Abstract
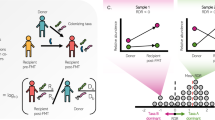
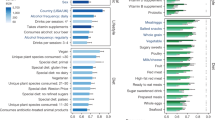
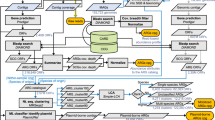
The intestinal microbiota is considered to be a major reservoir of antibiotic resistance determinants (ARDs) that could potentially be transferred to bacterial pathogens via mobile genetic elements. Yet, this assumption is poorly supported by empirical evidence due to the distant homologies between known ARDs (mostly from culturable bacteria) and ARDs from the intestinal microbiota. Consequently, an accurate census of intestinal ARDs (that is, the intestinal resistome) has not yet been fully determined. For this purpose, we developed and validated an annotation method (called pairwise comparative modelling) on the basis of a three-dimensional structure (homology comparative modelling), leading to the prediction of 6,095 ARDs in a catalogue of 3.9 million proteins from the human intestinal microbiota. We found that the majority of predicted ARDs (pdARDs) were distantly related to known ARDs (mean amino acid identity 29.8%) and found little evidence supporting their transfer between species. According to the composition of their resistome, we were able to cluster subjects from the MetaHIT cohort (n = 663) into six resistotypes that were connected to the previously described enterotypes. Finally, we found that the relative abundance of pdARDs was positively associated with gene richness, but not when subjects were exposed to antibiotics. Altogether, our results indicate that the majority of intestinal microbiota ARDs can be considered intrinsic to the dominant commensal microbiota and that these genes are rarely shared with bacterial pathogens.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The 6,095 pdARDs PDB files, nucleotide and amino acid sequences can be downloaded from http://mgps.eu/Mustard/. The 3.9 million gene catalogue and the MGS database are accessible at https://www.cbs.dtu.dk/projects/CAG/. The reads from the clinical samples generated in this study are available under the accession number PRJEB27799 at the European Nucleotide Archive.
References
United Nations High-Level Meeting on Antimicrobial Resistance (WHO, UN, 2016).
Ghosh, T. S., Gupta, S. S., Nair, G. B. & Mande, S. S. In silico analysis of antibiotic resistance genes in the gut microflora of individuals from diverse geographies and age-groups. PLoS ONE 8, e83823 (2013).
Hu, Y. et al. Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota. Nat. Commun. 4, 2151 (2013).
Forslund, K. et al. Country-specific antibiotic use practices impact the human gut resistome. Genome Res. 23, 1163–1169 (2013).
Sommer, M. O. A., Dantas, G. & Church, G. M. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science 325, 1128–1131 (2009).
Moore, A. M. et al. Pediatric fecal microbiota harbor diverse and novel antibiotic resistance genes. PLoS ONE 8, e78822 (2013).
Gupta, S. K. et al. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 58, 212–220 (2014).
McArthur, A. G. et al. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 57, 3348–3357 (2013).
Zankari, E. et al. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644 (2012).
Arango-Argoty, G. et al. DeepARG: a deep learning approach for predicting antibiotic resistance genes from metagenomic data. Microbiome 6, 23 (2018).
Gibson, M. K., Forsberg, K. J. & Dantas, G. Improved annotation of antibiotic resistance determinants reveals microbial resistomes cluster by ecology. ISME J. 9, 207–216 (2015).
Wright, G. D. The antibiotic resistome: the nexus of chemical and genetic diversity. Nat. Rev. Microbiol. 5, 175–186 (2007).
Salyers, A. A., Gupta, A. & Wang, Y. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 12, 412–416 (2004).
Ghosh, S., Sadowsky, M. J., Roberts, M. C., Gralnick, J. A. & LaPara, T. M. Sphingobacterium sp. strain PM2-P1-29 harbours a functional tet(X) gene encoding for the degradation of tetracycline. J. Appl. Microbiol. 106, 1336–1342 (2009).
Stinear, T. P., Olden, D. C., Johnson, P. D., Davies, J. K. & Grayson, M. L. Enterococcal vanB resistance locus in anaerobic bacteria in human faeces. Lancet 357, 855–856 (2001).
