Abstract
Cells can sense and respond to their environment. Central to this process is the formation of molecular clutches, which are dynamic linkages between the extracellular matrix and the actin cytoskeleton mediated by integrins and adaptor proteins. Although it is well known that force-dependent interactions between molecular-clutch components are essential for sensing substrate rigidity, the influence of nonlinear adaptor-protein elasticity is poorly understood. Here we show that adaptor-protein elasticity and local interactions between molecular clutches and the extracellular matrix are key to cellular perception of substrate stiffness. We present a semi-analytical theory that integrates experimentally measured force responses of adaptor proteins to describe cell-adhesion sensing. We propose that molecular clutches probe local mechanical substrate properties and collectively function as a differential that allows a retrograde actin flow and substrate movement to occur at different rates while maintaining a stable mechanical coupling between them. Our model reproduces experimentally measured force-loading rates of individual molecular clutches and correctly predicts cell-adhesion behaviour for a range of substrate stiffnesses. The framework presented can be extended to study complex phenomena such as focal-adhesion maturation and cell-type-specific mechanosensing.
Similar content being viewed by others
Main
The ability of cells to detect and respond to various mechanical signals, such as an external force1, substrate stiffness and topography2, is crucial for the regulation of many physiological processes, including cell migration, proliferation, differentiation and apoptosis3. Aberrant cell mechanosensing has been implicated in a number of pathological conditions, such as fibrosis, cancer and cardiovascular diseases4. Therefore, a better understanding of cell mechanosensing mechanisms is essential for elucidating both physiological and pathological processes.
Molecular linkages between the cell cytoskeleton and the extracellular matrix (ECM), often referred to as molecular clutches, are the primary sites where cells physically interact with their microenvironment and probe its mechanical properties5 (Fig. 1a). Molecular clutches experience mechanical tension from both external mechanical cues and intracellular molecular motors. Molecular-clutch theory, which proposes that the tension-dependent formation and dissociation of molecular clutches are integrated at the mesoscale level to guide cell-adhesion behaviour, has become the leading model for understanding cellular mechanosensing processes6,7,8,9,10,11,12,13,14,15,16,17,18. This theory has successfully explained many aspects of cell spreading, migration and traction force generation on substrates with different mechanical properties6,7,11,12,13,14,16,17,18.
a, Integrins and adaptor proteins such as talin are the main components of cell-adhesion linkages that form between the actin cytoskeleton and the substrate. The myosin II-generated retrograde flow of actin filaments leads to mechanical stretching of these links, causing local substrate deformations (shown in red). Force-transmitting units, each including an intracellular part (integrin and adaptor proteins) and an extracellular part (locally deformed substrate), are referred to in this study as ‘molecular clutches’. b, In the model, molecular clutches are represented by composite springs consisting of two parts: an intracellular one (kc) and an extracellular one (\({k}_{{\rm{s}}}^{{\prime} }\)). Although we assume the latter has a linear response to the applied load, we consider the former to be a linear spring in the linear clutch model and a nonlinear spring with a talin-like force response in the talin clutch model. The pulling force (Fm) generated by myosin II motors causes actin filaments to move at a speed v. This leads to a gradual stretching of molecular clutches along the x axis parallel to the substrate surface (contact angle θ ≈ 15°)65, resulting in their extension, l. This, in turn, allows transmission of the myosin II-generated force (Fm) to the substrate, leading to the cell traction force (Fc), which causes deformation of the substrate (xs) over the entire area of cell-adhesion complexes. The efficiency of the force transmission is mainly determined by the kinetic behaviour of molecular clutches under a mechanical load, which is described by their formation and dissociation rates, kon and koff, respectively. The viscoelastic behaviour of the substrate at the scale of cell-adhesion complexes is described in the study using the Kelvin–Voigt model, which represents substrate deformations as a purely elastic spring with stiffness ks, connected in parallel to a purely viscous damper characterized by viscosity constant ξs. In general, by replacing the Kelvin–Voigt model with the Maxwell model or the standard linear solid model, it is possible to describe the mechanosensitive behaviour of cell-adhesion complexes on liquid-like materials and on materials with more complex viscoelastic behaviour, respectively. SLS model, standard linear solid model.
Despite this, recent experimental studies using FRET tension sensor tools have revealed substantial discrepancies between measured force-loading rates of molecular clutches and those predicted by conventional molecular-clutch models, highlighting the need to revise the conceptual framework. For example, tension-gauge DNA sensors show that molecular clutches experience force-loading rates of 0.2–4 pN s−1 in living cells grown on rigid substrates19,20, which is several orders of magnitude lower than the 60–8,000 pN s−1 predicted by existing molecular-clutch models7,11,12,13,14 (Extended Data Table 1). Moreover, these experiments show that the force-induced binding of vinculin to talin considerably reduces the force-loading rate and prolongs the lifetime of molecular clutches, even when the actin-binding ability of vinculin is compromised20, challenging the widely accepted notion that the primary function of vinculin is to enhance focal adhesion (FA) by recruiting more actin filaments13,21.
An important aspect often overlooked in existing molecular-clutch models is the elastic properties of intracellular molecular-clutch components, which are typically approximated by linear springs with stiffness ranges of ~1–1,000 pN nm−1 (refs. 7,11,12,13,14). However, single-molecule measurements have shown that key adaptor proteins in molecular clutches, such as talin, are orders of magnitude more elastic due to the presence of long flexible peptide linkers and force-induced unfolding of protein domains, which are capable of extending beyond 200 nm under only a few piconewtons of force22,23. The nonlinear force response and high elasticity of talin effectively act as a ‘shock absorber’ that radically alters force transmission across cell-adhesion complexes22,23, potentially explaining the lower force-loading rates and longer lifetimes of molecular clutches observed in experiments19,20. Yet, it remains unclear how this behaviour affects cellular mechanosensitivity.
To address these gaps, we developed a semi-analytical model that integrates the experimentally measured force response of adaptor proteins together with local substrate deformations at the attachment sites of molecular clutches. By incorporating the nonlinear elasticity of adaptor proteins such as talin, the model was able to capture key experimental observations, including force-loading rates and the dynamics of molecular clutches. We found that molecular clutches act as a ‘differential’ that allows the actin cytoskeleton and substrate to have different movement rates while maintaining a stable mechanical connection between them. This framework reconciles discrepancies between previous molecular-clutch models and experimental observations and provides a basis for further study of the FA maturation process and cell-type-specific mechanosensing.
Results
Semi-analytical mechanical model of cell-adhesion complexes
To study the potential effect of the elastic properties of molecular clutches on the mechanosensing behaviour of cell adhesion, we developed a semi-analytical theoretical framework consistent with the experimentally observed force response of molecular-clutch components at the single-molecule level22,24,25 and experimentally measured tension26,27 and force-loading rates of molecular clutches19,20. This allowed a comprehensive study of the dynamic aspects of cell-adhesion mechanics, which provided new insights into the role of different cellular elements in shaping mechanosensitive cell-adhesion behaviour.
Notably, our model can be used to describe two types of cell-adhesion complexes: talin-based and non-talin-based. In the former case, cell-adhesion complexes correspond to individual nascent adhesions (NAs) and FAs, whereas in the latter case, they represent sets of integrin clusters observed in experiments on talin-deficient cells13.
In the model, large-scale substrate deformations at the scale of cell-adhesion complexes are described using the Kelvin–Voigt model (Fig. 1b), which depicts the behaviour of elastic-like substrates with residual viscosity commonly used in experiments, such as acrylamide hydrogel and polydimethylsiloxane28,29. However, in principle, our model also allows for the incorporation of any other physical model describing substrate deformations, including the Maxwell model for liquid-like substrates (Fig. 1b) or even the more general standard linear solid model.
Furthermore, individual molecular clutches are represented in our model by two-part springs: one part describes cell adaptor proteins connecting adhesion receptors to the cytoskeleton, and the other part depicts local deformations of the substrate at the attachment sites of the adhesion receptors (Methods and Fig. 1). The elasticity of adaptor proteins is approximated either by linear springs or by a more realistic nonlinear force response described by the worm-like chain model (Methods and Supplementary Appendix F), which better reflects the experimentally observed force-induced softening of adaptor proteins, such as talin, caused by the unfolding of mechanosensitive protein domains. The local substrate deformations are related to the substrate rigidity through a spring constant, the value of which can be obtained from the theory of elasticity (equation (1) and Supplementary Appendix A).
The model distinguishes between the ‘occupied’ (‘on’) and ‘free’ (‘off’) states of sites available for the formation of molecular clutches. It quantifies the deformation of each molecular clutch under the influence of a mechanical load created by the contractile activity of myosin II motors. The dynamic behaviour of cell-adhesion complexes in the model is determined by the balance between the force generated by myosin II motors, which pulls on actin filaments within adhesion complexes, and the traction force created by mechanically stretched molecular clutches in accordance with Newton’s third law. When this balance is disturbed (due to the formation or dissociation of molecular clutches), it is restored due to the convergence of the molecular-clutch system to a stable stationary state corresponding to a steady state of the system (Extended Data Fig. 1). Thus, stable stationary states provide information about the long-term behaviour of the molecular-clutch system. By finding the probability distribution for the extension of molecular clutches in stationary states for a given substrate rigidity and knowing how the tension of molecular clutches depends on their extension, one can then calculate the total traction force created by a cell-adhesion complex resulting from the molecular-clutch-mediated transmission of the pulling force generated by myosin II motors (Fig. 1b and equation (6)). The efficiency of this force transmission, which is determined by the dynamics of molecular-clutch formation and dissociation, is a key element for understanding the mechanosensitive behaviour of cell adhesion, as can be seen from the master equation describing the time evolution of the molecular-clutch system (Supplementary equation (B10)).
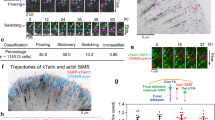
A key feature of our model, revealed by exploring the model parameter space, is that it can predict diverse dynamic behaviours of cell-adhesion complexes, which are jointly determined by a number of sensitive model parameters, including the rate of molecular-clutch formation, adaptor-protein elasticity and myosin II density. As shown in Fig. 2a, three characteristic types of cell-adhesion behaviour were observed on varying the rate of molecular-clutch formation and adaptor-protein elasticity. The first was the strong cell-adhesion regime, in which cell traction increases monotonically with the substrate rigidity (Fig. 2a). This monophasic behaviour is consistent with the behaviour of wild-type (WT) fibroblast cells observed in experiments13. This regime corresponds to a high rate of molecular-clutch formation and high adaptor-protein elasticity, which buffer the tension experienced by individual molecular clutches and keep it at a low level, so that a sufficiently large number of molecular clutches can be formed to maintain strong cell traction. By contrast, when the rate of molecular-clutch formation is low and the adaptor proteins are stiff, cell traction exhibits a biphasic behaviour, increasing on soft substrates and decreasing on stiffer ones due to the shorter lifetime of molecular clutches because of their higher tension (Fig. 2a), reminiscent of the behaviour of talin-knockout (KO) fibroblasts13. Interestingly, for a certain range of adaptor-protein stiffnesses and molecular-clutch formation rates, the model predicts a bistable region in which both strong and weak cell-adhesion regimes coexist on rigid substrates. The molecular-clutch system in this case resembles the clutch of a car, where the state of strong cell adhesion corresponds to a low gear and the state of weak cell adhesion to a high gear. The bistability mode is essentially a state with a ‘rev matching’ condition that allows cells to ‘switch gear’ and change between states with strong and weak adhesion.
