).
Birkenmeyer, R. D., Kroll, S. J., Lewis, C., Stern, K. F. & Zurenko, G. E. Synthesis and antimicrobial activity of clindamycin analogs: pirlimycin, a potent antibacterial agent. J. Med. Chem. 27, 216–223 (1984).
O’Dowd, H., Erwin, A. L. & Lewis, J. G. in Natural Products in Medicinal Chemistry (ed. Hanessian, S.) 251–270 (Wiley-VCH, 2014).
Hirai, Y. et al. Characterization of compound A, a novel lincomycin derivative active against methicillin-resistant Staphylococcus aureus. J. Antibiot. 74, 124–132 (2021).
Mitcheltree, M. J. et al. A synthetic antibiotic class overcoming bacterial multidrug resistance. Nature 599, 507–512 (2021).
Wu, K. J. Y. et al. An antibiotic preorganized for ribosomal binding overcomes antimicrobial resistance. Science 383, 721–726 (2024).
Wynalda, M. A., Hutzler, J. M., Koets, M. D., Podoll, T. & Wienkers, L. C. In vitro metabolism of clindamycin in human liver and intestinal microsomes. Drug Metab. Dispos. 31, 878–887 (2003).
Doyle, L. M. et al. Stereoselective epimerizations of glycosyl thiols. Org. Lett. 19, 5802–5805 (2017).
Ding, P. et al. Syntheses of conformationally constricted molecules as potential NAALADase/PSMA inhibitors. Org. Lett. 6, 1805–1808 (2004).
Orita, A., Hamada, Y., Nakano, T., Toyoshima, S. & Otera, J. Highly efficient deacetylation by use of the neutral organotin catalyst [tBu2SnOH(Cl)]2. Chem. Eur. J. 7, 3321–3327 (2001).
Dess, D. B. & Martin, J. C. Readily accessible 12-I-5 oxidant for the conversion of primary and secondary alcohols to aldehydes and ketones. J. Org. Chem. 48, 4155–4156 (1983).
Liu, G., Cogan, D. A., Owens, T. D., Tang, T. P. & Ellman, J. A. Synthesis of enantiomerically pure N-tert-butanesulfinyl imines (tert-butanesulfinimines) by the direct condensation of tert-butanesulfinamide with aldehydes and ketones. J. Org. Chem. 64, 1278–1284 (1999).
Yamamoto, K., Biswas, K., Gaul, C. & Danishefsky, S. J. Effects of temperature and concentration in some ring closing metathesis reactions. Tetrahedron Lett. 44, 3297–3299 (2003).
Middleton, W. J. New fluorinating reagents. Dialkylaminosulfur fluorides. J. Org. Chem. 40, 574–578 (1975).
Mason, J. D., Terwilliger, D. W., Pote, A. R. & Myers, A. G. Practical gram-scale synthesis of iboxamycin, a potent antibiotic candidate. J. Am. Chem. Soc. 143, 11019–11025 (2021).
Carpino, L. A. 1-Hydroxy-7-azabenzotriazole. An efficient peptide coupling additive. J. Am. Chem. Soc. 115, 4397–4398 (1993).
Wu, K. J. Y., Tresco, B. I. C., Xiao, J., See, D. N. Y. & Myers, A. G. A. Practical synthesis of macrobicyclic thiolincosamines. J. Am. Chem. Soc. 146, 29135–29139 (2024).
WHO Bacterial Priority Pathogens List, 2024: Bacterial Pathogens of Public Health Importance to Guide Research, Development and Strategies to Prevent and Control Antimicrobial Resistance (World Health Organization, 2024).
Dunkle, J. A., Xiong, L., Mankin, A. S. & Cate, J. H. D. Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc. Natl Acad. Sci. USA 107, 17152–17157 (2010).
Miertuš, S., Scrocco, E. & Tomasi, J. Electrostatic interaction of a solute with a continuum. A direct utilization of ab initio molecular potentials for the prevision of solvent effects. Chem. Phys. 55, 117–129 (1981).
Miertuš, S. & Tomasi, J. Approximate evaluations of the electrostatic free energy and internal energy changes in solution processes. Chem. Phys. 65, 239–245 (1982).
Becke, A. D. Density‐functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 98, 5648–5652 (1993).
