Abstract
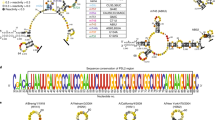
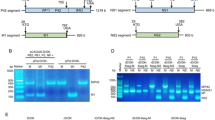
Influenza A viruses (IAVs) constitute a major threat to human health. The IAV genome consists of eight single-stranded viral RNA segments contained in separate viral ribonucleoprotein (vRNP) complexes that are packaged together into a single virus particle. The structure of viral RNA is believed to play a role in assembling the different vRNPs into budding virions1,2,3,4,5,6,7,8 and in directing reassortment between IAVs9. Reassortment between established human IAVs and IAVs harboured in the animal reservoir can lead to the emergence of pandemic influenza strains to which there is little pre-existing immunity in the human population10,11. While previous studies have revealed the overall organization of the proteins within vRNPs, characterization of viral RNA structure using conventional structural methods is hampered by limited resolution and an inability to resolve dynamic components12,13. Here, we employ multiple high-throughput sequencing approaches to generate a global high-resolution structure of the IAV genome. We show that different IAV genome segments acquire distinct RNA conformations and form both intra- and intersegment RNA interactions inside influenza virions. We use our detailed map of IAV genome structure to provide direct evidence for how intersegment RNA interactions drive vRNP cosegregation during reassortment between different IAV strains. The work presented here is a roadmap both for the development of improved vaccine strains and for the creation of a framework to ‘risk assess’ reassortment potential to better predict the emergence of new pandemic influenza strains.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
References
Gerber, M., Isel, C., Moules, V. & Marquet, R. Selective packaging of the influenza A genome and consequences for genetic reassortment. Trends Microbiol. 22, 446–455 (2014).
Hutchinson, E. C., von Kirchbach, J. C., Gog, J. R. & Digard, P. Genome packaging in influenza A virus. J. Gen. Virol. 91, 313–328 (2010).
Gavazzi, C. et al. A functional sequence-specific interaction between influenza A virus genomic RNA segments. Proc. Natl Acad. Sci. USA 110, 16604–16609 (2013).
Fournier, E. et al. A supramolecular assembly formed by influenza A virus genomic RNA segments. Nucleic Acids Res. 40, 2197–2209 (2012).
Marsh, G. A., Rabadán, R., Levine, A. J. & Palese, P. Highly conserved regions of influenza A virus polymerase gene segments are critical for efficient viral RNA packaging. J. Virol. 82, 2295–2304 (2008).
Gog, J. R. et al. Codon conservation in the influenza A virus genome defines RNA packaging signals. Nucleic Acids Res. 35, 1897–1907 (2007).
Hutchinson, E. C., Curran, M. D., Read, E. K., Gog, J. R. & Digard, P. Mutational analysis of cis-acting RNA signals in segment 7 of influenza A virus. J. Virol. 82, 11869–11879 (2008).
Hutchinson, E. C., Wise, H. M., Kudryavtseva, K., Curran, M. D. & Digard, P. Characterisation of influenza A viruses with mutations in segment 5 packaging signals. Vaccine 27, 6270–6275 (2009).
Gilbertson, B. et al. Influenza NA and PB1 gene segments interact during the formation of viral progeny: localization of the binding region within the PB1 gene. Viruses 8, E238 (2016).
Lowen, A. C. Constraints, drivers, and implications of influenza A virus reassortment. Annu. Rev. Virol. 4, 105–121 (2017).
Taubenberger, J. K. & Kash, J. C. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe 7, 440–451 (2010).
Arranz, R. et al. The structure of native influenza virion ribonucleoproteins. Science 338, 1634–1637 (2012).
Moeller, A., Kirchdoerfer, R. N., Potter, C. S., Carragher, B. & Wilson, I. A. Organization of the influenza virus replication machinery. Science 338, 1631–1634 (2012).
Te Velthuis, A. J. & Fodor, E. Influenza virus RNA polymerase: insights into the mechanisms of viral RNA synthesis. Nat. Rev. Microbiol. 14, 479–493 (2016).
Pflug, A., Lukarska, M., Resa-Infante, P., Reich, S. & Cusack, S. Structural insights into RNA synthesis by the influenza virus transcription-replication machine. Virus Res. 234, 103–117 (2017).
