Abstract
Numerous important environments harbour low levels of microbial biomass, including certain human tissues, the atmosphere, plant seeds, treated drinking water, hyper-arid soils and the deep subsurface, with some environments lacking resident microbes altogether. These low microbial biomass environments pose unique challenges for standard DNA-based sequencing approaches, as the inevitability of contamination from external sources becomes a critical concern when working near the limits of detection. Likewise, lower-biomass samples can be disproportionately impacted by cross-contamination and practices suitable for handling higher-biomass samples may produce misleading results when applied to lower microbial biomass samples. This Consensus Statement outlines strategies to reduce contamination and cross-contamination, focusing on marker gene and metagenomic analyses. We also provide minimal standards for reporting contamination information and removal workflows. Considerations must be made at every study stage, from sample collection and handling through data analysis and reporting to reduce and identify contaminants. We urge researchers to adopt these recommendations when designing, implementing and reporting microbiome studies, especially those conducted in low-biomass systems.
Similar content being viewed by others
Main
The past two decades have seen a surge in microbiome studies driven by the adoption of cultivation-independent approaches. Most notably, sequencing of targeted marker genes and metagenomes are now widely used to explore the diversity and capabilities of bacteria, archaea, fungi, protists and viruses in different environments. As microbiome research continues to expand, the delineation of best practices has also expanded, with the research community outlining how to best design, implement and report microbiome studies1,2,3,4,5,6,7. However, such recommendations are generally based on practices for studying systems with high levels of microbial biomass, for example, surface soil, wastewater and human stool samples. Microbial DNA yields from these environments can be sufficiently high that contamination is less likely to lead to spurious results, as the target DNA ‘signal’ is far larger than the contaminant ‘noise’8. Yet many systems harbour such low levels of microbial biomass that they approach the limits of detection using standard DNA-based sequencing approaches (Fig. 1). Given the proportional nature of sequence-based datasets, even small amounts of microbial DNA contaminants can strongly influence study results and their interpretation, with this problem becoming particularly relevant when studying low-biomass systems. Such low-biomass systems can include the atmosphere9,10,11, poorly preserved ancient samples12, the deep subsurface13, hyper-arid soils14,15, dry permafrost16, drinking water17, metal surfaces18, rocks19, hypersaline brines20, snow21 and ice cores22,23. Likewise, despite often containing high amounts of host DNA, certain host-associated systems may also harbour minimal amounts of microbial DNA. This includes the respiratory tract24, breastmilk25, fetal tissues26 and blood of humans27, as well as certain plant tissues (for example, seeds)28,29 and certain animal guts (for example, caterpillars)30. Some environments are reported to lack detectable resident microorganisms altogether, including the human placenta, certain animal guts and some polyextreme environments31,32,33,34.
Estimates were obtained using cultivation-independent methods (primarily direct cell counts), but we note that these are average approximations and there can be considerable variability in cell numbers across samples collected from a given environment. Cell numbers/counts were obtained from the published literature and adjusted to account for the sample amounts (volumes or weights) typically used for DNA extractions. See Supplementary Table 1 for details. Points of different colours in this plot indicate general categories of environments.
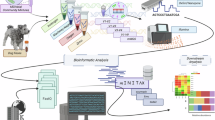
Studying low microbial biomass environments requires careful consideration of methods, including the approaches used for sample collection, laboratory processing and data analysis, to reduce and identify contaminants. Contaminants can be introduced from various sources—notably human sources, sampling equipment, reagents/kits and laboratory environments—and can be introduced at many stages such as sampling, storage, DNA extraction, sequencing and other processing steps8,35,36,37,38. Another persistent problem is cross-contamination, that is, the transfer of DNA or sequence reads between samples, for example, due to well-to-well leakage of DNA39,40,41,42 (Fig. 2). Various post hoc approaches have been developed to remove contaminants from sequence datasets, but such approaches often struggle to accurately distinguish signal from noise, especially for extensively and variably contaminated datasets43,44,45. Concerns regarding contamination in microbiome studies are widely noted, with both refs. 26,41 having detailed a set of guidelines to reduce potential contamination. However, contamination issues persist, and the use of appropriate controls has not increased over the past decade46. Researchers thus remain justifiably skeptical of some published microbiome studies, especially those focused on low-biomass systems26,47. At best, failure to follow suitable practices can cast doubt on the quality of published studies or reduce comparability of results. At worst, there is a risk that inaccurate results may contribute to incorrect conclusions and misinform applications of the research. For example, contamination can distort ecological patterns and evolutionary signatures40,48,49, cause false attribution of pathogen exposure pathways9,50, or lead to inaccurate claims of the presence of microbes in various environments. Consider the debate surrounding the ‘placental microbiome’32,51,52,53, which raised awareness of contamination issues and ‘best practices’ to reduce potential contamination. This is not an isolated example: there have been ongoing debates about contamination issues in other systems, ranging from human blood27,54, brains55 and cancerous tumours47, to the deep subsurface56,57,58 and the upper atmosphere59,60,61.
The target sample (indicated by red smooth shapes) can be contaminated by external contaminants (indicated by grey-shaded sharp shapes). These external contaminants could be cells or DNA from sources other than the sampled community (for example, laboratory reagents, sampling equipment). In addition, the target sample may also be affected by cross-contamination, where cells or DNA are inadvertently exchanged from other samples (indicated by brown-shaded smooth shapes) during sampling, laboratory processing and/or via ‘tag switching’ (as can occur when barcoded reads are misassigned to the incorrect sample). Furthermore, contaminants and cross-contaminants can accumulate throughout the workflow. Prep., preparation.