a, Stability diagram of cell-adhesion complexes. Solid curves (black, red and blue) show bifurcations that lead to the formation of a bistability region (red area) and a biphase–monophase boundary (green dotted curve), which affect the mechanosensitive behaviour of cell adhesion (‘Steady-state behaviour and bifurcation of the clutch system’). The insets around the bistability region illustrate representative cell traction curves within each domain. It is evident that increasing the stiffness of the intracellular part of molecular clutches (kc) changes the shape of the cell traction curve, shifting cells from the soft-clutch behaviour of MEF WT cells to the rigid-clutch behaviour of MEF Talin 1 KO, Talin 2 shRNA cells. b, Ensemble-averaged force–extension curves of nonlinear talin-based molecular clutches (talin WT model) for different values of Young’s modulus for the substrate (E). Dashed lines represent the force–extension curves of molecular clutches with folded and unfolded mechanosensitive R3 domains of talin at the corresponding value of Young’s modulus for the substrate. Inset: close-up of curves for 10 kPa. c,d, Fitting of experimental data for cell traction (c) and retrograde actin flow (d) from refs. 13,21 collected for MEF WT cells (red circles) and MEF Talin 1 KO, Talin 2 shRNA cells (green triangles) to the linear clutch model (linear WT and linear KD model, solid curves) and to the nonlinear talin clutch model (talin WT model, dotted curves), respectively. Both WT models reproduce the WT data, indicating that talin can be approximated as a linear spring for the substrate elasticities used in the experiment. The fitted parameter values of the linear models (Extended Data Table 2) reveal two molecular-clutch archetypes: a soft, talin-dominant one, prevalent in WT cells, and a more rigid, talin-depleted one, prevalent in talin-deficient cells.
In addition to the rate of molecular-clutch formation and adaptor-protein elasticity, these three types of cell-adhesion behaviour also depend on the densities of myosin II motors and ECM ligands, which will be discussed in more detail in the following sections. Because most of these parameters can be modulated by cells through regulating cell contractility and the composition of FAs, cells can potentially switch between the weak and strong adhesion states.
Effect of elasticity of adaptor proteins on cell adhesion
Using a few physiologically relevant fitting parameters (Extended Data Table 2), we found that our model could fit well previously published experimental data, such as retrograde actin flow and cell traction measured at different values of Young’s modulus for the substrate13,21,30 (Fig. 2c,d). This makes it possible for the model to be used to plot cell-adhesion stability diagrams and extract detailed information about molecular-clutch dynamics at the single-molecule level from experimental data obtained from cells, including the average tension and conformational changes of molecular clutches, which may provide new insights into the key factors regulating FA behaviour.
Previous single-molecule experiments showed that the mechanical responses of adaptor proteins, such as talin, are highly nonlinear and subject to force-induced softening due to the unfolding of their force-bearing protein domains. Therefore, we first investigated how the nonlinear elasticity of adaptor proteins affects the mechanosensitivity of cell adhesion. To do this, we treated the intracellular part of molecular clutches as a nonlinear spring with a force response corresponding to that of talin measured in single-molecule studies22, including the force-dependent unfolding of the mechanosensitive R3 domain of talin. See the talin WT model described in Supplementary Appendix F.
Fitting experimental data about the mechanosensitive behaviour of mouse embryonic fibroblast (MEF) WT cells from ref. 13 to the steady-state solution of the talin WT model (Fig. 2c,d) using the model-fitting parameters listed in Extended Data Table 2 revealed that even at the highest substrate stiffness, most talin-based molecular clutches experienced relatively low tension in the range 0.7–1.3 pN (Supplementary Fig. 1b). This tension was below the force level at which the nonlinear elastic properties of talin become strongly pronounced (Fig. 2b). In addition, the model predicted that on rigid substrates, only a small percentage (1–2%) of talin-based molecular clutches would be subject to forces sufficient to substantially stretch talin molecules and trigger the unfolding of their mechanosensitive domain (Supplementary Fig. 1a). This finding is in good agreement with a recent experimental study performed on Xenopus laevis cells grown on a fibronectin-coated glass substrate, in which only about ~4% of talin molecules underwent substantial conformational changes upon stretching the molecular clutch31.
These results indicate that if the elasticity of adaptor proteins has any effect on cell traction and retrograde actin flow, it is the passive linear elasticity of adaptor proteins that is important. Accordingly, the experimental data from ref. 13 could be fitted to a simplified model in which the intracellular part of molecular clutches is represented by a linear spring (the linear WT model) as accurately as the talin WT model with a more realistic nonlinear force response of the intracellular part of molecular clutches (Fig. 2c,d).
Further comparison of model parameter values obtained by fitting experimental data collected from WT and talin-depleted MEF cells to the linear molecular-clutch model confirmed that the elasticity of adaptor proteins is, indeed, one of the key factors influencing cell-adhesion behaviour. For MEF WT cells, the fitted elasticity of the intracellular parts of molecular clutches (kc = 0.05 pN nm−1) was found to be close to the elasticity of talin estimated from previous single-molecule measurements (0.09 pN nm−1; Supplementary Table 3). On the other hand, the intracellular parts of molecular clutches in MEF Talin 1 KO, Talin 2 shRNA cells were predicted to be more than ten times stiffer (kc = 1.6 pN nm−1) compared to MEF WT cells. Because previous single-molecule studies have shown that typical cytoskeletal proteins found in cell-adhesion complexes have elasticities less than 0.3 pN nm−1 (Supplementary Table 3), these data indicate that rigid molecular clutches may form through the clustering of adaptor proteins. Indeed, FA proteins such as kindlin and α-actinin have been shown to bind to FAs before talin and form clusters or condensates32,33,34. Because these proteins are also present in MEF Talin 1 KO, Talin 2 shRNA cells, they could potentially contribute to the increase in molecular-clutch rigidity predicted by the model. This result is supported by the much larger characteristic size of the adhesion site of molecular clutches in talin-KO cells (r = 26 nm) compared to WT cells (r = 0.6–0.8 nm), which was obtained by fitting experimental data (Extended Data Table 2). In the future, super-resolution microscopy could be used to test the model-predicted difference between the characteristic sizes of adhesion sites of talin-based and non-talin-based molecular clutches. Overall, our model indicates that there are two types of molecular clutches in MEF cells—soft, talin-based and rigid, non-talin-based—which are formed by clustered adaptor proteins. Indeed, a model describing the competitive formation of these two types of molecular clutches (WT-2 model; Supplementary Appendix G) was found to more closely fit the experimental data obtained from WT and talin-depleted MEF cells (Extended Data Fig. 2a,b).
Finally, a more detailed analysis of the force–extension curves of nonlinear talin-based molecular clutches at different values of Young’s modulus for the substrate (E) showed that on soft substrates (E ≤ 10 kPa), the mechanosensitive behaviour of cell adhesion is determined primarily by the elastic characteristics of the extracellular part of the molecular clutches (local substrate deformations), whereas on rigid substrates (E > 10 kPa), the elasticity of the intracellular part of the molecular clutches begins to play a more important role in this process (Fig. 2b). This result is in good agreement with the graphs presented in Extended Data Fig. 3a,b, which show that the spring constant of the intracellular part of molecular clutches (kc) affects cell behaviour only on rigid substrates, but not as much on soft ones. Thus, the elasticity of both the intracellular and extracellular parts of molecular clutches is a key factor influencing the mechanosensitive cell-adhesion behaviour on substrates with different stiffnesses. Because many adaptor proteins contain long stretches of conserved intrinsically disordered regions, such as tensin, these results imply that such flexible domains may play an important role in regulating the mechanosensitive behaviour of cell adhesion.
Ligand density affects cell adhesion through myosin II feedback
To investigate the effect of the density of molecular clutches on cell adhesion, we fitted previously published experimental data on MEF WT cells and MEF Talin 1 KO, Talin 2 shRNA cells plated on elastic substrates that were coated with different fibronectin concentrations13. Because the density of the fibronectin network affects the rate of molecular-clutch formation and the distribution of the mechanical load near adhesion sites of molecular clutches, the data were fitted by varying the clutch formation rate and the characteristic size of adhesion sites of molecular clutches. The density of myosin II motors was also varied. Using this approach, the experimental data could be fitted fairly well to the linear knockdown (KD), linear WT and talin WT models (Fig. 3a,b).
a,b, Cell traction measured on MEF WT cells (a) and MEF WT and MEF Talin 1 KO, Talin 2 shRNA cells (b) plated on substrates coated with increasing concentrations of fibronectin13. The experimental data were fitted to the talin WT model (dotted curves) and the linear WT and KD models (solid curves) by varying the rate of molecular-clutch formation (\({k}_{{\rm{on}}}^{0}\)), the characteristic size of adhesion sites of molecular clutches (r) and the density of myosin II motors (σm). Other model parameters were held fixed (Supplementary Table 1). c, The best-fitting parameter values as a function of fibronectin density show that enriching the ligand density accelerates molecular-clutch formation, reduces the characteristic size of adhesion sites of molecular clutches and increases the density of myosin II, thereby enhancing actin–substrate force transmission. d, Structured illumination microscopy image of an NIH-3T3 fibroblast expressing myosin RLC-GFP. Inset: close-up of the boxed region. e,f, Quantification of myosin II filament stacks in NIH-3T3 and HeLa cells. Increasing the fibronectin density did not alter the density of myosin II stacks (e) but did alter their projected area in a cell-type-dependent manner (f). The response of NIH-3T3 cells is consistent with model predictions for MEF WT cells. In e from left to right, N = 13, 11, 14, 13, 12 and 16 cells were processed under the corresponding experimental conditions. In f from left to right, N = 2,534, 846, 3,836, 834, 2,305 and 928 stacks of myosin II filaments were collected and processed under the corresponding experimental conditions from the same number of cells as indicated for e. Boxes and error bars in e represent mean ± standard errors of the mean (s.e.m.) and standard deviation, respectively. The circles in f show the median, and boxes represent the mean ± s.e.m. The pairwise statistical difference between the corresponding datasets was calculated using one-way ANOVA with post-hoc Tukey’s honestly significant difference test (no adjustments).
Interestingly, the fitted data showed a correlation between myosin II motor density (σm) and fibronectin concentration, as the fitting did not converge when the myosin II density was fixed. According to the model, the density of myosin II motors pulling on actin filaments decreased by ~25% on substrates coated with 1 μg ml−1 fibronectin solution compared to those coated with 100 μg ml−1 fibronectin solution (Fig. 3c). To test this model prediction, we transfected NIH-3T3 cells with the myosin regulatory light chain (RLC)-GFP35 and measured the density and the average size of stacks of myosin II filaments labelled with this construct as a function of the fibronectin concentration (Fig. 3d–f). As a control, we also performed similar measurements with HeLa cells.
We found that, although in HeLa cells myosin II motors formed more or less uniform assemblies resembling beads on a string (Supplementary Fig. 2), in NIH-3T3 cells, they formed stacks that were more irregularly shaped (Fig. 3d). More importantly, although the fibronectin concentration was found to have little effect on the density of stacks of myosin II filaments in both NIH-3T3 and HeLa cells (Fig. 3e), it had a strong effect on their average size in NIH-3T3 cells (~35% drop for a 1 μg ml−1 fibronectin solution compared to a 100 μg ml−1 solution) but not in HeLa cells (Fig. 3f), indicating that this effect may depend on cell type. Thus, the model developed in our study can accurately capture the mechanical states of fibroblast cells and make physiologically relevant predictions.