Wilkinson, K. A. et al. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 6, e96 (2008).
Smola, M. J., Rice, G. M., Busan, S., Siegfried, N. A. & Weeks, K. M. Selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) for direct, versatile and accurate RNA structure analysis. Nat. Protoc. 10, 1643–1669 (2015).
Watts, J. M. et al. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 460, 711–716 (2009).
Lee, N. et al. Genome-wide analysis of influenza viral RNA and nucleoprotein association. Nucleic Acids Res. 45, 8968–8977 (2017).
Williams, G. D. et al. Nucleotide resolution mapping of influenza A virus nucleoprotein-RNA interactions reveals RNA features required for replication. Nat. Commun. 9, 465 (2018).
Baudin, F., Bach, C., Cusack, S. & Ruigrok, R. W. Structure of influenza virus RNP. I. influenza virus nucleoprotein melts secondary structure in panhandle RNA and exposes the bases to the solvent. EMBO J. 13, 3158–3165 (1994).
Kobayashi, Y. et al. Computational and molecular analysis of conserved influenza A virus RNA secondary structures involved in infectious virion production. RNA Biol. 13, 883–894 (2016).
Gultyaev, A. P. et al. RNA structural constraints in the evolution of the influenza A virus genome NP segment. RNA Biol. 11, 942–952 (2014).
Aw, J. G. A. et al. In vivo mapping of eukaryotic RNA interactomes reveals principles of higher-order organization and regulation. Mol. Cell 62, 603–617 (2016).
Cobbin, J. C., Verity, E. E., Gilbertson, B. P., Rockman, S. P. & Brown, L. E. The source of the PB1 gene in influenza vaccine reassortants selectively alters the hemagglutinin content of the resulting seed virus. J. Virol. 87, 5577–5585 (2013).
Cobbin, J. C. et al. Influenza virus PB1 and neuraminidase gene segments can cosegregate during vaccine reassortment driven by interactions in the PB1 coding region. J. Virol. 88, 8971–8980 (2014).
Moreira, É. A. et al. A conserved influenza A virus nucleoprotein code controls specific viral genome packaging. Nat. Commun. 7, 12861 (2016).
Saira, K. et al. Sequence analysis of in vivo defective interfering-like RNA of influenza A H1N1 pandemic virus. J. Virol. 87, 8064–8074 (2013).
Noda, T. et al. Three-dimensional analysis of ribonucleoprotein complexes in influenza A virus. Nat. Commun. 3, 639 (2012).
Hutchinson, E. C. et al. Mapping the phosphoproteome of influenza A and B viruses by mass spectrometry. PLoS Pathog. 8, e1002993 (2012).
Hutchinson, E. C. et al. Conserved and host-specific features of influenza virion architecture. Nat. Commun. 5, 4816 (2014).
Mitra, S. Detecting RNA tertiary folding by sedimentation velocity analytical ultracentrifugation. Methods Mol. Biol. 1086, 265–288 (2014).
Turner, R., Shefer, K. & Ares, M. Jr. Safer one-pot synthesis of the ‘SHAPE’ reagent 1-methyl-7-nitroisatoic anhydride (1m7). RNA 19, 1857–1863 (2013).
Wilkinson, K. A., Merino, E. J. & Weeks, K. M. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 1, 1610–1616 (2006).
Aw, J. G. A., Shen, Y., Nagarajan, N. & Wan, Y. Mapping RNA-RNA interactions globally using biotinylated psoralen. J. Vis. Exp. 123, 55255 (2017).
Jiang, H., Lei, R., Ding, S. & Zhu, S. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics 15, 182 (2014).
Busan, S. & Weeks, K. M. Accurate detection of chemical modifications in RNA by mutational profiling (MaP) with ShapeMapper 2. RNA 24, 143–148 (2018).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Gu, Z., Gu, L., Eils, R., Schlesner, M. & Brors, B. circlize implements and enhances circular visualization in R. Bioinformatics 30, 2811–2812 (2014).
Busch, A., Richter, A. S. & Backofen, R. IntaRNA: efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinformatics 24, 2849–2856 (2008).