Here, we propose a series of recommendations for minimizing contamination along with minimal standard guidelines for reporting contamination in microbiome studies. Developed through consensus with leaders in the microbiome field (Supplementary Information), many of our collective recommendations reiterate and refine those described previously36,41. These recommendations and guidelines have been developed so that they are broadly applicable to all microbiome studies, including those focused on host, natural and built environment systems. However, they are particularly important for low microbial biomass environments, as well as any studies where low-level contamination can distort conclusions (for example, pathogen tracking, forensics). Notably, although we focus on marker gene (for example, 16S rRNA gene sequencing) and metagenomic sequencing, these guidelines are also relevant for avoiding contamination across other microbiome methods, for example, metatranscriptomics, DNA stable isotope probing, quantitative PCR and cultivation. We anticipate that careful consideration of these recommendations will ultimately improve the quality of microbiome research and limit some of the more persistent, but often avoidable, problems encountered when studying low-biomass systems. Indeed, while contamination cannot be fully eliminated, these steps enable contamination to be minimized and detected.
Sampling strategies in low-biomass systems
Contamination of a sample can occur at any point in the workflow, from the moment a sample is collected to the generation of the sequence data37,41 (Fig. 2). Major contamination sources during sampling include human operators, sampling equipment and adjacent environments (for example, exposure of a patient’s blood sample to their skin, or a sediment sample to overlying water)62. Due to the largely untargeted nature of most DNA-based approaches, any microbial DNA introduced during sampling can be challenging to distinguish from DNA originating from the sample of interest. A contamination-informed sampling design is therefore recommended to minimize and identify contamination26. The appropriate measures for reducing contamination at the time of sampling will depend on the nature of the system, although there are some core principles that apply. Researchers should consider all possible contamination sources the sample will be exposed to, from the in situ environment to the collection vessel, and take measures to avoid contamination from these sources both before and during sampling26. Before sampling, researchers should take extensive steps to identify and reduce potential contaminants, for example, checking that sampling reagents (for example, sample preservation solutions) are DNA free, and conduct test runs to identify issues and optimize procedures. During sampling, consistent awareness of the objects and environments the sample may be exposed to will enable identification of contamination sources that can be handled by appropriate decontamination or the introduction of barriers. Importantly, training or instruction should be provided to personnel conducting the sampling to ensure procedures are followed. Researchers should be aware that the lower the amount of microbial biomass in the initial sample, the larger the proportional potential impact of contamination on the final sequence-based datasets. Wherever possible, researchers should incorporate the following (see also Table 1).
Decontaminate sources of contaminant cells or DNA
This applies to equipment, tools, vessels and gloves. Ideally, single-use DNA-free objects should be used (such as swabs and collection vessels), but where this is not practical, thorough decontamination is required. For example, decontamination of objects or surfaces with 80% ethanol (to kill contaminating organisms) followed by a nucleic acid degrading solution (to remove traces of their DNA) will minimize contamination from sampling equipment, especially if the same equipment must be used for consecutive samples. Gloves should be similarly decontaminated and should not touch anything before sample collection. Plasticware or glassware used to collect or store samples should be pre-treated by autoclaving or ultraviolet (UV-C) light sterilization, and remain sealed until sample collection to ensure sterility before sampling. It is important to note that sterility is not the same as DNA free: even if viable cells are removed, cell-free DNA can remain on surfaces even after autoclaving or ethanol treatment. Thus, we recommend removing DNA via sodium hypochlorite (bleach), UV-C exposure, hydrogen peroxide, ethylene oxide gas or commercially available DNA removal solutions where safe and practical63.
Use PPE or other barriers to limit contact between samples and contamination sources
Samples should not be handled more than is necessary. If a human operator is taking a sample, they should cover exposed body parts with personal protective equipment (PPE) (including gloves, goggles, coveralls or cleansuits, and shoe covers, as appropriate for the sampling environment). PPE can protect the sample from human aerosol droplets generated while breathing or talking64, as well as from cells shed from clothing, skin and hair65. Some leading examples can be found in cleanroom studies and ancient DNA laboratories. For example, ref. 66 outlined a protocol for spacecraft cleanroom sampling that required all exposed human surfaces to be covered with PPE. Reference 67 described standard ultra-clean laboratory PPE, which includes face masks, suits, visors and three layers of gloves to enable frequent changes while eliminating skin exposure within the lab. While such extensive PPE is only necessary under extreme circumstances, using moderate PPE for all sample collection procedures is a relatively straightforward and inexpensive way to substantially reduce human-derived contamination.