Previous studies have shown that fibronectin density can influence the activation of rho kinase (ROCK) in endothelial cells. ROCK controls the assembly of myosin II filaments36. Thus, MEF cells could potentially use a similar mechanism. However, the precise molecular pathways responsible for the fibronectin-dependent activation of ROCK remain unknown. Because our observations indicate that such a feedback loop depends on cell type, it may be possible to gain detailed insights into the origin of this phenomenon by comparing normal and transformed cells.
Vinculin modulates force-loading of molecular clutches
Having established that the model can reproduce the mechanosensitive behaviour of cell adhesion, such as traction force and retrograde actin flow, we next tested whether the model can describe the mechanical dynamics of individual molecular clutches. Recent advances in the development of tension-gauge DNA sensors have made it possible to directly measure the force-loading rate and lifetime of integrin–ECM adhesion bonds. Experimental studies have shown that individual molecular clutches can experience forces above 40 pN, whereas measured force-loading rates are in the range 0.2–4 pN s−1 (refs. 19,20). Furthermore, the force-loading rate seems to vary with the molecular-clutch tension20. For example, it takes much longer for molecular clutches to reach a tension of 7–19 pN than to subsequently achieve a tension of 19–41 pN, a phenomenon that cannot be explained by existing models in which molecular clutches are represented by linear springs, because the force-loading rate of such molecular clutches does not depend on the applied tension. In addition, Hu et al. demonstrated that vinculin plays an important role in stabilizing the tension experienced by molecular clutches, as MEF vinculin-KO cells showed an increased force-loading rate and reduced lifetime of adhesion bonds20. The latter result is counter-intuitive, as the measured maximum tension experienced by molecular clutches in MEF vinculin-KO cells was lower than that in MEF WT cells, which is in good agreement with the experimentally observed reduction in cell traction in MEF vinculin-KO cells21. Because the binding of vinculin to talin, on the one hand, stabilizes molecular clutches37,38 and, on the other hand, keeps talin in an unfolded state22 causing an increase in the elasticity of molecular clutches, we tested whether these two factors are the main ones responsible for the observed effect of vinculin on cell traction and force-loading rates of molecular clutches.
To do this, we first tested the effect of vinculin KO by fitting cell traction and retrograde actin flow measured in MEF vinculin-KO cells21 to the WT-2 model. For this purpose, we used the dissociation rate of talin-based molecular clutches as a model-fitting parameter, because molecular clutches in MEF vinculin-KO cells are expected to be less stable due to the loss of vinculin–actin interactions that stabilize talin association with the actin cytoskeleton39,40,41. As shown in Fig. 4c,d, the WT-2 model correctly predicted a decrease in cell traction and an increase in retrograde actin flow compared to MEF WT cells in this case (blue curve). The fitted dissociation rate of molecular clutches showed a 30% increase compared to MEF WT cells, consistent with vinculin having a role in stabilizing molecular clutches. The fitting further improved when we added the myosin II density (σm) as another model-fitting parameter to phenomenologically account for the loss of tension transferred by vinculin molecules to talin (Fig. 4c,d, magenta curve). In this case, the model predicted a 60% increase in the dissociation rate of molecular clutches and a 20% decrease in the number of myosin II molecules exerting a mechanical load on molecular clutches. The decrease in myosin II density compensates for the increases in retrograde actin flow caused by the increase in the dissociation rate of molecular clutches and at the same time reduces cell traction, thus providing a better fit to the experimental data obtained in MEF vinculin-KO cells. Thus, the fitting results indicate that vinculin influences both the mechanical load of molecular clutches and their stability, which is in good agreement with published experimental data21.
a, Numerical solution of the master equation describing NA maturation (Supplementary equation (I1)). The calculations show that the occupancy probability of sites available for molecular-clutch formation by molecular clutches with folded talin (blue curve) reaches a steady-state level within 10–20 s, whereas the amount of vinculin-bound molecular clutches (green curve) grows more slowly due to the limited pool of molecular clutches with force-unfolded talin (red curve). b, Total internal reflection fluorescence measurements of NA area (blue) and also talin (yellow) and vinculin (green) intensities in NAs in NIH-3T3 cells (mean ± s.e.m., N = 437 peripheral NAs pooled from four cells), fitted to the talin WT model. Talin accumulates first, because for vinculin to bind to it requires the unfolding of the mechanosensitive R3 domain of talin. c,d, Retrograde actin flow (d) and cell traction (c) data for WT (red circles), vinculin-KO (green triangles) and vinculin VD1-expressing MEF cells (blue triangles)13,21 fitted to the WT-2 model, which accounts for vinculin-dependent changes in the dissociation rate and tension of molecular clutches. The fit shows that vinculin enhances the stability and mechanical load of molecular clutches. e, Force-loading rates of molecular clutches predicted by the model at their different tensions in mature FAs. Higher force-loading rates were predicted for molecular clutches under higher tension. f, Force-loading rates of molecular clutches in the presence of different vinculin mutants predicted by the talin WT model. KO of vinculin (blue) or vinculin with reduced talin binding affinity (vinA50I, red) markedly accelerates the force-loading rate of molecular clutches and increases the sensitivity to the density of sites available for molecular-clutch formation, whereas vinculin with reduced actin-binding affinity (vinI997A, yellow) has little effect compared to MEF WT cells (green). Experimental data were digitalized from ref. 20 (mean ± 95% confidence interval for three independent experiments). g, Force–extension curves of talin alone (no VD1) and in the presence of vinculin VD1 (cycles 1–3)22. Binding of vinculin VD1 to force-unfolded talin domains prevents their refolding, increasing talin extension in the 5–10 pN range by almost threefold, thereby softening talin. Vcl, vinculin. Panel g adapted from ref. 22 under a Creative Commons license CC BY 4.0.
We then calculated the dynamics of the talin-based molecular-clutch system in the presence of vinculin–talin interactions. To do this, we added three kinetic rates to the talin WT model, shown in Supplementary Table 4. These rates describe the kinetics of vinculin association or dissociation from the unfolded mechanosensitive R3 domain of talin (\({k}_{{\rm{on}}}^{{\rm{R3v}}}\) and \({k}_{{\rm{off}}}^{{\rm{R3v}}}\), respectively) and the formation of molecular clutches by vinculin-bound talins (\({k}_{{\rm{on,v}}}^{0}\); Supplementary Appendix I). The values for the first two rates were taken from previously published single-molecule experimental measurements25. The value of the \({k}_{{\rm{on,v}}}^{0}\) rate was chosen based on the assumption that vinculin-bound talins remain inside FAs even after the dissociation of molecular clutches from the substrate, so that they can readily form new molecular clutches compared to vinculin-unbound talins. Furthermore, based on previously published experimental data22, it was assumed that by binding to mechanically unfolded talin, vinculin locks it in the unfolded, more elastic state until it dissociates from talin. Finally, the effect of vinculin-mediated stabilization of talin-based molecular clutches was modelled by increasing the dissociation rate of such molecular clutches when vinculin is not bound to them to a value found by fitting the cell traction and retrograde actin flow data shown in Fig. 4c,d.
As shown in Fig. 4a,b, the evolution of the molecular-clutch system predicted by the model in this case has dynamics like those in the formation of NAs, as the kinetics of talin and vinculin accumulation within NAs and changes in NA area are like those observed in experiments performed on NIH-3T3 cells. The total amount of talin in NAs was found to increase rapidly, reaching within seconds ~50% of the total change in integral talin intensity in NAs observed during the first 3 min, whereas vinculin recruitment due to the binding to talin had a slower trend. Further calculations showed that the average force-loading rate of molecular clutches at various tension values in the range 0–30 pN also agrees well with experimental observations that demonstrate a highly nonlinear change in the force-loading rate with the tension experienced by molecular clutches20 (Fig. 4e). Furthermore, the average force-loading rate of molecular clutches is in the range 0.2–5 pN s−1, which is in good agreement with the experimental results obtained using tension-gauge DNA sensors19,20.
We then tested the effects of vinculin-KO and vinculin A50I and I997A mutants with reduced binding affinity to talin and actin, respectively, on the force-loading rate of molecular clutches. To do this, vinculin-KO/A50I was treated in the model by setting \({k}_{{\rm{on}}}^{{\rm{R3v}}}=0\) (no vinculin binding to talin). Additionally, the model parameter α, which describes the average number of new sites available for molecular clutch formation that are added to NAs and FAs upon unfolding of the mechanosensitive R3 domain of talin and subsequent binding of vinculin to it, was set to zero: α = 0 (no vinculin-mediated force-induced enhancement of NAs and FAs). As for the vinculin I997A mutant, the same \({k}_{{\rm{on}}}^{{\rm{R3v}}}\) and α values were used as in the case of MEF WT cells, but \({k}_{{\rm{on,v}}}^{0}\) was set to zero: \({k}_{{\rm{on,v}}}^{0}=0\) (as there is no rebinding of vinculin-associated molecular clutches after their dissociation from the substrate, because the vinculin I997A mutant is potentially unable to retain talin in NAs and FAs due to its reduced binding affinity for actin). In addition, for both vinculin-KO/A50I and vinculin I997A, the dissociation rate of molecular clutches was set to a higher level and the myosin II density was set to a lower level in the model calculations compared to MEF WT cells, according to the results shown in Fig. 4c,d. We calculated the effects of these mutants on the molecular-clutch dynamics on a rigid substrate coated with different ligand densities. The results showed that vinculin-KO and vinculin mutants strongly affected the force-loading dynamics of molecular clutches (Fig. 4f). In good agreement with experimental observations20, an increase in the average force-loading rate was observed for the vinculin-KO/A50I mutant, whereas the vinculin I997A mutant resulted in a negligible change in the force-loading rate (Fig. 4f). Furthermore, our model also correctly predicted the dependence of the force-loading rate of molecular clutches on the substrate ligand density, as there was an increase in the force-loading rate at lower ligand densities in all experimental conditions, with the most pronounced effect observed under vinculin KO conditions (Fig. 4f).
Overall, these results indicate that the mechanical stability of molecular clutches and the nonlinear force response of talin regulated by vinculin binding strongly affect the dynamics of molecular clutches. On the one hand, vinculin depletion reduces the stability of molecular clutches, leading to a decrease in cell traction under vinculin KO conditions. On the other hand, the binding of vinculin to the mechanosensitive R3 domain of talin and the locking of the latter in the unfolded state leads to an overall softening of talin, reducing the build-up of talin tension. In addition, due to the nonlinear force response of talin, which exhibits higher elasticity at lower stretching forces, the tension build-up in talin will be slower in the lower force range compared to the higher force range, in good agreement with experimental observations (Fig. 4e,g).
To illustrate the contribution of talin elasticity to the force-loading dynamics of molecular clutches, we produced qualitative estimates based on single-molecule measurements of the force response of talin in the absence and presence of vinculin binding22. As shown in Fig. 4g, the measured end-to-end extension of the folded talin under a tension of 10 pN is ~40 nm greater than that under a tension of 5 pN. For talin with all nine domains containing unfolded vinculin binding sites, the difference in the end-to-end extension of talin at the same tension values is ~110 nm. Considering the retrograde actin flow of ~7–10 nm s−1 obtained from fitting experimental data to the talin WT model with rigid substrates (E ≥ 1 MPa; Fig. 2d), we easily found the values of the force-loading rate in both cases to be ~0.9–1.3 and ~0.3–0.5 pN s−1 in the absence and presence of vinculin binding to talin, respectively, which are in good agreement with experimental measurements20 (Fig. 4f, experiment). This effect of vinculin on talin stiffness and force-loading rates of molecular clutches is mediated solely by vinculin binding to talin and does not require vinculin to bind to actin. Accordingly, our model predicts that the force-loading rates of molecular clutches in MEF WT cells and MEF vinculin I997A cells should be very similar, which is also in good agreement with experimental data (Fig. 4f).