Leontis, N. B. & Zirbel, C. L. in RNA 3D Structure Analysis and Prediction (eds Leontis, N. & Westhof, E.) 281–298 (Springer, 2012).
Natchiar, S. K., Myasnikov, A. G., Kratzat, H., Hazemann, I. & Klaholz, B. P. Visualization of chemical modifications in the human 80S ribosome structure. Nature 551, 472–477 (2017).
Nguyen, T. H. D. et al. The architecture of the spliceosomal U4/U6.U5 tri-snRNP. Nature 523, 47–52 (2015).
Te Velthuis, A. J. W. et al. Mini viral RNAs act as innate immune agonists during influenza virus infection. Nat. Microbiol. 3, 1234–1242 (2018).
Lorenz, R. et al. ViennaRNA Package 2.0. Algorithms Mol. Biol. 6, 26 (2011).
Hoffmann, E., Neumann, G., Kawaoka, Y., Hobom, G. & Webster, R. G. DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl Acad. Sci. USA 97, 6108–6113 (2000).
Hoffmann, E., Stech, J., Guan, Y., Webster, R. & Perez, D. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 146, 2275–2289 (2001).
Tannock, G. A., Paul, J. A. & Barry, R. D. Relative immunogenicity of the cold-adapted influenza virus A/Ann Arbor/6/60 (A/AA/6/60-ca), recombinants of A/AA/6/60-ca, and parental strains with similar surface antigens. Infect. Immun. 43, 457–462 (1984).
Acknowledgements
We thank G.G. Brownlee and J. Kenyon for helpful discussions, J. Kenyon, J.G. Aw, and Y. Wan for sharing protocols, J. Robertson for making the 1M7 reagent, J. Sharps for technical assistance and St Jude Children’s Research Hospital for providing the pHW2000 plasmid. This work was supported by a Wellcome Trust studentship (no. 105399/Z/14/Z to B.D.), a UK Biotechnology and Biological Sciences Research Council studentship (no. BB/M011224/1 to M.L.K.), grants from the National Institutes of Health (nos. HL111527, GM101237 and HG008133 to A.L.), a National Health and Medical Research Council of Australia programme grant (no. ID1071916 to L.E.B.), UK Medical Research Council programme grants (nos. MR/K000241/1 and MR/R009945/1 to E.F.) and a Sir Edward Penley Abraham Cephalosporin Junior Research Fellowship to D.L.V.B.
Author information
Authors and Affiliations
Contributions
B.D. designed, performed and analysed the SHAPE-MaP and SPLASH experiments and associated virus replication and purification work. M.L.K. performed and analysed the SPLASH experiments on reassortant viruses and associated virus replication and purification experiments. D.L.V.B. designed the SPLASH experiments and analysed the resulting data. B.G. designed and performed the competitive reassortant virus rescue experiments, created the reassortant viruses and analysed the data. S.T. and B.G. performed the viral replication experiments with the reassortant viruses. S.R., A.L., L.E.B., E.F. and D.L.V.B. supervised the work. B.D., B.G. and D.L.V.B. wrote the paper with contributions from the other authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–11.
Supplementary Table 1
Complete SHAPE-MaP data in SNRNASM format.
Supplementary Table 2
Complete SPLASH data and reference genome sequences.
Supplementary Table 3
Sequences of primers used.
Rights and permissions
About this article
Cite this article
Dadonaite, B., Gilbertson, B., Knight, M.L. et al. The structure of the influenza A virus genome. Nat Microbiol 4, 1781–1789 (2019). https://doi.org/10.1038/s41564-019-0513-7
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41564-019-0513-7
This article is cited by
-
Nucleoprotein S69 phosphorylation regulates influenza A virus replication
Animal Diseases (2025)
-
A Benchmark for Virus Infection Reporter Virtual Staining in Fluorescence and Brightfield Microscopy
Scientific Data (2025)
-
Coupling of polymerase-nucleoprotein-RNA in an influenza virus mini ribonucleoprotein complex
Nature Communications (2025)
-
Antiviral potential of diosmin against influenza A virus
Scientific Reports (2025)
-
Designing a multi-epitope vaccine candidate against pandemic influenza a virus: an immunoinformatics and structural vaccinology approach
Molecular Diversity (2025)