Abstract
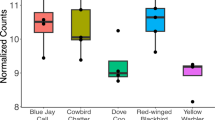
Uncovering the genomic bases of phenotypic adaptation is a major goal in biology, but this has been hard to achieve for complex behavioural traits. Here we leverage the repeated, independent evolution of obligate cavity nesting in birds to test the hypothesis that pressure to compete for a limited breeding resource has facilitated convergent evolution in behaviour, hormones and gene expression. We used an integrative approach, combining aggression assays in the field, testosterone measures and transcriptome-wide analyses of the brain in wild-captured females and males. Our experimental design compared species pairs across five avian families, each including one obligate cavity-nesting species and a related species with a more flexible nest strategy. We find behavioural convergence, with higher levels of territorial aggression in obligate cavity nesters, particularly among females. Across species, levels of testosterone in circulation were not associated with nest strategy nor aggression. Phylogenetic analyses of individual genes and co-regulated gene networks revealed more shared patterns of brain gene expression than expected by drift, although the scope of convergent gene expression evolution was limited to a small percentage of the genome. When comparing our results to other studies that did not use phylogenetic methods, we suggest that accounting for shared evolutionary history may reduce the number of genes inferred as convergently evolving. Altogether, we find that behavioural convergence in response to shared ecological pressures is associated with largely independent evolution of gene expression across different avian families, punctuated by a narrow set of convergently evolving genes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data that support the findings of this study are available in the Supplementary Information, on Github via Zenodo at https://doi.org/10.5281/zenodo.14715061 (ref. 86), and via Figshare repository at https://doi.org/10.6084/m9.figshare.26367061 (ref. 87). Raw sequence reads and count data can be obtained from the Gene Expression Omnibus database (GEO accession number GSE244480).
Code availability
Scripts for processing and analysing data are available on Github via Zenodo at https://doi.org/10.5281/zenodo.14715061 (ref. 86).
References
Darwin, C. R. On the Origin of Species (Harvard Univ. Press, 1859).
Rosenblum, E. B., Parent, C. E. & Brandt, E. E. The molecular basis of phenotypic convergence. Annu. Rev. Ecol. Evol. Syst. 45, 203–226 (2014).
Arendt, J. & Reznick, D. Convergence and parallelism reconsidered: what have we learned about the genetics of adaptation? Trends Ecol. Evol. 23, 26–32 (2008).
Gallant, J. R. & O’Connell, L. A. Studying convergent evolution to relate genotype to behavioral phenotype. J. Exp. Biol. 223, jeb213447 (2020).
Jourjine, N. & Hoekstra, H. E. Expanding evolutionary neuroscience: insights from comparing variation in behavior. Neuron 109, 1084–1099 (2021).
Rosvall, K. A. Evolutionary endocrinology and the problem of Darwin’s tangled bank. Horm. Behav. 146, 105246 (2022).
Rittschof, C. C. et al. Neuromolecular responses to social challenge: common mechanisms across mouse, stickleback fish, and honey bee. Proc. Natl Acad. Sci. USA 111, 17929–17934 (2014).
Toth, A. L. et al. Brain transcriptomic analysis in paper wasps identifies genes associated with behaviour across social insect lineages. Proc. Biol. Sci. 277, 2139–2148 (2010).
Pfenning, A. R. et al. Convergent transcriptional specializations in the brains of humans and song-learning birds. Science 346, 1256846 (2014).
Mikheyev, A. S. & Linksvayer, T. A. Genes associated with ant social behavior show distinct transcriptional and evolutionary patterns. Elife 4, e04775 (2015).
Warner, M. R., Qiu, L., Holmes, M. J., Mikheyev, A. S. & Linksvayer, T. A. Convergent eusocial evolution is based on a shared reproductive groundplan plus lineage-specific plastic genes. Nat. Commun. 10, 2651 (2019).
Boyle, E. A., Li, Y. I. & Pritchard, J. K. An expanded view of complex traits: from polygenic to omnigenic. Cell 169, 1177–1186 (2017).
Young, R. L. et al. Conserved transcriptomic profiles underpin monogamy across vertebrates. Proc. Natl Acad. Sci. USA 116, 1331–1336 (2019).
Pease, J. B. et al. Layered evolution of gene expression in ‘superfast’ muscles for courtship. Proc. Natl Acad. Sci. USA 119, e2119671119 (2022).
Harrison, M. C. et al. Hemimetabolous genomes reveal molecular basis of termite eusociality. Nat. Ecol. Evol. 2, 557–566 (2018).
Johnson, Z. V. et al. Cellular profiling of a recently-evolved social behavior in cichlid fishes. Nat. Commun. 14, 4891 (2023).
Nelson, R. J. & Trainor, B. C. Neural mechanisms of aggression. Nat. Rev. Neurosci. 8, 536–546 (2007).