Collect and process samples from potential contamination sources
The inclusion of sampling controls is important for determining the identity and sources of potential contaminants, to evaluate the effectiveness of prevention measures, and interpret the data in context. Sampling controls may include an empty collection vessel, a swab exposed to the air in the sampling environment, swabs of PPE, a swab of surfaces that the sample may come into contact with during sample collection, or an aliquot of sample preservation solution or sampling fluid. Environmental microbiome studies that involve drilling or cutting often include the drilling or cutting fluid as a negative control68, and some studies place a tracer dye within the fluid to indicate contamination of the sample with the fluid69,70. For example, in a fetal meconium study, ref. 71 swabbed decontaminated maternal skin before the procedure and used additional swabs exposed to the operating theatre air to identify sources of contamination, determining that the fetal meconium microbiome is indistinguishable from negative controls. Sampling controls should be included alongside the samples through all processing steps to account for any contaminants introduced during sample collection and downstream processing. Multiple sampling controls should be included to accurately quantify the nature and extent of contamination, and we recommend including at least one control sample for every four samples when possible. These multiple sampling controls can be analysed in conjunction with negative controls from other processing steps, including DNA extraction and library preparation steps, to specifically identify the steps at which any contaminants may have been introduced (see below). All controls should be documented and reported (Table 2 and Box 1).
Laboratory practices to minimize and identify potential contamination
Laboratory procedures, including DNA extraction, PCR amplification, library preparation and sequencing, can both introduce and amplify contaminants. Laboratory reagents and consumables, including extraction and PCR kits, preservation solutions, plastic tubes and even purified water, often contain amplifiable cellular or cell-free DNA from notoriously persistent bacteria (for example, Ralstonia, Pseudomonas)8,72. Contamination is also possible from various other sources, including human operators, laboratory surfaces or air, and other samples or cultures. For example, on the basis of the sequencing of 144 negative controls, the contaminant profile of one laboratory was shown to vary by month, season and researcher67. The accidental mixing or aerosolization of DNA between different tubes or wells during extraction or other processing steps is a major cause of cross-contamination in microbiome studies40,41. Cross-contamination can even occur via ‘tag jumping’, leading to the erroneous assignment of sequences to samples42. Contamination-aware laboratory setup, study design and experimental practices are all necessary to minimize contamination and cross-contamination. Appropriate steps include maintaining suitable and clean workspaces, wearing PPE, confirming and maintaining reagent integrity, and carefully considering how samples are arranged during processing. Despite this, contamination and cross-contamination may still occur (Fig. 2). Thus, ensuring that multiple negative and positive experimental controls are included in the study design and then sequenced alongside the original samples is essential for identifying the extent and nature of contamination before conducting downstream computational analyses. The following steps are recommended (see also Table 1).
Maintain pristine and physically isolated molecular facilities
The physical characteristics of the laboratory workspace are important to consider when attempting to minimize contamination. There should be physical separation between pre- and post-extraction workspaces, as well as pre- and post-PCR workspaces, with a unidirectional workflow between each workspace. Such separation is important given that a single PCR run can produce trillions of DNA molecules—both from samples and contaminants—that, upon aerosolization (for example, due to handling and pipetting), can contaminate the environment and other samples40,73. Molecular work should be performed in enclosed hoods to limit the possibility that contaminants are introduced from the surrounding air. To reduce contamination of reagents, the setup of PCR or other master mixes should be performed in clean hoods absent of template DNA using a dedicated set of pipettors that are never used with DNA samples. Pipetting and handling of samples or reagents should be performed using filtered tips to prevent aerosolized DNA contamination of pipettors. Hoods should be thoroughly decontaminated before and after each use with an appropriate DNA-degrading solution to prevent cross-sample contamination and limit the magnitude of laboratory-derived contamination. In addition, hoods and equipment can be irradiated with UV-C light to further reduce exogenous laboratory contamination. As with sampling, laboratory personnel should have skin surfaces covered with PPE and frequently change gloves, especially when handling different types of sample and reagent. Laboratory PPE, equipment and supplies should be dedicated to each space, and if disposable PPE is used, it should only be worn across rooms in a ‘clean’ to ‘dirty’ direction. In addition, floors and other horizontal surfaces should be regularly cleaned to limit dust accumulation.
Confirm and maintain the integrity of reagents
Ultrapure reagents and consumables, for example, DNA-free tubes and PCR-grade water, should be sourced from trusted suppliers. Their integrity should be validated before use on low microbial biomass samples to confirm that they lack DNA, for example, through PCR amplification followed by agarose gel electrophoresis (low sensitivity) or, ideally, DNA sequencing or quantitative PCR (higher sensitivity). Never assume that a reagent or consumable is truly ‘DNA free’ even if advertised as such. Ideally, stock reagents should only be handled at the start of each batch (before opening any sample tubes) and aliquoted into small volumes to minimize repeated exposure to potential contaminants. As different batches of reagents may contain different amounts and types of contaminant8, aliquots of each lot should be included as negative controls to check for potential contamination. In addition, plastic consumables (for example, pipette tips, tubes) should be sterilized with UV-C before use, and tubes containing samples or DNA entering workspace hoods should be thoroughly decontaminated to remove surface contaminants. If any steps during microbiome processing are outsourced (for example, library preparation and sequencing), we recommend confirming whether providers have experience handling low microbial biomass samples and conducting a test run if possible; after all, most commercial sequencing facilities are established for sequencing high-biomass samples such as stool, human or bacterial isolate DNA.