Finally, using the same parameter values fitted for MEF WT cells, our model was also able to reproduce experimental observations of the mechanosensitive cell-adhesion behaviour on liquid-like viscoelastic substrates, which in the simplest case can be described by the Maxwell model, thus highlighting the robustness of our model predictions (Extended Data Fig. 4).
Steady-state behaviour and bifurcation of the clutch system
To further investigate the dynamic behaviour and stability of cell-adhesion complexes under different physiological conditions, we used the linear KD and linear WT models, which were found to accurately fit cell traction and retrograde actin flow measurements made in cells plated on substrates of different elasticities.
First, we mapped the bistability region in the space of model parameters. To do this, we varied the values of the spring constant of the intracellular part of molecular clutches (kc) and the myosin II density (σm) in the linear KD and linear WT models (Fig. 5). As a result, for the MEF Talin 1 KO, Talin 2 shRNA cells, the bistability region was found to be localized at moderate values of the myosin II density, close to the parameter range of the linear KD model corresponding to MEF Talin 1 KO, Talin 2 shRNA cells (Fig. 5a,e). This finding indicates that molecular clutches probably experience the bistability in myosin II-depleted regions of such cells that can often be found at the cell periphery42. Indeed, experiments have recently shown that filopodia exhibit both strong and weak adhesion to fibronectin-coated substrates in a myosin II-dependent manner42.
a,b, Stability diagrams of the linear KD (a) and linear WT (b) models. Three-dimensional red shapes and triangles denote the bistability regions and their projections, respectively. Yellow cylinders represent the model parameter ranges corresponding to MEF Talin 1 KO, Talin 2 shRNA and MEF WT cells. The blue dotted contour in a indicates a cross section of the bistability region detailed in c. Notably, talin-deficient MEF cells lie close to this region, whereas MEF WT cells are far from it, indicating that the high elasticity of talin stabilizes NAs and FAs. c, Close-up of a cross section of the bistability region indicated in a. Its boundaries are formed by the trajectories of the two saddle-node points shown in d. The bistability region arises from a cusp-type catastrophe when the rigidity of the intracellular part of molecular clutches is sufficiently large. d, Cusp catastrophe experienced by the molecular-clutch system. The pale green surface depicts cell traction (P) as a function of the retrograde actin flow (v) and substrate elasticity (E) in the linear KD model. The solid and dashed curves denote the stable and unstable branches of the solution for different myosin II densities (σm). Reducing σm to ~44 μm−2 results in the appearance of two saddle-node bifurcation points, a typical sign of a cusp catastrophe. Further decreasing σm shifts these points along the surface, outlining an instability region that includes only the unstable branches of cell traction curves. e,f, Stability diagrams of cell adhesion complexes in the case of MEF Talin 1 KO, Talin 2 shRNA cells (e) and MEF WT cells (f) obtained by projecting the bistability regions shown in a and b onto the horizontal plane. The black, red and blue solid curves mark the onset of the cusp catastrophe and bifurcations due to saddle-node points moving to infinity, which lead to the formation of biphase–monophase boundaries (green dotted curves), which affect the shapes of cell traction curves both within and beyond the bistability region. The insets around the bistability region illustrate representative cell traction curves within each domain. The graphs show that the elasticity of the intracellular part of molecular clutches and the myosin II density have a profound effect on the mechanosensitive behaviour of cell adhesion.
In addition, we fitted previously published experimental data collected for MEF Talin 1 KO, Talin 2 shRNA cells treated with a myosin II inhibitor, blebbistatin13, to the linear KD model. We found that these cells begin to develop bistable cell-adhesion behaviour when the density of myosin II motors in them decreases by ≳1.6 times (15 μM blebbistatin case; Extended Data Fig. 5a), with a further decrease in the myosin II density leading to a more pronounced bistability of cell adhesion (50 μM blebbistatin case; Extended Data Fig. 5a). On the other hand, blebbistatin-treated MEF WT cells did not seem to exhibit bistable cell-adhesion behaviour (Extended Data Fig. 5b), mainly due to the very high elasticity of talin-based molecular clutches, which results in MEF WT cells being far from the corresponding bistability region in the model parameter space (Fig. 5b,f). This finding indicates that talin molecules could potentially play an important role in stabilizing NAs and FAs during their maturation.
The existence of stable stationary states at each value of Young’s modulus for the substrate indicates that the molecular-clutch system should converge over time to one of these states, which, thus, represent possible steady states of the system. To find out how quickly the molecular-clutch system reaches these stationary states starting from an initial configuration with zero adhesion bonds between a cell and the substrate, we used the finite-difference method to numerically solve the master equation describing the time evolution of the molecular-clutch system (Supplementary equation (B10)). Both the linear WT and linear KD models predicted the rapid convergence of key experimental observables, such as cell traction and retrograde actin flow, to their stationary values within ~10–20 s (Extended Data Figs. 6a,b and 7a,b). The probability distribution of molecular-clutch extension also demonstrated rapid convergence to the steady-state distribution given by equation (9) in as little as 20 s (Extended Data Figs. 6c and 7c). These results are in good agreement with previous experimental observations showing that the retrograde flow of actin filaments in filopodia or near the edges of living cells occurs at an almost constant rate (v(t) ≈ const.)7,13,21,42,43, indicating that cell-adhesion complexes should function close to a steady state.
Furthermore, the characteristic relaxation time of the molecular-clutch system to the steady state predicted by our model for elastic-like substrates was two to four orders of magnitude less than the average lifetime of FAs (Extended Data Figs. 6e and 7e and Supplementary Table 5), indicating that they spend more than 98% of their time in a steady or quasi-steady state. In addition, from Supplementary Table 5, it is clear that the experimentally measured characteristic binding and dissociation times of vinculin and paxillin to or from FAs are an order of magnitude greater than the characteristic relaxation time of the molecular-clutch system to a steady state. This result indicates that talin may serve as a rapid response protein that helps establish the initial landscape of molecular clutches in a substrate-mechanosensitive manner, which can subsequently be reinforced and shaped by vinculin or paxillin into matured FAs, in good agreement with previous experimental data13,21,26,27,44.
Discussion
In this work, we developed a semi-analytical model of cell adhesion based on the molecular-clutch theory to examine the potential roles of adaptor-protein elasticity, local substrate deformations, and myosin II and ligand densities on the mechanosensitive behaviour of cell adhesion. One of the main differences between our model and previous studies is that we took a more explicit approach to modelling the elastic properties of adaptor proteins and substrate deformations. Then, by deriving a semi-analytical steady-state solution to the differential master equation of the model, we were able to evaluate experimental observables characterizing the mechanosensitive behaviour of cell-adhesion complexes, such as cell traction force and the rate of retrograde actin flow, using experimentally measured parameters, such as the stiffness of adaptor proteins and the dissociation rate of molecular clutches. This allowed us to identify several important factors that have a major impact on the mechanosensing behaviour of molecular clutches but which are not discussed in detail in the literature.
First, our work highlights the important role of the elastic properties of adaptor proteins, in particular talin, in the mechanosensory behaviour of cells. We showed that the difference in elasticity of talin and other adaptor proteins is one of the key parameters responsible for the different mechanosensitive adhesion behaviour of WT cells and talin-deficient cells. Increasing the stiffness of adaptor proteins, mimicking talin KO and KD conditions, results in decreased molecular-clutch density and reduced cell traction, altering cell behaviour in line with experimental observations13.
The inclusion of elastic adaptor proteins results in the molecular-clutch system behaving fundamentally differently from descriptions in previously published studies. Conventional molecular-clutch models assume that molecular clutches have a sufficiently high stiffness (1 pN nm−1 to 1,000 pN nm−1 range7,11,12,13,14), which results in tight coupling between the substrate and the retrograde actin flow, leading to the ‘load-and-fail’ mechanism on soft substrates and the ‘frictional slippage’ mechanism on rigid substrates. In both cases, when the mechanical stress of the substrate reaches a certain point and some of the engaged molecular clutches dissociate due to the mechanical load, there is virtually no relaxation of the substrate mechanical stress, which redistributes among a smaller number of the remaining molecular clutches engaged to the substrate. Therefore, the tension of the remaining molecular clutches becomes higher, which leads to their rapid dissociation, the ‘fail’ event, when all molecular clutches dissociate and the substrate returns to its relaxed conformation (Fig. 6a,c).
a,b, Schematic representations of the load-and-fail mechanism (a) and the differential-transmission mechanism (b). The formation of molecular clutches results in transmission of the force generated by myosin II motors to the substrate, causing its deformation. In the load-and-fail mechanism, when the increasing tension of molecular clutches reaches a certain value, the molecular clutches dissociate in a highly cooperative manner as a result of their high rigidity, which is usually used in conventional molecular-clutch models (a). By contrast, in the models developed in our study (b), molecular clutches exhibit highly elastic properties, which is in good agreement with experiments. This leads to a loss of synchronization of the dissociation of molecular clutches under the myosin II-generated load, and as a result, the molecular-clutch system exhibits a more uniform behaviour, functioning as a differential transmission that mechanically couples the substrate and actin cytoskeleton, which are moving at different speeds. c–e, Stochastic kinetic simulations of the conventional molecular-clutch model (c) and our linear KD (d) and linear WT (e) models. The conventional molecular-clutch model predicts stochastic oscillations between zero and the maximum cell traction on soft substrates (load-and-fail mechanism). By contrast, the linear WT and linear KD models predict much milder fluctuations of the molecular-clutch system around its steady state, with the amplitude of fluctuations being smaller for softer molecular-clutch components (linear WT model) due to the differential-transmission mechanism schematically shown in b.
By contrast, in our model, molecular clutches are represented by highly elastic adaptor proteins (0.05–1.6 pN nm−1) mechanically coupled through adhesion receptors to local substrate deformations at the adhesion sites of the molecular clutches. Cell traction and retrograde actin flow fluctuate around their steady-state values, which are determined by the force balance between the tension of adaptor proteins, local substrate deformations and the mechanical load created by myosin II motors (Fig. 6d,e). This behaviour of cell traction and retrograde actin flow is in better agreement with previous experiments compared to the load-and-fail and frictional slippage mechanisms45. Indeed, the high elasticity of molecular clutches in our model results in dynamic frictional coupling between the substrate and retrograde actin flow for both soft and rigid substrates. Thus, the dissociation of some of the molecular clutches leads to a rapid redistribution of forces and a partial release of the substrate tension, so that the tension in the remaining molecular clutches is less than in the load-and-fail scenario. This makes it possible for the remaining molecular clutches to withstand the load long enough to allow new molecular clutches to form (Fig. 6b). As a result, the molecular-clutch system functions as a differential transmission that enables the retrograde actin flow and substrate to move at different speeds, while maintaining stable mechanical coupling between them. Notably, the amplitude of fluctuations experienced by the molecular-clutch system in this case strongly depends on both the elastic properties of the adaptor proteins and the assembly rate of molecular clutches, with the more elastic talin molecules demonstrating a greater ability to buffer substrate stress and deformations, resulting in fairly stable cell traction and retrograde actin flow, even on very rigid substrates, in good agreement with experimental observations7,13,21,30,42,43.