Wingfield, J. C., Hegner, R. E., Dufty, J., Alfred, M. & Ball, G. F. The ‘Challenge Hypothesis’: theoretical implications for patterns of testosterone secretion, mating systems, and breeding strategies. Am. Nat. 136, 829–846 (1990).
Horton, B. M. et al. Estrogen receptor alpha polymorphism in a species with alternative behavioral phenotypes. Proc. Natl Acad. Sci. USA 111, 1443–1448 (2014).
Rosvall, K. A. et al. Neural sensitivity to sex steroids predicts individual differences in aggression: implications for behavioural evolution. Proc. Biol. Sci. 279, 3547–3555 (2012).
Saul, M. C. et al. Cross-species systems analysis of evolutionary toolkits of neurogenomic response to social challenge. Genes Brain Behav. 18, e12502 (2019).
Zhang-James, Y. et al. An integrated analysis of genes and functional pathways for aggression in human and rodent models. Mol. Psychiatry 24, 1655–1667 (2019).
Kelly, A. M. & Wilson, L. C. Aggression: perspectives from social and systems neuroscience. Horm. Behav. 123, 104523 (2020).
O’Connell, L. A. & Hofmann, H. A. Evolution of a vertebrate social decision-making network. Science 336, 1154–1157 (2012).
Rosvall, K. A. Proximate perspectives on the evolution of female aggression: good for the gander, good for the goose? Philos. Trans. R. Soc. Lond. B 368, 20130083 (2013).
Clutton-Brock, T. Sexual selection in males and females. Science 318, 1882–1885 (2007).
Rosvall, K. A. Intrasexual competition in females: evidence for sexual selection? Behav. Ecol. 22, 1131–1140 (2011).
Wright, A. E. & Mank, J. E. The scope and strength of sex-specific selection in genome evolution. J. Evol. Biol. 26, 1841–1853 (2013).
Goymann, W. & Wingfield, J. C. Male-to-female testosterone ratios, dimorphism, and life history—what does it really tell us? Behav. Ecol. 25, 685–699 (2014).
Rosvall, K. A., Bentz, A. B. & George, E. M. How research on female vertebrates contributes to an expanded challenge hypothesis. Horm. Behav. 123, 104565 (2020).
Smiley, K. O. et al. Beyond a biased binary: a perspective on the misconceptions, challenges, and implications of studying females in avian behavioral endocrinology. Front. Physiol. 13, 970603 (2022).
Duckworth, R. A. Adaptive dispersal strategies and the dynamics of a range expansion. Am. Nat. 172, S4–S17 (2008).
Leffelaar, D. & Robertson, R. J. Nest usurpation and female competition for breeding opportunities by tree swallows. Wilson Bull. 97, 221–224 (1985).
Merilä, J. & Wiggins, D. A. Interspecific competition for nest holes causes adult mortality in the collared flycatcher. Condor 97, 445–450 (1995).
Albers, A. N., Jones, J. A. & Siefferman, L. Behavioral differences among eastern bluebird populations could be a consequence of tree swallow presence: a pilot study. Front. Ecol. Evol 5 (2017).
Krieg, C. A. & Getty, T. Fitness benefits to intrasexual aggression in female house wrens, Troglodytes aedon. Anim. Behav. 160, 79–90 (2020).
Rosvall, K. A. Sexual selection on aggressiveness in females: evidence from an experimental test with tree swallows. Anim. Behav. 75, 1603–1610 (2008).
Rubenstein, D. R. & Lovette, I. J. Reproductive skew and selection on female ornamentation in social species. Nature 462, 786–789 (2009).
Schuppe, E. R., Sanin, G. D. & Fuxjager, M. J. The social context of a territorial dispute differentially influences the way individuals in breeding pairs coordinate their aggressive tactics. Behav. Ecol. Sociobiol. 70, 673–682 (2016).
Lipshutz, S. E. & Rosvall, K. A. Nesting strategy shapes territorial aggression but not testosterone: a comparative approach in female and male birds. Horm. Behav. 133, 104995 (2021).
Fang, Y. T., Tuanmu, M. N. & Hung, C. M. Asynchronous evolution of interdependent nest characters across the avian phylogeny. Nat. Commun. 9, 1863 (2018).
Zenil-Ferguson, R., McEntee, J. P., Burleigh, J. G. & Duckworth, R. A. Linking ecological specialization to its macroevolutionary consequences: an example with passerine nest type. Syst. Biol. 72, 294–306 (2023).
Jawor, J. M. et al. Seasonal and individual variation in response to GnRH challenge in male dark-eyed juncos (Junco hyemalis). Gen. Comp. Endocrinol. 149, 182–189 (2006).