Collect and sequence multiple negative and positive controls
It is critical to include numerous controls in any given batch of samples to determine the nature and extent of contamination that may have been introduced at any steps, from sample collection to sequencing. Such controls must be collected, processed and reported alongside the samples, not post hoc, as the sources and extent of contamination may have changed (Table 2 and Box 1). In addition to sampling controls, the inclusion of additional negative controls at other processing steps can allow monitoring of exogenous contamination present in reagents or introduced during specific laboratory processing steps8. Such negative controls should include DNA extraction controls (DNA extractions without input sample) and non-template controls (PCR or library preparations without input sample DNA). Sequencing these controls alongside samples makes it possible to identify contamination points and provides a baseline detection level. For example, ref. 52 used negative controls with quantitative PCR to determine that placental samples had no more bacterial DNA than the negative controls. Even if negative controls yield insufficient DNA for equimolar pooling, they should still be sequenced to identify contaminants or validate their absence. The absence of a visible band after PCR-based amplification and agarose gel electrophoresis of negative controls is insufficient to confirm the absence of contamination due to the low sensitivity of this method. As noted previously, these types of negative control are needed for every separate batch of reagents that are used in a specific experiment, due to the potential for varying contaminant profiles between different batches. Positive controls are also valuable for calibrating detection limits, monitoring cross-sample contamination and detecting laboratory-introduced contaminants32,66. These positive controls can also be diluted to span a range in concentrations to identify the effective detection limit; if sample results look similar to results from diluted positive controls, this can indicate that contaminants have obscured true biological signals26. Commercially available mock community standards are recommended as positive controls, composed of either whole cells, which are useful for evaluating DNA extraction procedures, or purified DNA, which is useful for evaluating library preparation and sequencing steps. Positive control spike-ins, such as cross-contamination-checking oligonucleotides (coligos) described in ref. 74, can also be added during DNA extraction and library preparation to monitor and quantify cross-sample contamination.
Barcoding and distributing samples to reduce and detect cross-contamination
Careful attention should be paid to the spatial arrangement of samples and controls during laboratory processing to reduce the risk of systematic contamination75,76. For example, if a given study includes both low- and high-biomass samples, these should be processed separately to minimize cross-contamination of DNA from the higher- to lower-biomass samples40. Likewise, negative and positive controls should be placed in different well positions if working in multiwell plates, as the potential for contamination may not be equivalent across all well positions (for example, well-to-well cross-contamination may be higher in the middle of 96-well plates than on the edges due to the proximity to more samples). There is generally a lower likelihood of cross-contamination when doing DNA extractions in single tubes instead of multiwell plates, but this often comes with a trade-off in sample throughput. Unique barcodes should be used for each sample, ideally dual-index error-correcting barcodes, to detect and correct for cross-contamination during sequencing. It is also possible to chemically tag sample DNA before extraction and library preparation (for example, through conversion of cytosines to uracils via bisulfite salts)77, enabling discrimination of contaminating DNA introduced after tagging.
Detection and potential removal of contaminants from sequence data
Contamination-aware data analysis is critical when analysing microbiome datasets, particularly those derived from low microbial biomass environments. Even with the most stringent sampling and laboratory techniques, the risk of sample contamination and cross-contamination is never eliminated. As such, it is crucial to analyse sequencing data to evaluate how much contamination has occurred and if it can be reliably removed using post hoc approaches. The best way to achieve this is to systematically compare the sequences, taxa and/or genes detected in positive and negative controls to those in samples. In addition, a range of decontamination software allow detection of potential contaminants through statistical approaches (see Table 3). However, metagenomic datasets are challenging to decontaminate due to their inherent complexity and the limited availability of decontamination pipelines compared with those for marker gene datasets. In certain cases, it may be justified to remove contaminants from the processed datasets if potential contaminants are minimal or constitute a small proportion of the dataset; however, ensuring transparency throughout this process remains essential. Any decontamination steps should be recorded, and the original datasets should still be reported and deposited (see Table 2 and Box 1). In many cases, the signal-to-noise ratio will be too low to disentangle what are contaminants from targets for detection, meaning decontamination steps could distort information. In these cases, it is usually necessary to discard entire samples or datasets, although the data obtained can still be helpful for troubleshooting where contamination occurred. A priori knowledge of both study systems and potential contaminants is useful to contextualize and evaluate samples and controls, as well as outputs from decontamination software. With low-biomass samples, the process of using negative controls to differentiate biological signal from contamination is often not straightforward. Therefore, removal of contaminants from sequence data is often challenging to do with absolute certainty, making transparency in the reporting of data and associated analyses even more critical (see Table 2 and Box 1).
Analyse sequenced controls and check for unexpected taxa
After initial processing of sequence data, quality control of the data should always be the next step. Controls should be analysed to check the quality of the dataset. Determine (1) what off-target DNA is present in negative and positive controls to identify contamination, (2) whether taxa from positive controls are also present in negative controls and samples to identify cross-contamination and (3) what proportion of the sequencing reads in the samples originate from contaminants or cross-contaminants to determine the magnitude of the problem. For marker gene sequencing, it is often feasible to identify the specific taxa shared between controls and samples, especially if analysing amplicon sequencing variants (ASVs). These taxa can be potentially removed from the processed dataset. For metagenomes, it is usually necessary to map sample reads to metagenome-assembled genomes (MAGs) generated from the corresponding controls78. Mapping of ASVs and reads to databases of likely contaminants, such as common reagent contaminants8,41, the human microbiome79 and the human genome, can also be helpful. We note that the post hoc detection and removal of external contaminants may not always be straightforward, given that particular taxa may be both in the controls and part of the actual biological signal. For example, taxa expected to be abundant in skin microbiome studies can also be common contaminants44,78. We also recommend carefully evaluating the specific taxa observed in samples to help assess plausibility of results. Common sense questions to ask are: do the taxa observed align with expectations? Do the dominant taxa in samples correspond to known groups of common contaminants?8 The presence of specific taxa in environments that are unexpected based on their known ecologies should immediately raise concern. Notable examples include the reports of abundant human commensals in high-altitude air above Antarctica48, photosynthetic cyanobacteria being major members of human brain tissues80, reagent contaminants being abundant in deep subsurface samples58, and the presence of extremophiles in cancerous tumours81. Although subjective and conservative, these initial steps can provide a useful means of assessment.