Furthermore, the force-loading rate of talin-based molecular clutches predicted by the model (0.2–5.5 pN s−1, rigid substrate) is in good agreement with experimental data (0.2–4 pN s−1)19,20. In addition, because the force-loading rate of molecular clutches is determined by the elasticity of their components, the model predicts that the nonlinear force response of talin will result in a higher force-loading rate of molecular clutches at higher tensions, whereas the vinculin-mediated locking of talin in the softer unfolded state is predicted to result in a low force-loading rate of molecular clutches even for the vinculin I997A mutant, which has reduced actin-binding ability. Both of these model predictions are in good agreement with direct experiment observations19,20. These results demonstrate that the elasticity of adaptor proteins is one of the key factors that must be taken into account when relating the force response of molecular clutches at the single-molecule level to mesoscale cell behaviour.
In addition to the elasticity of molecular-clutch components, another important aspect considered in our model is the local deformation of the substrate at the attachment sites of molecular clutches (Figs. 1 and 6b). When individual integrins or small clusters of them exert a mechanical force on the substrate, elasticity theory predicts that only a small region of the substrate in the immediate vicinity will be most strongly deformed46. Consistently, it has been widely reported in the literature that local changes in the traction force or substrate deformation on scales smaller than cell-adhesion complexes play a critical role in regulating the mechanosensitive behaviour of cell adhesion20,45,47,48.
From a modelling perspective, if the substrate elasticity is represented by a spring describing its global deformation at the scale of cell-adhesion complexes, then it can be theoretically shown that for soft molecular clutches, the system will lose its ability to sense substrate rigidity and will always reach the same steady state (the same cell traction and retrograde actin flow), regardless of the substrate stiffness, unless some parameters of the system, such as cell contractility, are adaptively changed49. In this scenario, the substrate remains relatively stationary at a nearly constant steady-state tension. As a result, the molecular clutches have no information about the degree of substrate deformation required to achieve the steady-state tension, and therefore, they cannot distinguish between different substrate stiffnesses.
By contrast, if local substrate deformations at the attachment sites of molecular clutches are considered, then the intracellular part (adaptor proteins) and the extracellular part (local substrate deformation) of each molecular clutch will establish their own individual force balance, which collectively determines the state of larger cell-adhesion complexes. In this case, the overall rigidity of local substrate deformations and adaptor proteins will influence the force-loading dynamics and the lifetime of molecular clutches, allowing the system to reach different substrate-dependent steady states. Consequently, our model can robustly predict the mechanosensitive response of cell-adhesion complexes over a wide range of substrate rigidities without the need for any rigidity-dependent adjustments to the physiological parameters of the system. Thus, we propose that the sensing of substrate rigidity at the molecular level, where each molecular clutch probes the mechanical properties of its local substrate by establishing its own force balance, is critical for the mechanosensitive cell-adhesion response.
Our model also predicts that when the adaptor proteins and substrate have high stiffnesses (kc ≳ 0.5–0.8 pN nm−1 and E ≳ 100 kPa), the molecular-clutch system exhibits bistable behaviour, in which both weak and strong cell-adhesion states are stable at the same value of substrate elasticity. From a physical point of view, this is reminiscent of the previously reported bistable cytoadhesion properties of malaria-infected red blood cells50,51. Bistability can potentially arise at the level of a single cell or individual FAs. It is influenced by the heterogeneity and topography of the ECM ligand network, which affect the local density of sites available for forming molecular clutches and the rate of molecular-clutch assembly. This, in turn, influences the tension experienced by molecular clutches and actin filaments, also potentially altering the mechano-dependent cooperative assembly of myosin IIA and myosin IIB stacks regulated by ROCK, MLCK52 and PKCz53.
The quasi-static nature of molecular-clutch kinetics and the emergence of bistable cell-adhesion behaviour revealed in our model indicate that, for a given substrate rigidity, several stable states of molecular clutches can coexist. This means that our model can, in principle, be used as a theoretical basis to describe the maturation process of FAs, in which small NAs are gradually transformed into mature FAs with a different molecular composition, molecular-clutch density and traction force, despite being in the same extracellular environment. In this context, the maturation of FAs can be viewed as a series of quasi-steady states, each of which represents a snapshot of maturing cell adhesion. Changes in the physiological parameters describing mechanical or biochemical processes, such as local molecular-clutch density and elasticity and myosin II contractility, collectively model the evolution of the quasi-steady-state behaviour of FAs towards more mature states. In addition, the bistability of cell adhesion found in our work may also explain why only a small fraction of NAs mature on rigid substrates whereas the rest disassemble, which warrants future study.
Note that in our study, we considered the unfolding of the mechanosensitive R3 domain of talin. Yet, the force-induced unfolding of other domains may also stabilize the tension experienced by talin and influence molecular-clutch stability, indicating a broader role for adaptor-protein elasticity54,55. Understanding how the unfolding of talin domains and other adaptor proteins regulates mechanosensitive cell-adhesion behaviour remains an important question that may be potentially clarified by integrating our model with molecular dynamics simulations56.
Similarly, one of the critical missing links in existing molecular-clutch models is how the dynamic behaviour of cell-adhesion complexes is involved in the interplay with other cell mechanosensing mechanisms that influence cell-adhesion behaviour. For example, mechanosensitive ion channels57,58,59, formins1,60,61,62,63 and LIM domain proteins64 have been shown to potentially play important roles in regulating the force response of FAs or the cell-signalling cascades involved in remodelling cell-adhesion complexes. With more accurate and realistic modelling of the conformational states of molecular-clutch proteins, which is possible within our model, one can begin to quantitatively investigate the complex interactions between these mechanosensitive processes at the molecular level, as they have important consequences for many physiological processes, from morphogenesis to cancer progression.
Finally, in addition to substrate stiffness, the rheological properties of the ECM, such as viscoelasticity, have also been shown to be an important regulator of cellular functions and are known to be related to pathological processes of tumour growth and metastasis. There have been several interesting attempts to model the mechanosensitive behaviour of cells on viscoelastic substrates based on the conventional molecular-clutch theory. These have successfully reproduced various cell behaviours, such as periodic oscillations in cell spreading and cell migration10,14,16,17,18. In the future, it will be interesting to combine our semi-analytic model with these previous studies to explore how physiological factors, such as the stiffness of molecular-clutch components, regulate these important cell behaviours.
Methods
Linear molecular-clutch model
In this study, we use the model shown schematically in Fig. 1b. Molecular clutches are represented by springs consisting of two parts: (1) cellular, corresponding to adaptor proteins, and (2) extracellular, describing local substrate deformations at the adhesion sites of molecular clutches. In the simplest case, these two parts of the molecular clutches can be described by linear springs with spring constants kc and \({k}_{{\rm{s}}}^{{\prime} }\), respectively.
In addition to local substrate deformation at the adhesion sites of molecular clutches, the model also takes into account the average substrate deformation over a much larger area of cell-adhesion complexes with a characteristic size R. This type of substrate deformation is described in the model by the spring constant ks shown in Fig. 1b.
Using the theory of elasticity, it can be shown that the spring constant \({k}_{{\rm{s}}}^{{\prime} }\) describing local substrate deformations is related to the Young’s modulus of the substrate (E) as (Supplementary Appendix A):
where r is the characteristic size of the adhesion site of a single molecular clutch.
The spring constant ks describes large-scale substrate deformations. In general, it depends both on the Young’s modulus of the substrate and on the shape of cell-adhesion complexes: ks = ks(E, shape). For example, if the shapes of cell-adhesion complexes can be approximated by ellipses, then ks(E) = 4πRE/9, where R = a/Ie(ec) is the characteristic size of the cell-adhesion complexes. Here a and ec are the major axis and eccentricity of the ellipse representing the shape of the cell-adhesion complexes, and Ie is an elliptic integral whose value as a function of the ellipse eccentricity is shown in Supplementary Fig. 3a (see also Supplementary Appendix A). Thus, like \({k}_{{\rm{s}}}^{{\prime} }\), ks in this case is proportional to the characteristic size of cell-adhesion complexes, R. As a result, taking into account the experimentally measured characteristic size of FAs (R ≈ 1.7 μm)13 and the typical size of the binding sites of adhesion receptors (up to several nanometres), it follows that local substrate deformations at the adhesion sites of molecular clutches should be much greater than the average deformation of the substrate on the FA scale and, consequently, have a much stronger effect on the dissociation kinetics of molecular clutches compared to the average substrate deformation. Moreover, as shown below, the main results of our study are essentially independent of the spring constant ks and its exact functional form, because it is absent from the main formulae determining the steady-state behaviour of the molecular-clutch system.
To describe the dynamic state of the system, the sites available for molecular-clutch formation are divided into two groups in the model: those that are occupied by molecular clutches (on state) and those that are not (off state).
In addition, each molecular clutch is characterized by its own deformation resulting from the mechanical load Fm generated by myosin II motors (Fig. 1b). Super-resolution images of the nanoscale architecture of FAs show that the average contact angle of the intracellular part of molecular clutches with the cell membrane or substrate surface, θ, is very small (θ ≈ 15°; ref. 65; see the schematic in Fig. 1b). Thus, molecular clutches experience much greater deformation along the x axis parallel to the substrate surface compared to the z axis perpendicular to it. As a result, the physical state of each molecular clutch can be described to a first approximation by its extension, l, along the x axis parallel to the substrate surface, which includes the mechanical deformation of both the intracellular and extracellular parts of the molecular clutch (Fig. 1b). Accordingly, the state of the entire system can be represented by a time-dependent probability distribution pon(l, t) of observing a molecular clutch with extension l at time t, for each site available for molecular-clutch formation. This distribution satisfies the following normalization condition:
where poff(t) is the time-dependent probability of the off state. The extension of a molecular clutch can be positive or negative, depending on the orientation of the molecular clutch along the x axis.
When formed, molecular clutches should have an extension randomly distributed around the mechanically relaxed state, with a peak corresponding to zero extension along the x axis (l = 0). As a result, the extension-dependent assembly rate of molecular clutches kon(l) can be approximated using the Dirac δ function δ(l):
where \({k}_{{\rm{on}}}^{0}\) is a constant.
A retrograde flow of actin filaments created by myosin II motors results in a gradual stretching of molecular clutches. This, in turn, leads to an increase in their deformation, causing subsequent dissociation. We assumed in this study that the dissociation of molecular clutches is mainly driven by the detachment of integrins from their ECM ligands, such as fibronectin. Indeed, previous studies have indicated that this may be the weakest point of molecular clutches, as the bonds between other molecular-clutch components seem to be substantially strengthened by an applied mechanical load37,41,66. For example, talin has a multivalent association with the actin cytoskeleton mediated by talin-bound vinculins, which form catch bonds with F-actin that exhibit force-dependent enhancement41. This model assumption is further supported by recent DNA tension-gauge tether experiments showing that individual molecular clutches can withstand forces >10 pN for tens of seconds, far exceeding previous estimates based on the experimentally measured strength of talin–actin bonds39,41. Because integrin–ECM bonds have previously been shown to exhibit catch-bond behaviour, exhibiting force-dependent bond enhancement in the range 0–30 pN and force-induced bond dissociation at higher forces67, the dissociation rate of molecular clutches (koff) was approximated by the following two-term exponential formula, which was found to fit the experimental data very well (Supplementary Fig. 4):
where xt and \({x}_{{\rm{t}}}^{{\prime} }\) are two transition state distances characterizing the force-dependent behaviour of the integrin–ECM catch bond, and \({k}_{{\rm{off}}}^{0}\) and \({k}_{{\rm{off}}}^{{0}^{{\prime} }}\) are the corresponding dissociation rates at zero mechanical load. β = 1/kBT is the reciprocal of the thermodynamic temperature, where kB is the Boltzmann constant. ∣F∣ is the absolute value of the tension applied to the bond.