Rosvall, K. A., Bergeon Burns, C. M., Jayaratna, S. P. & Ketterson, E. D. Divergence along the gonadal steroidogenic pathway: implications for hormone-mediated phenotypic evolution. Horm. Behav. 84, 1–8 (2016).
Bukhari, S. A. et al. Temporal dynamics of neurogenomic plasticity in response to social interactions in male threespined sticklebacks. PLoS Genet. 13, e1006840 (2017).
Goodson, J. L. The vertebrate social behavior network: evolutionary themes and variations. Horm. Behav. 48, 11–22 (2005).
Stelzer, G. et al.The GeneCards Suite: from gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinform. 54, 1.30.31–31.30.33 (2016).
Ghanem, N. et al. The Rb/E2F pathway modulates neurogenesis through direct regulation of the Dlx1/Dlx2 bigene cluster. J. Neurosci. 32, 8219–8230 (2012).
Chandra, B., Voas, M. G., Davies, E. L. & Roberts-Galbraith, R. H. Ets-1 transcription factor regulates glial cell regeneration and function in planarians. Development 150, dev201666 (2023).
Hung, H. H. et al. Selective involvement of UGGT variant: UGGT2 in protecting mouse embryonic fibroblasts from saturated lipid-induced ER stress. Proc. Natl Acad. Sci. USA 119, e2214957119 (2022).
Yan, C. et al. Correction: IRE1 promotes neurodegeneration through autophagy-dependent neuron death in the Drosophila model of Parkinson’s disease. Cell Death Dis. 11, 150 (2020).
Zhu, J. Y., Chen, M., Mu, W. J., Luo, H. Y. & Guo, L. The functional role of Higd1a in mitochondrial homeostasis and in multiple disease processes. Genes Dis. 10, 1833–1845 (2023).
Crombie, E. M., Cleverley, K., Timmers, H. T. M. & Fisher, E. M. C. The roles of TAF1 in neuroscience and beyond. R. Soc. Open Sci. 11, 240790 (2024).
Baran, N. M., McGrath, P. T. & Streelman, J. T. Applying gene regulatory network logic to the evolution of social behavior. Proc. Natl Acad. Sci. USA 114, 5886–5893 (2017).
Bentz, A. B. et al. Experimental competition induces immediate and lasting effects on the neurogenome in free-living female birds. Proc. Natl Acad. Sci. USA 118, e2016154118 (2021).
Filby, A. L., Paull, G. C., Hickmore, T. F. & Tyler, C. R. Unravelling the neurophysiological basis of aggression in a fish model. BMC Genomics 11, 498 (2010).
Storz, J. F. Causes of molecular convergence and parallelism in protein evolution. Nat. Rev. Genet. 17, 239–250 (2016).
West-Eberhard, M. J. Sexual selection, social competition, and speciation. Q. Rev. Biol. 58, 155–183 (1983).
Harts, A. M., Booksmythe, I. & Jennions, M. D. Mate guarding and frequent copulation in birds: a meta-analysis of their relationship to paternity and male phenotype. Evolution 70, 2789–2808 (2016).
Drury, J. P., Cowen, M. C. & Grether, G. F. Competition and hybridization drive interspecific territoriality in birds. Proc. Natl Acad. Sci. USA 117, 12923–12930 (2020).
Husak, J. F. et al. Life history and environment predict variation in testosterone across vertebrates. Evolution 75, 1003–1010 (2021).
Goymann, W., Moore, I. T. & Oliveira, R. F. Challenge hypothesis 2.0: a fresh look at an established idea. BioScience 69, 432–442 (2019).
Wingfield, J. C., Ramenofsky, M., Hegner, R. E. & Ball, G. F. Whither the challenge hypothesis? Horm. Behav. 123, 104588 (2020).
Lischinsky, J. E. & Lin, D. Neural mechanisms of aggression across species. Nat. Neurosci. 23, 1317–1328 (2020).
Lipshutz, S. E., George, E. M., Bentz, A. B. & Rosvall, K. A. Evaluating testosterone as a phenotypic integrator: from tissues to individuals to species. Mol. Cell. Endocrinol. 496, 110531 (2019).
Fischer, E. K., Song, Y., Hughes, K. A., Zhou, W. & Hoke, K. L. Nonparallel transcriptional divergence during parallel adaptation. Mol. Ecol. 30, 1516–1530 (2021).
Renn, S. C. P. et al. Gene expression signatures of mating system evolution. Genome 61, 287–297 (2018).
Alaux, C. et al. Honey bee aggression supports a link between gene regulation and behavioral evolution. Proc. Natl Acad. Sci. USA 106, 15400–15405 (2009).
Mullon, C., Wright, A. E., Reuter, M., Pomiankowski, A. & Mank, J. E. Evolution of dosage compensation under sexual selection differs between X and Z chromosomes. Nat. Commun. 6, 7720 (2015).