Consider using decontamination software but be aware of their limitations
Various software packages are available to aid in the detection and removal of reads originating from external contaminants, with some of the more popular packages summarized in Table 3. Generally, decontamination software use quantitative approaches to identify contaminants and are not solely reliant on taxa identified in control samples. For example, the widely used decontam R package identifies and removes potential contaminants on the basis of either their prevalence (for instance, taxa present in negative controls compared to samples) or frequency (for instance, taxa that are more abundant in lower-biomass samples) in datasets43. A limitation of most decontamination tools is that they target externally introduced contamination rather than cross-contamination (Fig. 2). Exceptions include SCRuB, which incorporates information regarding the spatial position of samples during processing to detect cross-contamination44. Decontamination tools should not be used indiscriminately. As evaluated in Table 3, each decontamination tool is designed on the basis of a set of assumptions regarding contaminants that may not always be valid, and each have both strengths and weaknesses depending on the dataset and purpose. Strong performance of these tools often depends on high-quality controls, sometimes reference databases, and sometimes measures of microbial biomass (for example, DNA concentrations, direct cell counts, qPCR). Some taxa may be incorrectly flagged as contaminants (false positives), or actual contaminants may be missed (false negatives), especially when the contamination profile is complex, variable or overlaps significantly with true sample sequences.
Additional considerations for metagenomic sequencing
The aforementioned statistical approaches for identifying and removing contaminant sequences are primarily useful for marker gene sequence datasets. There are few software packages for screening contaminants in metagenomic data and most have major limitations (Table 3). Nevertheless, SCRuB44, decontam43 and the recently developed tool Squeegee82 can also be applied to metagenomic data. When conducting metagenomic analyses, the inclusion and sequencing of appropriate negative controls alongside samples is critical. The sequence data from these controls should be examined carefully and any reads or MAGs recovered from these controls should be tracked back to the contamination source and, if necessary, removed from the dataset. However, such approaches will only be effective for contaminants with high sequence coverage in control samples, meaning some contaminant reads may remain in the samples. MAG-based mapping is also limited by MAGs generated from short-read sequencing typically being incomplete consensus assemblies that represent only a proportion of the metagenomic reads. Moreover, it can be highly challenging to discriminate between sequences from closely related true and contaminant species, leading to false positives and negatives.
Prevention is always better than cure
We advise researchers to thoroughly invest time into minimizing contamination before engaging in extensive sampling and sequencing campaigns. Although some contamination is inevitable, extensive contamination is not: we and others have produced high-quality datasets from some of the lowest-biomass ecosystems83 and even demonstrated that resident microbes are absent from certain environments32. However, in each case, this required an extensive process to reduce sources of contamination, including developing contaminant-free sampling procedures, ensuring reagent and water integrity, and carefully analysing the resulting sequence data. With the aid of extensive controls, it is possible to forensically identify the likely sources of contamination (for example, sampling, reagents and human operators), noting that datasets are often compromised by contamination from multiple sources. This information can then be used to iteratively improve practices to minimize or eliminate contamination.
For datasets with appreciable contamination, post hoc approaches to identify and remove sequences originating from contaminants will rarely be effective. For example, a recent survey of the global atmospheric microbiome required the post hoc removal of approximately half of all sequences, including entire genera known to be reagent contaminants (for example, Pseudomonas) that may also occur in the atmosphere61. Such extensive but variable contamination limited the inferences that can be made regarding the composition and drivers of these communities, as the signal-to-noise ratio was uncertain. Ultimately, no decontamination pipeline is perfect and retrospective decontamination of sequence data may result in false inclusions or exclusions of data. The relative impact of contamination, subsequent decontamination strategies and the ultimate utility of the dataset depend on the research questions and the extent of contamination. If a substantial number of sequences need to be removed from marker gene or metagenomic data, the integrity of the sample data should be called into question, and researchers should query the integrity of whole datasets if contamination problems are persistent and substantial. While it may not be logistically or financially feasible in all studies, if the question of contamination is pressing enough, the best available method of verification is to obtain identical results independent of the laboratory of origin84,85.
Conclusions
When using DNA-based approaches to analyse microbiomes, it is always best to assume that contamination and cross-contamination is inevitable, particularly when working with samples from lower-biomass systems. Thus, researchers should aim to minimize contamination and use appropriate controls to check the nature and extent of potential contamination. Perhaps most importantly, it is essential to report the procedures used to minimize contamination, what contaminants may have been detected, and how any potential contaminants were handled in downstream analyses. Doing so will improve transparency and provide the scientific community with more confidence in reported findings. The suggestions provided here are not intended to be an exhaustive list of procedures to follow, and we do not imply that all of the suggestions are compulsory. However, our hope is that a more careful consideration of contamination and cross-contamination issues will improve the overall quality of microbiome studies and avoid some of the more persistent sources of uncertainty in previously published work.