Pascual-ahuir, J. L., Silla, E. & Tuñon, I. GEPOL: an improved description of molecular surfaces. III. A new algorithm for the computation of a solvent-excluding surface. J. Comput. Chem. 15, 1127–1138 (1994).
Gauss, J. Calculation of NMR chemical shifts at second-order many-body perturbation theory using gauge-including atomic orbitals. Chem. Phys. Lett. 191, 614–620 (1992).
Gauss, J. Effects of electron correlation in the calculation of nuclear magnetic resonance chemical shifts. J. Chem. Phys. 99, 3629–3643 (1993).
Gauss, J. Accurate calculation of NMR chemical shifts. Ber. Bunsenges. Phys. Chem. 99, 1001–1008 (1995).
Cheeseman, J. R., Frisch, M. J., Devlin, F. J. & Stephens, P. J. Ab initio calculation of atomic axial tensors and vibrational rotational strengths using density functional theory. Chem. Phys. Lett. 252, 211–220 (1996).
Yu, H. S., He, X., Li, S. L. & Truhlar, D. G. MN15: a Kohn–Sham global-hybrid exchange–correlation density functional with broad accuracy for multi-reference and single-reference systems and noncovalent interactions. Chem. Sci. 7, 5032–5051 (2016).
Mazzeo, G., Abbate, S., Longhi, G., Castiglioni, E. & Villani, C. Vibrational circular dichroism (VCD) reveals subtle conformational aspects and intermolecular interactions in the carnitine family. Chirality 27, 907–913 (2015).
Performance Standards for Antimicrobial Susceptibility Testing 33rd edn (Clinical and Laboratory Standards Institute, 2023).
Ito, A. et al. Siderophore cephalosporin cefiderocol utilizes ferric iron transporter systems for antibacterial activity against Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 60, 7396–7401 (2016).
Krause, L., Herbst-Irmer, R., Sheldrick, G. M. & Stalke, D. Comparison of silver and molybdenum microfocus X-ray sources for single-crystal structure determination. J. Appl. Crystallogr. 48, 3–10 (2015).
Sheldrick, G. M. Crystal structure refinement with SHELXL. Acta Cryst. C Struct. Chem. 71, 3–8 (2015).
Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. & Puschmann, H. OLEX2: a complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 42, 339–341 (2009).
Polikanov, Y. S., Steitz, T. A. & Innis, C. A. A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome. Nat. Struct. Mol. Biol. 21, 787–793 (2014).
Polikanov, Y. S., Melnikov, S. V., Soll, D. & Steitz, T. A. Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly. Nat. Struct. Mol. Biol. 22, 342–344 (2015).
Syroegin, E. A., Aleksandrova, E. V. & Polikanov, Y. S. Insights into the ribosome function from the structures of non-arrested ribosome-nascent chain complexes. Nat. Chem. 15, 143–153 (2023).
Kabsch, W. XDS. Acta Cryst. D Biol. Crystallogr. 66, 125–132 (2010).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Svetlov, M. S. et al. Structure of Erm-modified 70S ribosome reveals the mechanism of macrolide resistance. Nat. Chem. Biol. 17, 412–420 (2021).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. D Biol. Crystallogr. 66, 213–221 (2010).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Cryst. D Biol. Crystallogr. 60, 2126–2132 (2004).
Mohamadi, F. et al. Macromodel—an integrated software system for modeling organic and bioorganic molecules using molecular mechanics. J. Comp. Chem. 11, 440–467 (1990).
Watts, K. S., Dalal, P., Tebben, A. J., Cheney, D. L. & Shelley, J. C. Macrocycle conformational sampling with MacroModel. J. Chem. Inf. Model. 54, 2680–2696 (2014).
Banks, J. L. et al. Integrated Modeling Program, Applied Chemical Theory (IMPACT). J. Comp.Chem. 26, 1752–1780 (2005).
Frisch, M. J. et al. Gaussian 16 revision A.03 (Gaussian, Inc., 2016).
GaussView version 6.1.1 (Semichem, 2019).
CYLview20 (Université de Sherbrooke, 2020).
Cheeseman, J. R., Trucks, G. W., Keith, T. A. & Frisch, M. J. A comparison of models for calculating nuclear magnetic resonance shielding tensors. J. Chem. Phys. 104, 5497–5509 (1996).