For molecular clutches represented by two-part linear springs, the force–extension behaviour of each molecular clutch is described by
where \(k={k}_{{\rm{c}}}{k}_{{\rm{s}}}^{{\prime} }/({k}_{{\rm{c}}}+{k}_{{\rm{s}}}^{{\prime} })\) is the resulting spring constant of molecular clutches consisting of the two parts: extracellular (\({k}_{{\rm{s}}}^{{\prime} }\)) and intracellular (kc) (Fig. 1b).
Given the probability distribution pon(l, t), then the total tension of molecular clutches or, what is the same, the traction force created by molecular clutches within a single cell-adhesion complex, Fc(t) (Fig. 1b), can be found as:
where Nc is the number of sites available for the formation of molecular clutches in the cell-adhesion complex. Recent experimental studies show that the average density of talin and, probably therefore, the density of sites available for molecular-clutch formation are very similar in both small and large FAs27, indicating that these are independent of the size of the FAs. As a result, we concluded that Nc is proportional to the area of the cell-adhesion complexes S: Nc = σcS, where σc is the average density of sites available for the formation of molecular clutches in a cell-adhesion complex.
According to the Newton’s third law, the total tension in engaged molecular clutches (Fc) is equal to the force generated by myosin II motors: Fm(t) = Fc(t). Following previous studies7,11,12,13,14, the resisting effect of such a load on the movement rate of myosin II motors, v(t), is approximated by a linear force–velocity relation:
where Fst is the stalling force of a single myosin II motor. v0 is the movement rate of myosin II motors at zero mechanical load. Nm is the number of myosin motors pulling actin filaments within a cell-adhesion complex. Because previous studies indicated that myosin II motors exert a local pulling effect on the actin cytoskeleton in the cell cortex42, we assumed in the model that Nm is proportional to the area of cell-adhesion complexes: Nm = σmS, where σm is the average surface density of myosin II motors near the cell–substrate interface.
Experimental studies show that actin filaments exhibit retrograde movement at nearly constant rates (v(t) ≈ const.)7,13,21,42,43. Thus, it can be concluded that myosin II motors and the entire ensemble of molecular clutches function close to a steady state in living cells. This makes it possible to derive a system of simple equations that describe the long-term steady-state behaviour of the entire ensemble of molecular clutches and myosin II motor proteins based on the above formulae (Supplementary Appendix B):
The first line of the equation describes the time-averaged probability distribution pon(l) = 〈pon(l,t)〉T of molecular-clutch extension, which is determined by the force-dependent dissociation rate of molecular clutches, koff(l) = koff(F(l)), and the retrograde actin flow, v. The second line simply states that in a steady state, molecular clutches are stretched in one direction by retrograde actin flow. The third line indicates that in a steady state, the total probability flux due to the dissociation of molecular clutches is balanced by the probability flux due to their formation, which provides the normalization condition for the pon(l) probability distribution. The fourth line represents Newton’s third law, which states that the total tension in engaged molecular clutches (Fc) is equal to the force generated by myosin II motors (Fm). Finally, the fifth line of the equation shows the average mechanical deformation of the substrate (xs) in a steady state due to the traction force (Fc) (Fig. 1b).
To determine the long-term behaviour of the molecular-clutch system and fit the experimental data, we solved equation (8) in terms of pon, Fc and v and then compared the calculated values of cell traction (Fc) and retrograde actin flow (v) to experimental measurements. To do this, equation (8) was solved by finding the intersections of the graphs of Fc(v) and Fm(v). This allowed us not only to construct a simple geometric interpretation of solutions to equation (8) but also to assess their stability (Extended Data Fig. 1).
Specifically, Fm(v) can be directly calculated from the fourth line of equation (8). On the other hand, the computation of Fc(v) requires knowledge of the probability distribution pon(l), which can be found from the first three lines of equation (8) (Supplementary Appendix C):
Knowing the pon(l) distribution, the total tension of molecular clutches (Fc) can easily be obtained from equation (8) as a function of the retrograde actin flow speed (v), which leads to the solution of equation (8).
To compare the cell traction force predicted by the model (Fc) with experimental measurements, we calculated the traction stress (P) exerted by cell-adhesion complexes on the substrate:
where S is the area of the cell-adhesion complexes.
Notably, as can be seen from equations (4)–(10), in the steady state, cell traction and the retrograde actin flow are determined by local substrate deformations at the sites of molecular-clutch attachment, which are described in the model by the spring constant \({k}_{{\rm{s}}}^{{\prime} }\) (Supplementary Appendix D) and do not depend on the spring constant ks describing large-scale substrate deformations on the scale of cell-adhesion complexes. Thus, we concluded that cell traction, retrograde actin flow and the average tension of molecular clutches do not depend on the size and shape of cell-adhesion complexes in the steady state (Supplementary Fig. 3b–e and Supplementary Appendix E), which is in good agreement with experimental data showing that the average density of talin molecules in FAs and their average tension are, indeed, independent of FA size27.
Finally, note that although in the above equations we considered molecular clutches to be simply represented by linear springs, the resulting equations (8)–(10) are very general, as an arbitrary force response of molecular clutches can be used in the model. For example, to describe nonlinear deformations of molecular clutches caused by reversible force-induced unfolding of their globular protein domains or the desorption of ECM from the substrate68,69, another parameter can be introduced into the model, the extension ly of molecular clutches, at which their linear behaviour switches to nonlinear behaviour. Thus, we change equation (5) to:
This coarse-grained approach is most suitable for describing molecular clutches with an unknown architecture or protein structure.
On the other hand, if the structure and mechanical stability of the main molecular-clutch components are known, more detailed information can be obtained compared to the above approach by considering all possible transitions between different folded and unfolded states of molecular clutches. We used this method to account for the highly nonlinear behaviour of talin molecules and the force-induced unfolding of the mechanosensitive talin domain (R3) observed in single-molecule experiments22 (Supplementary Appendix F).
Importantly, almost all model parameters can be measured experimentally (Supplementary Table 1). Regarding the remaining parameters, by simultaneously fitting experimental data obtained under different experimental conditions (WT or talin-deficient cells that are either treated with myosin II inhibitor or grown on substrates coated with different concentrations of fibronectin, as described in ‘Results’), the average number of fitting parameters per curve can be reduced to one or two, so that fitting the experimental data to the model is very robust. A list of key model-fitting parameters used in this study can be found in Extended Data Table 2.
Cell lines, constructs and experimental data processing
Cell culture, constructs and transfection
NIH-3T3 and HeLa cells were obtained from the laboratory of Q. Peng (Shenzhen Bay Laboratory, China). All cell lines were grown in Dulbecco’s modified Eagle’s medium (Gibco, C11995500BT) supplemented with 10% fetal bovine serum (Gibco, 10099-141C) and 1% penicillin/streptomycin (Gibco, 15070-063) at 37 °C under 5% CO2.
To visualize myosin II motors, cells were transfected with a plasmid encoding myosin RLC-GFP (gift from A. Bershadsky, Mechanobiology Institute, Singapore) using jetPRIME buffer (Polyplus, 712-60) and jetPRIME reagent (Polyplus, 114-15) according to the manufacturer’s instructions. Following the transfection, cells were seeded onto 35-mm glass-bottomed dishes coated with 1, 10 or 100 μg ml−1 fibronectin (Solarbio, F8180) dissolved in Dulbecco’s phosphate-buffered saline (DPBS, Gibco, C14190500BT). The day after transfection, cells were fixed for 15 min with 4% paraformaldehyde solution (Beyotime, P0099), followed by washing with DPBS (three times), after which the cells were imaged.
To visualize vinculin and talin, NIH-3T3 cells were transfected with plasmids encoding mCherry-Vinculin (gift from K. Yamada, Addgene 50527) and GFP-Talin 1 (gift from A. Huttenlocher, Addgene 26724) using Lipofectamine 3000 Transfection Reagent (Thermo Fisher Scientific). The next day, the cells were replated into glass-bottomed dishes coated with 10 μg ml−1 fibronectin (Corning) and imaged.
Fluorescence microscopy
Cells transfected with the myosin RLC-GFP construct were imaged using an ultra-high resolution microscope (Elyra 7, Zeiss) equipped with Plan Apochromatic ×63/1.4 oil differential interference contrast objective, which was controlled by software ZEN (black edition). During the experiment, the microscope was focused on the ventral side of the transfected cells, resulting in images of predominantly ventral myosin II motors. The images were captured with a pco.edge sCMOS camera (Lattice SIM module) and processed by the Lattice SIM2 structured illumination technique (horizontal resolution of 60 nm and axial resolution of 200 nm). Fluorescence was excited using optically pumped semiconductor lasers (488 nm at 500 mW and 561 nm at 500 mW). Cells transfected with mCherry-Vinculin and GFP-Talin 1 were imaged every 2 s for 4,000 s after 6 h of spreading using a total internal reflection fluorescence microscope (Nikon Ti2) equipped with a standard set of lasers and filters.
Image analysis and statistics
Collected images of RLC-GFP-labelled myosin II stacks were analysed with the help of ImageJ-win64 software. To do this, several random regions near the cell corners were selected for each cell and automatic thresholding was applied using the built-in Auto Threshold function of ImageJ to map the contours of myosin II filament stacks. The resulting images were further processed with the Analyze Particles function, which provided the total number and sizes of the detected myosin II filament stacks. After discarding all particles with an area less than 0.01 μm2, the average density and the average size of myosin II filament stacks as well as the corresponding s.e.m. were calculated. All experimental results presented in the article are representative of three independent experiments. Statistical analyses of the data were carried out using the one-way ANOVA with post-hoc Tukey’s honestly significant difference test.
Images of mCherry-Vinculin and GFP-Talin 1 cells were registered in ImageJ using the descriptor-based registration plugin70. The images were then cropped and scaled down by a factor of 0.75 in the x and y directions and by a factor of 0.5 in time. Segmenting, tracking and measuring the area of FAs and measuring the average intensities of talin and vinculin were carried out using the FAA server71 with a minimum FA size of 0.1 μm2 (Extended Data Fig. 8a). The tracked FAs were then filtered in MATLAB (Mathworks) to remove tracks with lifetimes of less than 5 min and tracks in which the size of the FAs exceeded 0.5 μm2 in more than three of the first five frames to remove tracks in which FAs were already mature when detected. This left only tracks corresponding to NAs. After this, all NAs were divided into two groups: peripheral NAs and central NAs. To identify peripheral and central NAs, the mask of each cell was eroded using a 4.3-μm-diameter disc. Those NAs whose centroid fell into this remaining region in the first frame they appeared in were considered central, and those that formed outside this region were considered to be peripheral (Extended Data Fig. 8b). The resulting mCherry-Vinculin and GFP-Talin 1 signal intensities measured within peripheral NAs were normalized to the first frame for each NA and then averaged over all peripheral NAs (Extended Data Fig. 8d,e). The mean integral intensity as a function of time was calculated by multiplying the mean normalized intensity by the mean area of NAs (Extended Data Fig. 8c). To estimate the mean integral intensity of talin-bound vinculins in peripheral NAs, the mean integral intensity of vinculin in each NA was offset by the initial value, because experimental studies show that the initial accumulation of vinculin in NAs occurs predominantly due to the binding of vinculin to paxillin and not to mechanically unfolded talin72. The resulting integral intensities of talin and talin-bound vinculin measured in peripheral NAs in the experiments were then compared with model predictions (Fig. 4b).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data supporting the findings of this study are presented in the main text and Supplementary Information.