References
Mallick, H. et al. Experimental design and quantitative analysis of microbial community multiomics. Genome Biol. 18, 228 (2017).
Widder, S. et al. Challenges in microbial ecology: building predictive understanding of community function and dynamics. ISME J. 10, 2557–2568 (2016).
Knight, R. et al. Best practices for analysing microbiomes. Nat. Rev. Microbiol. 16, 410–422 (2018).
Mirzayi, C. et al. Reporting guidelines for human microbiome research: the STORMS checklist. Nat. Med. 27, 1885–1892 (2021).
Pollock, J., Glendinning, L., Wisedchanwet, T. & Watson, M. The madness of microbiome: attempting to find consensus ‘best practice’ for 16S microbiome studies. Appl. Environ. Microbiol. 84, e02627-17 (2018).
Bharti, R. & Grimm, D. G. Current challenges and best-practice protocols for microbiome analysis. Brief. Bioinform. 22, 178–193 (2021).
Costea, P. I. et al. Towards standards for human fecal sample processing in metagenomic studies. Nat. Biotechnol. 35, 1069–1076 (2017).
Salter, S. J. et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 12, 87 (2014).
Rodó, X. et al. Microbial richness and air chemistry in aerosols above the PBL confirm 2,000-km long-distance transport of potential human pathogens. Proc. Natl Acad. Sci. USA 121, e2404191121 (2024).
Bowers, R. M., McLetchie, S., Knight, R. & Fierer, N. Spatial variability in airborne bacterial communities across land-use types and their relationship to the bacterial communities of potential source environments. ISME J. 5, 601–612 (2011).
Lappan, R. et al. The atmosphere: a transport medium or an active microbial ecosystem? ISME J. 18, wrae092 (2024).
Weyrich, L. S. et al. Neanderthal behaviour, diet, and disease inferred from ancient DNA in dental calculus. Nature 544, 357–361 (2017).
Heuer, V. B. et al. Temperature limits to deep subseafloor life in the Nankai Trough subduction zone. Science 370, 1230–1234 (2020).
Goordial, J. et al. Nearing the cold-arid limits of microbial life in permafrost of an upper dry valley, Antarctica. ISME J. 10, 1613–1624 (2016).
Schulze-Makuch, D. et al. Transitory microbial habitat in the hyperarid Atacama Desert. Proc. Natl Acad. Sci. USA 115, 2670–2675 (2018).
Wood, C. et al. Active microbiota persist in dry permafrost and active layer from Elephant Head, Antarctica. ISME Commun. 4, ycad002 (2024).
Ling, F., Whitaker, R., LeChevallier, M. W. & Liu, W.-T. Drinking water microbiome assembly induced by water stagnation. ISME J. 12, 1520–1531 (2018).
Lang, J. M. et al. A microbial survey of the International Space Station (ISS). PeerJ 5, e4029 (2017).
Tait, A. W., Gagen, E. J., Wilson, S., Tomkins, A. G. & Southam, G. Microbial populations of stony meteorites: substrate controls on first colonizers. Front. Microbiol. 8, 1227 (2017).
Cubillos, C. F., Aguilar, P., Grágeda, M. & Dorador, C. Microbial communities from the world’s largest lithium reserve, Salar de Atacama, Chile: life at high LiCl concentrations. J. Geophys. Res. Biogeosci. 123, 3668–3681 (2018).
Napoli, A. et al. Snow surface microbial diversity at the detection limit within the vicinity of the Concordia Station, Antarctica. Life 13, 113 (2022).
Zhong, Z.-P. et al. Clean low-biomass procedures and their application to ancient ice core microorganisms. Front. Microbiol. 9, 344419 (2018).
Shivaji, S. et al. Antarctic ice core samples: culturable bacterial diversity. Res. Microbiol. 164, 70–82 (2013).
Segal, L. N. & Blaser, M. J. A brave new world: the lung microbiota in an era of change. Ann. Am. Thorac. Soc. 11, S21–S27 (2014).
Stinson, L. F., Ma, J., Sindi, A. S. & Geddes, D. T. Methodological approaches for studying the human milk microbiome. Nutr. Rev. 81, 705–715 (2023).
Kennedy, K. M. et al. Questioning the fetal microbiome illustrates pitfalls of low-biomass microbial studies. Nature 613, 639–649 (2023).
Tan, C. C. S. et al. No evidence for a common blood microbiome based on a population study of 9,770 healthy humans. Nat. Microbiol. 8, 973–985 (2023).
Bintarti, A. F., Sulesky-Grieb, A., Stopnisek, N. & Shade, A. Endophytic microbiome variation among single plant seeds. Phytobiomes J. 6, 45–55 (2022).
Walsh, C. M., Becker-Uncapher, I., Carlson, M. & Fierer, N. Variable influences of soil and seed-associated bacterial communities on the assembly of seedling microbiomes. ISME J. 15, 2748–2762 (2021).
Hammer, T. J., Janzen, D. H., Hallwachs, W., Jaffe, S. P. & Fierer, N. Caterpillars lack a resident gut microbiome. Proc. Natl Acad. Sci. USA 114, 9641–9646 (2017).
Belilla, J. et al. Active microbial airborne dispersal and biomorphs as confounding factors for life detection in the cell-degrading brines of the polyextreme Dallol Geothermal Field. mBio 13, e0030722 (2022).
de Goffau, M. C. et al. Human placenta has no microbiome but can contain potential pathogens. Nature 572, 329–334 (2019).