Grath, S. & Parsch, J. Sex-biased gene expression. Annu Rev. Genet 50, 29–44 (2016).
Fridolfsson, A.-K. & Ellegren, H. A simple and universal method for molecular sexing of non-ratite birds. J. Avian Biol. 30, 116–121 (1999).
Soma, K. K., Schlinger, B. A., Wingfield, J. C. & Saldanha, C. J. Brain aromatase, 5 alpha-reductase, and 5 beta-reductase change seasonally in wild male song sparrows: relationship to aggressive and sexual behavior. J. Neurobiol. 56, 209–221 (2003).
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011).
Rhie, A. et al. Towards complete and error-free genome assemblies of all vertebrate species. Nature 592, 737–746 (2021).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Plaisier, S. B., Taschereau, R., Wong, J. A. & Graeber, T. G. Rank–rank hypergeometric overlap: identification of statistically significant overlap between gene-expression signatures. Nucleic Acids Res. 38, e169–e169 (2010).
Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 9, 559 (2008).
Mi, H., Muruganujan, A., Ebert, D., Huang, X. & Thomas, P. D. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 47, D419–D426 (2018).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504 (2003).
Hadfield, J. D. MCMC methods for multi-response generalized linear mixed models: the MCMCglmm R package. J. Stat. Softw. 33, 1–22 (2010).
Jetz, W., Thomas, G. H., Joy, J. B., Hartmann, K. & Mooers, A. O. The global diversity of birds in space and time. Nature 491, 444–448 (2012).
Paradis, E. & Schliep, K. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526–528 (2019).
Revell, L. J. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223 (2011).
Seabold, S. & Perktold, J. Statsmodels: Econometric and Statistical Modeling with Python. In Proc. 9th Python in Science Conference (eds van der Walt, S. & Millman, J.) 92–96 (2010).
Kolberg, L. et al. g:Profiler—interoperable web service for functional enrichment analysis and gene identifier mapping (2023 update). Nucleic Acids Res. 51, W207–W212 (2023).
Lipshutz, S. & Hibbins, M. slipshut/CavityNesting: cavity nesting github release (v1.1.1). Zenodo https://doi.org/10.5281/zenodo.14715061 (2025).
Lipshutz, S. et al. Repeated behavioral evolution is associated with convergence of gene expression in cavity-nesting songbirds. Figshare https://doi.org/10.6084/m9.figshare.26367061 (2024).
Kumar, S., Stecher, G., Suleski, M. & Hedges, S. B. TimeTree: a resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 34, 1812–1819 (2017).
Acknowledgements
For facilitating fieldwork, we thank the Indiana Department of Natural Resources; Indiana University’s Research and Teaching Preserve; T. Rothfus and C. Hagie at Therkilsden Field station; A. Fournier and S. Havera at Forbes Field Station; and J. Hoover, N. Antonson, H. Scharf and A. Funk. For vocal stimuli, we thank M. Wilkins, D. Reichard, C. Krieg and E. Chen. Thanks to E. Mueller for help with 3D printing; S. Nallu and J. Huang for TapeStation and library preparation; E. D. Curole and J. K. for assistance in the lab; and B. Young for assistance with data analysis. This work was funded by the US National Science Foundation grant DBI-1907134 (S.E.L.), DBI-2146866 (M.W.H.), IOS-1953226 (M.E.H.) and CAREER grant IOS-1942192 (K.A.R.), as well as support from Indiana University Biology, Loyola Biology and Duke Biology.
Author information
Authors and Affiliations
Contributions
S.E.L. and K.A.R. conceived of the study; S.E.L. led the data collection, supported by A.B.B., A.M.T., E.M.G, M.E.H., M.J.W., S.J.T, S.E.W., T.A.E. and W.M.S.; S.E.L., M.S.H., A.B.B., A.M.B., D.B.R., Q.K.T., M.W.H. and K.A.R. analysed and interpreted the data; S.E.L., M.S.H. and K.A.R. wrote the paper, and all authors contributed to revisions. The manuscript reflects the contributions and ideas of all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Ecology & Evolution thanks Natasha Bloch, Chih-Ming Hung and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Sections 1–12, Tables 1–13 and Figs. 1–20.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lipshutz, S.E., Hibbins, M.S., Bentz, A.B. et al. Repeated behavioural evolution is associated with convergence of gene expression in cavity-nesting songbirds. Nat Ecol Evol 9, 845–856 (2025). https://doi.org/10.1038/s41559-025-02675-x
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41559-025-02675-x
This article is cited by
-
The genetic foundations of convergent traits
Nature Reviews Genetics (2026)
-
Repeated evolution of a complex behaviour
Nature Ecology & Evolution (2025)