Code availability
The main program codes can be downloaded from websites artem-efremov.org or artemefremovlab.com. Other programs are available upon request.
References
Riveline, D. et al. Focal contacts as mechanosensors: externally applied local mechanical force induces growth of focal contacts by an mDia1-dependent and ROCK-independent mechanism. J. Cell Biol. 153, 1175–1186 (2001).
Vogel, V. & Sheetz, M. Local force and geometry sensing regulate cell functions. Nat. Rev. Mol. Cell Biol. 7, 265–275 (2006).
Wolfenson, H., Yang, B. & Sheetz, M. P. Steps in mechanotransduction pathways that control cell morphology. Annu. Rev. Physiol. 81, 585–605 (2019).
Nia, H. T., Munn, L. L. & Jain, R. K. Physical traits of cancer. Science 370, eaaz0868 (2020).
Kechagia, J. Z., Ivaska, J. & Roca-Cusachs, P. Integrins as biomechanical sensors of the microenvironment. Nat. Rev. Mol. Cell Biol. 20, 457–473 (2019).
Mitchison, T. & Kirschner, M. Cytoskeletal dynamics and nerve growth. Neuron 1, 761–772 (1988).
Chan, C. E. & Odde, D. J. Traction dynamics of filopodia on compliant substrates. Science 322, 1687–1691 (2008).
Srinivasan, M. & Walcott, S. Binding site models of friction due to the formation and rupture of bonds: state-function formalism, force-velocity relations, response to slip velocity transients, and slip stability. Phys. Rev. E 80, 046124 (2009).
Sabass, B. & Schwarz, U. S. Modeling cytoskeletal flow over adhesion sites: competition between stochastic bond dynamics and intracellular relaxation. J. Phys.: Condens. Matter 22, 194112 (2010).
Sens, P. Rigidity sensing by stochastic sliding friction. Europhys. Lett. 104, 38003 (2013).
Bangasser, B. L., Rosenfeld, S. S. & Odde, D. J. Determinants of maximal force transmission in a motor-clutch model of cell traction in a compliant microenvironment. Biophys. J. 105, 581–592 (2013).
Elosegui-Artola, A. et al. Rigidity sensing and adaptation through regulation of integrin types. Nat. Mater. 13, 631–637 (2014).
Elosegui-Artola, A. et al. Mechanical regulation of a molecular clutch defines force transmission and transduction in response to matrix rigidity. Nat. Cell Biol. 18, 540–548 (2016).
Gong, Z. et al. Matching material and cellular timescales maximizes cell spreading on viscoelastic substrates. Proc. Natl Acad. Sci. USA. 115, E2686–E2695 (2017).
Elosegui-Artola, A., Trepat, X. & Roca-Cusachs, P. Control of mechanotransduction by molecular clutch dynamics. Trends Cell Biol. 28, 356–367 (2018).
Sens, P. Stick-slip model for actin-driven cell protrusions, cell polarization, and crawling. Proc. Natl Acad. Sci. USA. 117, 24670–24678 (2020).
Ron, J. E., Monzo, P., Gauthier, N. C., Voituriez, R. & Gov, N. S. One-dimensional cell motility patterns. Phys. Rev. Res. 2, 033237 (2020).
Mukherjee, A. et al. Actin filaments couple the protrusive tips to the nucleus through the I-BAR domain protein IRSp53 during the migration of cells on 1D fibers. Adv. Sci. 10, e2207368 (2023).
Jo, M. H. et al. Determination of single-molecule loading rate during mechanotransduction in cell adhesion. Science 383, 1374–1379 (2024).
Hu, Y. et al. DNA-based ForceChrono probes for deciphering single-molecule force dynamics in living cells. Cell 187, 3445–3459 (2024).
Thievessen, I. et al. Vinculin-actin interaction couples actin retrograde flow to focal adhesions, but is dispensable for focal adhesion growth. J. Cell Biol. 202, 163–177 (2013).
Yao, M. et al. The mechanical response of talin. Nat. Commun. 7, 11966 (2016).
Margadant, F. et al. Mechanotransduction in vivo by repeated talin stretch-relaxation events depends upon vinculin. PLoS Biol. 9, e1001223 (2011).
Yao, M. et al. Mechanical activation of vinculin binding to talin locks talin in an unfolded conformation. Sci. Rep. 4, 4610 (2014).
Wang, Y. et al. Force-dependent interactions between talin and full-length vinculin. J. Am. Chem. Soc. 143, 14726–14737 (2021).
Austen, K. et al. Extracellular rigidity sensing by talin isoform-specific mechanical linkages. Nat. Cell Biol. 17, 1597–1606 (2015).
Kumar, A. et al. Talin tension sensor reveals novel features of focal adhesion force transmission and mechanosensitivity. J. Cell Biol. 213, 371–383 (2016).
Deguchi, S., Hotta, J., Yokoyama, S. & Matsui, T. S. Viscoelastic and optical properties of four different PDMS polymers. J. Micromech. Microeng. 25, 097002 (2015).
Williams, L. T. et al. Viscoelastic polyacrylamide MR elastography phantoms with tunable damping ratio independent of shear stiffness. J. Mech. Behav. Biomed. Mater. 154, 106522 (2024).
Driscoll, T. P., Ahn, S. J., Huang, B., Kumar, A. & Schwartz, M. A. Actin flow-dependent and -independent force transmission through integrins. Proc. Natl Acad. Sci. USA 117, 32413–32422 (2020).
Yamashiro, S. et al. Force transmission by retrograde actin flow-induced dynamic molecular stretching of talin. Nat. Commun. 14, 8468 (2023).
Bachir, A. I. et al. Integrin-associated complexes form hierarchically with variable stoichiometry in nascent adhesions. Curr. Biol. 24, 1845–1853 (2014).
Li, Y. et al. Kindlin2-mediated phase separation underlies integrin adhesion formation. Preprint at bioRxiv https://doi.org/10.1101/2020.07.10.197400 (2020).
Ma, N. et al. Kindlin-2 phase separation in response to flow controls vascular stability. Circ. Res. 135, 1141–1160 (2024).
Hu, S. Long-range self-organization of cytoskeletal myosin II filament stacks. Nat. Cell Biol. 19, 133–141 (2017).
Bhadriraju, K. et al. Activation of ROCK by RhoA is regulated by cell adhesion, shape, and cytoskeletal tension. Exp. Cell Res. 313, 3616–3623 (2007).
Huang, D. L., Bax, N. A., Buckley, C. D., Weis, W. I. & Dunn, A. R. Vinculin forms a directionally asymmetric catch bond with F-actin. Science 357, 703–706 (2017).
Le, S., Yu, M. & Yan, J. Direct single-molecule quantification reveals unexpectedly high mechanical stability of vinculin-talin/α-catenin linkages. Sci. Adv. 5, eaav2720 (2019).
Jiang, G., Giannone, G., Critchley, D. R., Fukumoto, E. & Sheetz, M. P. Two-piconewton slip bond between fibronectin and the cytoskeleton depends on talin. Nature 424, 334–337 (2003).
Hu, K., Ji, L., Applegate, K. T., Danuser, G. & Waterman-Storer, C. M. Differential transmission of actin motion within focal adhesions. Science 315, 111–115 (2007).
Owen, L. M., Bax, N. A., Weis, W. I. & Dunn, A. R. The C-terminal actin-binding domain of talin forms an asymmetric catch bond with F-actin. Proc. Natl Acad. Sci. USA. 119, e2109329119 (2022).
Alieva, N. O. et al. Myosin IIA and formin dependent mechanosensitivity of filopodia adhesion. Nat. Commun. 10, 3593 (2019).
Bornschlögl, T. et al. Filopodial retraction force is generated by cortical actin dynamics and controlled by reversible tethering at the tip. Proc. Natl Acad. Sci. USA. 110, 18928–18933 (2013).
Dumbauld, D. W. et al. How vinculin regulates force transmission. Proc. Natl Acad. Sci. USA. 110, 9788–9793 (2013).
Plotnikov, S. V., Pasapera, A. M., Sabass, B. & Waterman, C. M. Force fluctuations within focal adhesions mediate ECM-rigidity sensing to guide directed cell migration. Cell 151, 1513–1527 (2012).
Landau, L. D. & Lifshitz, E. M. Theory of Elasticity 2nd edn (Pergamon Press, 1970).
Ghassemi, S. et al. Cells test substrate rigidity by local contractions on submicrometer pillars. Proc. Natl Acad. Sci. USA 109, 5328–5333 (2012).
Liu, Y. et al. Nanoparticle tension probes patterned at the nanoscale: impact of integrin clustering on force transmission. Nano Lett. 14, 5539–5546 (2014).
Alonso-Matilla, R., Provenzano, P. P. & Odde, D. J. Optimal cell traction forces in a generalized motor-clutch model. Biophys. J. 122, 3369–3385 (2023).
Efremov, A. & Cao, J. Bistability of cell adhesion in shear flow. Biophys. J. 101, 1032–1040 (2011).
Xu, X. et al. Probing the cytoadherence of malaria infected red blood cells under flow. PLoS ONE 8, e64763 (2013).
Quintanilla, M. A. et al. Local monomer levels and established filaments potentiate non-muscle myosin 2 assembly. J. Cell Biol. 223, e202305023 (2024).
Schiffhauer, E. S. et al. Myosin IIB assembly state determines its mechanosensitive dynamics. J. Cell Biol. 218, 895–908 (2019).
Chen, H. et al. Dynamics of equilibrium folding and unfolding transitions of titin immunoglobulin domain under constant forces. J. Am. Chem. Soc. 137, 3540–3546 (2015).
Le, S. et al. Dystrophin as a molecular shock absorber. ACS Nano 12, 12140–12148 (2018).
Guo, S. et al. Structural-elastic determination of the force-dependent transition rate of biomolecules. Chem. Sci. 9, 5871–5882 (2018).
Coste, B. et al. Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science 330, 55–60 (2010).
Yao, M. et al. Force- and cell state-dependent recruitment of Piezo1 drives focal adhesion dynamics and calcium entry. Sci. Adv. 8, eabo1461 (2022).
Efremov, A. K. et al. Application of piconewton forces to individual filopodia reveals mechanosensory role of L-type Ca2+ channels. Biomaterials 284, 121477 (2022).
Kozlov, M. M. & Bershadsky, A. D. Processive capping by formin suggests a force-driven mechanism of actin polymerization. J. Cell Biol. 167, 1011–1017 (2004).
Yu, M. et al. mDia1 senses both force and torque during F-actin filament polymerization. Nat. Commun. 8, 1650 (2017).
Yu, M. et al. Effects of mechanical stimuli on profilin- and formin-mediated actin polymerization. Nano Lett. 18, 5239–5247 (2018).