Dragone, N. B. Exploring the boundaries of microbial habitability in soil. J. Geophys. Res. Biogeosci. 126, e2020JG006052362 (2021).
Hammer, T. J., Sanders, J. G. & Fierer, N. Not all animals need a microbiome. FEMS Microbiol. Lett. 366, fnz117 (2019).
Tanner, M. A., Goebel, B. M., Dojka, M. A. & Pace, N. R. Specific ribosomal DNA sequences from diverse environmental settings correlate with experimental contaminants. Appl. Env. Microbiol. 64, 3110–3113 (1998).
de Goffau, M. C. et al. Recognizing the reagent microbiome. Nat. Microbiol. 3, 851–853 (2018).
Weiss, S. et al. Tracking down the sources of experimental contamination in microbiome studies. Genome Biol. 15, 564 (2014).
Olm, M. R. et al. The source and evolutionary history of a microbial contaminant identified through soil metagenomic analysis. mBio 8, e01969-16 (2017).
Lou, Y. C. et al. Using strain-resolved analysis to identify contamination in metagenomics data. Microbiome 11, 36 (2023).
Minich, J. J. et al. Quantifying and understanding well-to-well contamination in microbiome research. mSystems 4, e00186-19 (2019).
Eisenhofer, R. et al. Contamination in low microbial biomass microbiome studies: issues and recommendations. Trends Microbiol. 27, 105–117 (2019).
Schnell, I. B., Bohmann, K. & Gilbert, M. T. P. Tag jumps illuminated – reducing sequence-to-sample misidentifications in metabarcoding studies. Mol. Ecol. Resour. 15, 1289–1303 (2015).
Davis, N. M., Proctor, D. M., Holmes, S. P., Relman, D. A. & Callahan, B. J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 6, 226 (2018).
Austin, G. I. et al. Contamination source modeling with SCRuB improves cancer phenotype prediction from microbiome data. Nat. Biotechnol. 41, 1820–1828 (2023).
Knights, D. et al. Bayesian community-wide culture-independent microbial source tracking. Nat. Methods 8, 761–763 (2011).
Welsh, B. L. & Eisenhofer, R. The prevalence of controls in phyllosphere microbiome research: a methodological review. New Phytol. 242, 23–29 (2024).
Gihawi, A. et al. Major data analysis errors invalidate cancer microbiome findings. mBio 14, e0160723 (2023).
Archer, S. D. et al. Airborne microbial transport limitation to isolated Antarctic soil habitats. Nat. Microbiol. 4, 925–932 (2019).
Archer, S. D. et al. Air mass source determines airborne microbial diversity at the ocean–atmosphere interface of the Great Barrier Reef marine ecosystem. ISME J. 14, 871–876 (2020).
Evans, G. E. et al. Contamination of Qiagen DNA extraction kits with Legionella DNA. J. Clin. Microbiol. 41, 3452 (2003).
Aagaard, K. et al. The placenta harbors a unique microbiome. Sci. Transl. Med. 6, 237ra65 (2014).
Leiby, J. S. et al. Lack of detection of a human placenta microbiome in samples from preterm and term deliveries. Microbiome 6, 196 (2018).
Lauder, A. P. et al. Comparison of placenta samples with contamination controls does not provide evidence for a distinct placenta microbiota. Microbiome 4, 29 (2016).
Castillo, D. J., Rifkin, R. F., Cowan, D. A. & Potgieter, M. The healthy human blood microbiome: fact or fiction? Front. Cell. Infect. Microbiol. 9, 148 (2019).
Bedarf, J. R. et al. Much ado about nothing? Off-target amplification can lead to false-positive bacterial brain microbiome detection in healthy and Parkinson’s disease individuals. Microbiome 9, 75 (2021).
Li, J. et al. Recycling and metabolic flexibility dictate life in the lower oceanic crust. Nature 579, 250–255 (2020).
Orsi, W. D. Contesting the evidence for gene expression in lower oceanic crust. Preprint at bioRxiv https://doi.org/10.1101/2020.03.25.005033 (2020).
Sheik, C. S. et al. Identification and removal of contaminant sequences from ribosomal gene databases: lessons from the Census of Deep Life. Front. Microbiol. 9, 840 (2018).
DeLeon-Rodriguez, N. et al. Microbiome of the upper troposphere: species composition and prevalence, effects of tropical storms, and atmospheric implications. Proc. Natl Acad. Sci. USA 110, 2575–2580 (2013).
Smith, D. J. & Griffin, D. W. Inadequate methods and questionable conclusions in atmospheric life study. Proc. Natl Acad. Sci. USA 110, E2084 (2013).
Archer, S. et al. Global biogeography of atmospheric microorganisms reflects diverse recruitment and environmental filtering. Preprint at Res. Square https://doi.org/10.21203/rs.3.rs-244923/v4 (2022).
Cando‐Dumancela, C., Liddicoat, C., McLeod, D., Young, J. M. & Breed, M. F. A guide to minimize contamination issues in microbiome restoration studies. Restor. Ecol. 29, e13358 (2021).
Nilsson, M., De Maeyer, H. & Allen, M. Evaluation of different cleaning strategies for removal of contaminating DNA molecules. Genes 13, 162 (2022).
Asadi, S. et al. Aerosol emission and superemission during human speech increase with voice loudness. Sci. Rep. 9, 2348 (2019).