Penders, J., Stobberingh, E. E., Savelkoul, P. H. M. & Wolffs, P. F. G. The human microbiome as a reservoir of antimicrobial resistance. Front. Microbiol. 4, 87 (2013).
Zhang, Y. & Skolnick, J. TM-Align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 33, 2302–2309 (2005).
Forsberg, K. J. et al. Bacterial phylogeny structures soil resistomes across habitats. Nature 509, 612–616 (2014).
Nielsen, H. B. et al. Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat. Biotechnol. 32, 822–828 (2014).
Goossens, H., Ferech, M., Van der Stichele, R. & Elseviers, M. & ESAC Project Group. Outpatient antibiotic use in Europe and association with resistance: a cross-national database study. Lancet 365, 579–587 (2005).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Berglund, F. et al. Identification of 76 novel B1 metallo-β-lactamases through large-scale screening of genomic and metagenomic data. Microbiome 5, 134 (2017).
Everard, A. et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl Acad. Sci. USA 110, 9066–9071 (2013).
Sokol, H. et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl Acad. Sci. USA 105, 16731–16736 (2008).
Leski, T. A. et al. Multidrug-resistant tet(X)-containing hospital isolates in Sierra Leone. Int. J. Antimicrob. Agents 42, 83–86 (2013).
Plaza Oñate, F. et al. MSPminer: abundance-based reconstitution of microbial pan-genomes from shotgun metagenomic data. Bioinformatics https://doi.org/10.1093/bioinformatics/bty830 (2018).
Li, J. et al. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 32, 834–841 (2014).
Arumugam, M. et al. Enterotypes of the human gut microbiome. Nature 473, 174–180 (2011).
de Smet, A. M. G. A. et al. Decontamination of the digestive tract and oropharynx in ICU patients. N. Engl. J. Med. 360, 20–31 (2009).
van Schaik, W. The human gut resistome. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370, (2015).
Le Chatelier, E. et al. Richness of human gut microbiome correlates with metabolic markers. Nature 500, 541–546 (2013).
Pehrsson, E. C. et al. Interconnected microbiomes and resistomes in low-income human habitats. Nature 533, 212–216 (2016).
Léonard, F., Andremont, A., Leclerq, B., Labia, R. & Tancrède, C. Use of β- lactamase-producing anaerobes to prevent ceftriaxone from degrading intestinal resistance to colonization. J. Infect. Dis. 160, 274–280 (1989).
Bilinski, J. et al. Fecal microbiota transplantation in patients with blood disorders inhibits gut colonization with antibiotic-resistant bacteria: results of a prospective, single-center study. Clin. Infect. Dis. 65, 364–370 (2017).
Lupo, A., Coyne, S. & Berendonk, T. U. Origin and evolution of antibiotic resistance: the common mechanisms of emergence and spread in water bodies. Front. Microbiol. 3, 18 (2012).
Lagier, J.-C. et al. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat. Microbiol. 1, 16203 (2016).
Illergård, K., Ardell, D. H. & Elofsson, A. Structure is three to ten times more conserved than sequence—a study of structural response in protein cores. Proteins 77, 499–508 (2009).
Baquero, F., Tedim, A. P. & Coque, T. M. Antibiotic resistance shaping multi-level population biology of bacteria. Front. Microbiol. 4, 15 (2013).
Martínez, J. L., Coque, T. M. & Baquero, F. What is a resistance gene? Ranking risk in resistomes. Nat. Rev. Microbiol. 13, 116–123 (2015).
Van Boeckel, T. P. et al. Global antibiotic consumption 2000 to 2010: an analysis of national pharmaceutical sales data. Lancet. Infect. Dis. 14, 742–750 (2014).
Allen, H. K., Moe, L. A., Rodbumrer, J., Gaarder, A. & Handelsman, J. Functional metagenomics reveals diverse β- lactamases in a remote Alaskan soil. ISME J. 3, 243–251 (2009).