Acknowledgements
We gratefully acknowledge M. J. Mitcheltree and G. Testolin for the design conception and first synthesis of (nine-membered) macrobicyclic thiolincosamines; S. L. Zheng for the collection of X-ray crystallographic data for intermediate 8 under the support of the Major Research Instrumentation (MRI) Program of the National Science Foundation (NSF), award 2216066; D. Cui for the support of variable-temperature 1H NMR experiments; J. X. Wang for the collection of mass spectrometry data; M. D. Cameron for the collection of liver microsomal stability data; Pharmaron (Ningbo, China) for collection of murine pharmacokinetic data; and D. R. Andes, L. Burrows, S. Chiang, P. Courvalin, T. J. Dougherty, R. Duggal, M. S. Gilmore, C. Grillot-Courvalin, T. Grossman, V. Hauryliuk, C. Keith, S. Lahiri, F. H. Lebreton, A. S. Mankin, H. E. Moser, D. P. Nicolau, K. M. Otte, A. Pisipati, S. Projan, N. C. Vazquez-Laslop and B. Wohl for their invaluable insights over the course of our research.
This work was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health, grants R01-AI168228 (A.G.M.) and R21-AI163466 (Y.S.P.); National Institute of General Medical Sciences, grants R35-GM151957 (Y.S.P.) and R01-GM132302 (Y.S.P.); Illinois State Startup Funds (Y.S.P.); and Corning Fund for Faculty Development (R.Y.L.).
This work is based upon research conducted at the Northeastern Collaborative Access Team (NE-CAT) beamlines, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (NIH; grant P30-GM124165 to NE-CAT). The Eiger 16M detector on the 24-ID-E beamline is funded by an NIH Office of Research Infrastructure Programs (ORIP) High-End Instrumentation (HEI) grant (grant S10-OD021527 to NE-CAT). This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under contract no. DE-AC02-06CH11357.
CARB-X funding for this research (A.G.M.) is supported by federal funds from the US Department of Health and Human Services (HHS), Administration for Strategic Preparedness and Response, Biomedical Advanced Research and Development Authority under agreement number 75A50122C00028, and by awards from Wellcome (WT224842), Germany’s Federal Ministry of Education and Research (BMBF) and the UK Department of Health and Social Care as part of the Global Antimicrobial Resistance Innovation Fund (GAMRIF). The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of CARB-X or its funders.
Author information
Authors and Affiliations
Contributions
B.I.C.T. designed, synthesized and characterized BT-33. B.I.C.T., K.J.Y.W. and D.N.Y.S. synthesized and characterized macrobicyclic oxepanoprolinamide analogues. A.R., B.I.C.T. and K.J.Y.W. designed and performed antimicrobial susceptibility testing assays for all analogues. E.V.A. and Y.S.P. designed and performed X-ray crystallography experiments. M.P. performed VCD measurements. R.Y.L. performed DFT calculations. A.G.M., Y.S.P. and R.Y.L. supervised the experiments. B.I.C.T. and A.G.M. wrote the manuscript. All authors interpreted the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
B.I.C.T., K.J.Y.W. and A.G.M. have filed international patent application WO/2023/205206, ‘Lincosamides and Uses Thereof’, and US provisional patent application US/63/676,017, ‘Synthesis of macrobicyclic Thiolincosamines’. B.I.C.T., K.J.Y.W. and A.G.M. are shareholders of Kinvard Bio, Inc. All other authors declare no competing interests.
Peer review
Peer review information
Nature Chemistry thanks Dale Boger and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–7, Tables 1–5, synthetic methods and characterization data for new OPP analogues and coordinates and energies for DFT-optimized structures.
Supplementary Video 1
Visualization of BT-33 binding to the T. thermophilus ribosome.
Supplementary Data 1
The CIF file for intermediate 8.
Source data
Source Data Fig. 4
MIC data for Fig. 4a,b and plasma concentrations for single-dose pharmacokinetics study.
Source Data Fig. 6
Calculated and experimental VCD spectra of 12.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tresco, B.I.C., Wu, K.J.Y., Ramkissoon, A. et al. Discovery of a fluorinated macrobicyclic antibiotic through chemical synthesis. Nat. Chem. 17, 582–589 (2025). https://doi.org/10.1038/s41557-025-01738-7
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41557-025-01738-7