Rutty, G. N., Hopwood, A. & Tucker, V. The effectiveness of protective clothing in the reduction of potential DNA contamination of the scene of crime. Int. J. Legal Med. 117, 170–174 (2003).
Minich, J. J. et al. KatharoSeq enables high-throughput microbiome analysis from low-biomass samples. mSystems 3, 00218-17 (2018).
Weyrich, L. S. et al. Laboratory contamination over time during low‐biomass sample analysis. Mol. Ecol. Resour. 19, 982–996 (2019).
Pendleton, H. L., Twing, K. I., Motamedi, S. & Brazelton, W. J. Potential microbial contamination from drilling lubricants into subseafloor rock cores. Sci. Drill. 29, 49–57 (2021).
Martínez-Pérez, C. et al. Phylogenetically and functionally diverse microorganisms reside under the Ross Ice Shelf. Nat. Commun. 13, 117 (2022).
Goordial, J. et al. Microbial diversity and function in shallow subsurface sediment and oceanic lithosphere of the Atlantis Massif. mBio 12, 0049021 (2021).
Kennedy, K. M. et al. Fetal meconium does not have a detectable microbiota before birth. Nat. Microbiol. 6, 865–873 (2021).
McFeters, G. A., Broadaway, S. C., Pyle, B. H. & Egozy, Y. Distribution of bacteria within operating laboratory water purification systems. Appl. Environ. Microb. 59, 1410–1415 (1993).
Walker, A. W. A lot on your plate? Well-to-well contamination as an additional confounder in microbiome sequence analyses. mSystems 4, e00362-19 (2019).
Harrison, J. G., Randolph, G. D. & Buerkle, C. A. Characterizing microbiomes via sequencing of marker loci: techniques to improve throughput, account for cross-contamination, and reduce cost. mSystems 6, 00294-21 (2021).
Leek, J. T. et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat. Rev. Genet. 11, 733–739 (2010).
Goh, W. W. B., Wang, W. & Wong, L. Why batch effects matter in omics data, and how to avoid them. Trends Biotechnol. 35, 498–507 (2017).
Mzava, O. et al. A metagenomic DNA sequencing assay that is robust against environmental DNA contamination. Nat. Commun. 13, 4197 (2022).
Saheb Kashaf, S. et al. Integrating cultivation and metagenomics for a multi-kingdom view of skin microbiome diversity and functions. Nat. Microbiol. 7, 169–179 (2022).
Turnbaugh, P. J. et al. The human microbiome project. Nature 449, 804–810 (2007).
Branton, W. G. et al. Brain microbiota disruption within inflammatory demyelinating lesions in multiple sclerosis. Sci. Rep. 6, 37344 (2016).
Gihawi, A., Cooper, C. S. & Brewer, D. S. Caution regarding the specificities of pan-cancer microbial structure. Microb. Genom. 9, mgen001088 (2023).
Liu, Y., Elworth, R. L., Jochum, M. D., Aagaard, K. M. & Treangen, T. J. De novo identification of microbial contaminants in low microbial biomass microbiomes with Squeegee. Nat. Commun. 13, 6799 (2022).
Bay, S. K. et al. Chemosynthetic and photosynthetic bacteria contribute differentially to primary production across a steep desert aridity gradient. ISME J. 15, 3339–3356 (2021).
Clausen, D. S. & Willis, A. D. Evaluating replicability in microbiome data. Biostatistics 23, 1099–1114 (2022).
Poinar, H. N. & Cooper, A. Ancient DNA: do it right or not at all. Science 5482, 416 (2000).
McKnight, D. T. et al. microDecon: a highly accurate read‐subtraction tool for the post‐sequencing removal of contamination in metabarcoding studies. Environ. DNA 1, 14–25 (2019).
Acknowledgements
We thank the late C. Cary for inspirational discussions and R. Amann for helpful comments. N.F. was supported by grants from the US National Science Foundation (BROADN Biology Integration Institute and Office of Polar Programs). C.G. was supported by grants from the Australian Research Council (FT240100502 and SR200100005) and Human Frontiers Science Program (RGY0058/2022). R.L. and P.M.L. were supported by fellowships from the Australian Research Council (DE230100542 and DE250101210).
Author information
Authors and Affiliations
Contributions
N.F. and C.G. conceived, designed and wrote this manuscript. P.M.L., R.L., R.E., F.R. and S.I.H. drafted sections and tables. All authors contributed best practice suggestions, edited the manuscript and endorsed the recommendations.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Microbiology thanks Julia Segre, Brent Christner and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Supplementary information
Supplementary Information
Supplementary Methods, Table 1 and References.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Fierer, N., Leung, P.M., Lappan, R. et al. Guidelines for preventing and reporting contamination in low-biomass microbiome studies. Nat Microbiol 10, 1570–1580 (2025). https://doi.org/10.1038/s41564-025-02035-2
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41564-025-02035-2
This article is cited by
-
Spatial and temporal patterns of public transit aerobiomes
Microbiome (2026)
-
Responses of skin and salivary microbiome to different environmental exposures
International Journal of Legal Medicine (2026)
-
Metagenomics and its impact on environmental and therapeutic microbiology
Archives of Microbiology (2026)
-
Gut microbial metabolites in cancer immunomodulation
Molecular Cancer (2025)
-
The multi-kingdom cancer microbiome
Nature Microbiology (2025)