Hyatt, D. et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119 (2010).
Fu, L., Niu, B., Zhu, Z., Wu, S. & Li, W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152 (2012).
Powell, S. et al. eggNOGv3.0: orthologous groups covering 1133 organisms at 41 different taxonomic ranges. Nucleic Acids Res. 40, D284–D289 (2012).
Liu, Y.-Y. et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168 (2016).
Kultima, J. R. et al. MOCAT: a metagenomics assembly and gene prediction toolkit. PLoS ONE 7, e47656 (2012).
Finn, R. D., Clements, J. & Eddy, S. R. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37 (2011).
Pearson, W. R. & Lipman, D. J. Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA 85, 2444–2448 (1988).
Berman, H. M. et al. The Protein Data Bank. Nucleic Acids Res. 28, 235–242 (2000).
Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011).
Jones, D. T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202 (1999).
Sali, A. & Blundell, T. L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 (1993).
Wallner, B. & Elofsson, A. Can correct protein models be identified?. Protein Sci. 12, 1073–1086 (2003).
Wiederstein, M. & Sippl, M. J. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 35, W407–W410 (2007).
Ortiz, A. R., Strauss, C. E. M. & Olmea, O. MAMMOTH (matching molecular models obtained from theory): an automated method for model comparison. Protein Sci. Publ. Protein Soc. 11, 2606–2621 (2002).
Tibshirani, R. Regression shrinkage and selection via the lasso. J R. Stat. Soc B 58, 267–288 (1996).
Fan, R. E., Chang, K. W., Hsieh, C. J., Wang, X. R. & Lin, C. J. A library for large linear classification. JMLR 9, 1871–1874 .
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J. & Chandler, M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36 (2006).
Guglielmini, J., Quintais, L., Garcillán-Barcia, M. P., de la Cruz, F. & Rocha, E. P. C. The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet. 7, e1002222 (2011).
Cury, J., Jové, T., Touchon, M., Néron, B. & Rocha, E. P. Identification and analysis of integrons and cassette arrays in bacterial genomes. Nucleic Acids Res. 44, 4539–4550 (2016).
Holmes, I., Harris, K. & Quince, C. Dirichlet multinomial mixtures: generative models for microbial metagenomics. PLoS ONE 7, e30126 (2012).
Ding, T. & Schloss, P. D. Dynamics and associations of microbial community types across the human body. Nature 509, 357–360 (2014).
Dray, S. & Legendre, P. Testing the species traits-environment relationships: the fourth-corner problem revisited. Ecology 89, 3400–3412 (2008).
Godon, J. J., Zumstein, E., Dabert, P., Habouzit, F. & Moletta, R. Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl. Environ. Microbiol. 63, 2802–2813 (1997).
Suau, A. et al. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65, 4799–4807 (1999).
Pons, N. et al. METEOR—A Platform for Quantitative Metagenomic Profiling of Complex Ecosystems (JOBIM Montpellier, 2010); http://www.jobim2010.fr/sites/default/files/presentations/27Pons.pdf
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Quereda, J. J. et al. Bacteriocin from epidemic Listeria strains alters the host intestinal microbiota to favor infection. Proc. Natl Acad. Sci. USA 113, 5706–5711 (2016).
Robert, P. & Escoufier, Y. A unifying tool for linear multivariate statistical methods: the RV-coefficient. Appl. Stat. 25, 257 (1976).
Acknowledgements
The authors are grateful to the GENOTOUL (Toulouse, France), GENOUEST (Rennes, France), ABIMS (Roscoff, France), MIGALE (Jouy-en-Josas) and TGCC-GENCI (Institut Curie) calculation clusters. The authors also thank B. Perichon (Institut Pasteur, Paris, France) for providing ARD sequences from Acinetobacter baumannii, P. Siguier (CNRS, Toulouse, France) for helping the search of insertion sequences with ISfinder, J. Guglielmini (Institut Pasteur, Paris, France) for his assistance in finding conjugative elements, S. Volant (Institut Pasteur, Paris, France) for the design of the statistical model in SHAMAN, T. Jové (University of Limoges, France) for his assistance in finding integrons, M. Petitjean (IAME Research Center, Paris, France) for her assistance in bioinformatic analyses, and F. Plaza-Oñate and M. Almeida for their help with MSPs. The project was funded in part by the European Union Seventh Framework Programme (FP7-HEALTH-2011-single-stage) under grant agreement number 282004, EvoTAR. IRYCIS authors acknowledge the European Development Regional Fund ‘A way to achieve Europe’ for co-founding the Spanish R&D National Plan 2012–2019 Work (PI15-0512), CIBER (CIBERESP; CB06/02/0053) and the Government of Madrid (InGeMICS- B2017/BMD-3691). V.F.L. was further funded by a Research Award Grant 2016 of the European Society for Clinical Microbiology and Infectious Diseases.
Author information
Authors and Affiliations
Contributions
E.R., A.G. and J.T. performed the analysis. E.R., A.G., J.T., W.v.S., A.d.B. and S.P.K. wrote the manuscript. A.S.A. and N.M. handled the data management. T.C., S.H.A., I.C. and J.L.M. performed the gene synthesis experiments. J.L.M., T.M.C., V.F.L., F.B., A.d.B., J.D., S.P.K., F.H. and S.D.E. discussed the protocol and results. L.M., T.G., V.d.L., N.A., B.F., I.W., A.A., W.v.S., M.R., X.Z. and R.J.L. recruited the patients and collected the samples. H.B., V.L., A.L. and F.L. handled the wet lab experiments. N.P., P.L. and J.M.B. managed the informatics and the calculation clusters. K.W. and N.P. designed the website (http://mgps.eu/Mustard/).
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–17, Supplementary Notes, Supplementary References.
Supplementary Table 1
The 6,095 pdARDs that were found in the 3.9 million protein catalogue. PCM score missing values means that the candidate could not be modelled with the negative template, so that the PCM score was considered to be over 50%.
Supplementary Table 2
Description of the 16 candidates sharing at least 40% amino acid identity with a reference ARD but being not predicted as an ARD by PCM. The TM score represents the degree of correct alignment of the structure generated by PCM and a reference structure (the highest score being 1).
Supplementary Table 3
Description of the 49 pdARDs found in plasmids and/or phages from GenBank.
Supplementary Table 4
Description of the 82 pdARDs shared by ≥2 species. Insertion sequences, conjugative elements and integrons were searched the same way as described in the Methods section.
Supplementary Table 5
Details on the 74 MGS that were found to be differentially abundant between subjects with no recent antibiotic exposure (n = 31) to antibiotics and subjects under chronic exposure to antibiotics (n = 30) using the Wald unpaired test. Padj, adjusted P-value (Benjamini–Hochberg correction).
Supplementary Table 6
Details on the 133 MGS that were found to be differentially abundant between subjects (n = 10) before and after SDD using the Wald paired test. Padj, adjusted P-value (Benjamini–Hochberg correction).
Supplementary Table 7
Predictions of ARDs in the functional metagenomics dataset from soils9 by the PCM method.
Rights and permissions
About this article
Cite this article
Ruppé, E., Ghozlane, A., Tap, J. et al. Prediction of the intestinal resistome by a three-dimensional structure-based method. Nat Microbiol 4, 112–123 (2019). https://doi.org/10.1038/s41564-018-0292-6
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41564-018-0292-6
This article is cited by
-
Towards unraveling antimicrobial resistance dynamics: a longitudinal exploration of rectal swab metagenomes
BMC Microbiology (2025)
-
Comprehensive genome catalog analysis of the resistome, virulome and mobilome in the wild rodent gut microbiota
npj Biofilms and Microbiomes (2025)
-
ProtAlign-ARG: antibiotic resistance gene characterization integrating protein language models and alignment-based scoring
Scientific Reports (2025)
-
Multi-dimensional metagenomics
Nature Reviews Bioengineering (2025)
-
Antibiotic-perturbed microbiota and the role of probiotics
Nature Reviews Gastroenterology & Hepatology (2025)