Nishimura, Y., Shi, S., Li, Q., Bershadsky, A. D. & Viasnoff, V. Crosstalk between myosin II and formin functions in the regulation of force generation and actomyosin dynamics in stress fibers. Cells Dev. 168, 203736 (2021).
Sun, X. et al. Mechanosensing through direct binding of tensed F-actin by LIM domains. Dev. Cell 55, 468–482 (2020).
Liu, J. et al. Talin determines the nanoscale architecture of focal adhesions. Proc. Natl Acad. Sci. USA. 112, E4864–E4873 (2015).
Neumann, J. & Gottschalk, K.-E. The integrin-talin complex under force. Protein Eng. Des. Sel. 29, 503–512 (2016).
Kong, F., García, A. J., Mould, A. P., Humphries, M. J. & Zhu, C. Demonstration of catch bonds between an integrin and its ligand. J. Cell Biol. 185, 1275–1284 (2009).
Paturej, J., Dubbeldam, J. L. A., Rostiashvili, V. G., Milchev, A. & Vilgis, T. A. Force spectroscopy of polymer desorption: theory and molecular dynamics simulations. Soft Matter 10, 2785–2799 (2014).
Weinberg, S. H., Mair, D. B. & Lemmon, C. A. Mechanotransduction dynamics at the cell-matrix interface. Biophys. J. 112, 1962–1974 (2017).
Preibisch, S., Saalfeld, S., Schindelin, J. & Tomancak, P. Software for bead-based registration of selective plane illumination microscopy data. Nat. Methods 7, 418–419 (2010).
Berginski, M. E. & Gomez, S. M. The Focal Adhesion Analysis Server: a web tool for analyzing focal adhesion dynamics. F1000Res 2, 68 (2013).
Case, L. B. et al. Molecular mechanism of vinculin activation and nanoscale spatial organization in focal adhesions. Nat. Cell Biol. 17, 880–892 (2015).
Bennett, M. et al. Molecular clutch drives cell response to surface viscosity. Proc. Natl Acad. Sci. USA. 115, 1192–1197 (2018).
Billington, N., Wang, A., Mao, J., Adelstein, R. S. & Sellers, J. R. Characterization of three full-length human nonmuscle myosin II paralogs. J. Biol. Chem. 288, 33398–33410 (2013).
Acknowledgements
We greatly appreciate the encouraging and insightful discussions with J. Yan (MBI, Singapore) and A. D. Bershadsky (MBI, Singapore) about the results of our study. We are grateful to A. D. Bershadsky for providing genetic constructs used in the study. We thank Q. Peng (Shenzhen Bay Laboratory, China) for NIH-3T3 and HeLa cell lines. Computation and data analysis were mainly carried out at Shenzhen Bay Laboratory High Performance Computing and Informatics Core Facility. We also thank the Bioimaging Core Facility of Shenzhen Bay Laboratory for providing cell imaging support. We are also grateful to Bioimaging Core engineer Y. Mei for assistance with SIM microscopy (Elyra 7). This study was supported by start-up funds from Shenzhen Bay Laboratory (A.K.E.) and the National Key R&D Programme of China (Grant No. 2024YFA0920100 to M.Y.). M.Y. acknowledges support from the Guangdong Basic and Applied Basic Research Foundation (Grant No. 2025Al515012151) and the Guangdong Provincial Key Laboratory of Advanced Biomaterials (Grant No. 2022B1212010003). H.E.J. acknowledges support from the HKU Seed Grant (Grant No. 2202100793).
Author information
Authors and Affiliations
Contributions
P.L. and Q.W. performed the experiments. P.L., Q.W., X.D., L.P., J.W., W.Z., H.E.J. and A.K.E. analysed the data. M.Y. and A.K.E. designed the research and wrote the paper. A.K.E. developed the theoretical framework, derived the formulae and carried out the computations.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Physics thanks Tristan Driscoll, Namrata Gundiah and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Geometric interpretation of the force balance in the molecular clutch system.
(a, b) Intersections of the cell traction force curve, Fc(v), and the myosin II force-velocity curve, Fm(v), correspond to the stationary points (S1 and S2) described by Eq. (8). Stability of such points is mainly determined by the slopes of Fc(v) and Fm(v) curves. If dFm/dv > dFc/dv [panel (a)], the corresponding stationary point (S1) is stable. On the other hand, if dFm/dv < dFc/dv [panel (b)], then the stationary point (S2) is unstable. As an example, consider point B in panel (a). An initial deviation of the system from point S1 (for example, due to dissociation of several molecular clutches) will first result in an increase in retrograde actin flow, since myosin II motors must work against a smaller resisting force. However, the system cannot remain in the new state (point B’) for long, since myosin II motors cannot generate enough force to balance the resisting traction force created by replenished molecular clutches (point B”). As a result, retrograde actin flow will slow down, bringing the system to stationary point S1. Similar considerations apply to the other points shown in panels (a, b). Numerical simulations of the molecular clutch system show good agreement with the above physical considerations, see Extended Data Fig. 3. (c) Example of 3D plots of cell traction force (Fc, colored surface) and myosin II-pulling force (Fm, gray surface) as a function of retrograde actin flow (v) and substrate elasticity (E). The black solid curves formed by the intersection of the two surfaces denote stationary points of the system that attract trajectories to their neighborhood (stable branches), whereas the black dashed curve denotes unstable stationary points (unstable branch). The white dot designates the saddle-node bifurcation point where the stable and unstable branches merge together. All cell traction and retrograde actin flow curves shown in Results were obtained by projecting the stable and unstable branches of 3D plots like those displayed in panel (c) onto the vertical and horizontal planes, respectively.
Extended Data Fig. 2 Experimental data fitting to a model describing the competitive formation of two types of molecular clutches (WT-2 model).
(a, b) Fitting of the cell traction and retrograde actin flow experimental data from refs. 13,21 collected on MEF WT cells (red circles) and MEF Talin 1 KO, Talin 2 shRNA cells (green triangles) to the WT-2 model describing the competitive formation of two types of molecular clutches: 1) soft, nonlinear, talin-dominant one, prevalent in WT cells, and 2) more rigid, linear, non-talin-based one, prevalent in talin-deficient cells. The WT-2 model fits experimental data obtained in MEF WT cells more accurately [red curves in panels (a, b)] compared to the linear WT and talin WT models [Fig. 2(c,d) in the main text], with most of the model fitting parameters being similar to those in the linear KD and talin WT models, see Tables T1 and T2, SI. Moreover, the WT-2 model predicts that even 10% of the maximum density of talin in FAs, and hence 10% of the rate of talin-based molecular clutch formation (\({k}_{{\rm{on}}}^{0}\)), is sufficient to stabilize cell adhesion to rigid substrates (E > 10 kPa), but not to soft substrates (5 kPa < E < 10 kPa), see the blue curves in panels (a, b). (c) Negative correlation between cell traction and retrograde actin flow. Existing experimental data and models tested in this study show a negative linear correlation between cell traction and retrograde actin flow, which arises from the linear force-velocity relationship of myosin II motors, see panel (d) and Appendix H, SI for details. (d) Schematic explanation of the graphs shown in panel (c). The physicochemical properties of molecular clutches do not affect the correlation between experimentally measured cell traction and retrograde actin flow, since stable stationary points, which represent the steady-state of the molecular clutch system, correspond to the intersection points of the cell traction force curve, Fc(v), and the myosin II force-velocity curve, Fm(v). Thus, these points always lie on the linear force-velocity curve of myosin II motors, resulting in a linear correlation between experimentally measured cell traction and retrograde actin flow.
Extended Data Fig. 3 Effect of the elasticity of the intracellular part of molecular clutches on cell adhesion.
(a, b) Graphs of the cell traction and retrograde actin flow curves calculated using the linear WT model for various values of the spring constant (kc) describing the elastic properties of the intracellular part of molecular clutches. The solid curves denote stable branches of the graphs, and the dashed curves indicate unstable branches, see Extended Data Fig. 1 for details. It can be seen from the plots that the spring constant kc has a strong influence on the cell adhesion behaviour in the case of rigid substrates (E > 10 kPa), while there is almost no change on soft substrates (E ≤ 10 kPa). In addition, the figure shows that in the case of stiff molecular clutches (kc > 2.6 pN/nm), the molecular clutch system undergoes bifurcation, leading to bistability of the cell traction and retrograde actin flow curves on rigid substrates. To demonstrate the bifurcation, the model parameter values used in the calculations were the same as for the case of low fibronectin density (1 μg/ml) shown in Fig. 3(a) in the main text. (c) Representative trajectories demonstrating the time evolution of the molecular clutch system obtained by solving the master equation [Eq. (B10), SI] for points A, B and C (kc = 5 pN/nm) shown in panel (b) by using stochastic simulations and finite-difference calculations. From the plot it can be seen that if at low values of Young’s modulus of the substrate (E < 2 MPa) there is a unique stationary solution that attracts trajectories to its neighborhood (point A), then at higher values of Young’s modulus (E > 2 MPa) there is a pair of such stationary solutions (points B and C), which indicates the bistable behaviour of the molecular clutch system. Notably, stochastic simulations demonstrate that both strong and weak cell adhesion states corresponding to points B and C are very stable and can last for many minutes without experiencing stochastic transitions between each other.
Extended Data Fig. 4 Cell traction, retrograde actin flow and time evolution of NAs in cells grown on elastic-like and liquid-like substrates.
(a-c) Quasi-steady-state cell traction (a) and retrograde actin flow (b), as well as the dynamics of NA growth (c) in the case where large-scale substrate deformations are represented by the Maxwell model (liquid-like substrate) and the Kelvin-Voigt model (elastic-like substrate), respectively. It can be seen that in the case of liquid-like substrates, the model predicts that cell traction increases, and retrograde actin flow decreases with increasing viscosity of liquid-like substrates, which is in good agreement with previously published experimental data shown in the figure that were digitalized from ref. 73. Furthermore, maturation of NAs is greatly hampered in the case of low-viscosity liquid-like substrates compared to elastic-like substrates due to the slower accumulation of vinculin in NAs, which is also consistent with previous experimental data73 (from left to right N = 11, 12, 17 samples, data are presented as mean ± SD). The calculations were performed for E = 1 MPa Young’s modulus of the substrate. The results shown in panels (a-c) were obtained by numerically solving the master equation [Eq. (I1) and Eq. (I3), SI] describing the time evolution of the molecular clutch system using the finite-difference method. The values of the model parameters used in the calculations were the same as in Table T1 and Table T4, SI.
Extended Data Fig. 5 The role of myosin II density in the mechanosensitive cell adhesion behaviour.
(a, b) Fitting of experimental data collected on MEF Talin 1 KO, Talin 2 shRNA cells (a) and MEF WT cells (b) treated with different concentrations of the myosin II inhibitor, blebbistatin [13]. The data were fitted to the linear KD model [solid curves, panel (a)], linear WT model [solid curves, panel (b)] and talin WT model [dotted curves, panel (b)], respectively, by varying only the density of myosin II motors. The remaining model parameters were fixed at the values shown in Table T1, SI. The dashed segments of the cell traction curves in panel (a) indicate unstable branches of the corresponding graphs. Panels (a, b) show that while MEF Talin 1 KO, Talin 2 shRNA cells are predicted to have bistable behaviour on rigid substrates at low myosin II densities, MEF WT cells lack this type of behaviour due to the high elasticity of talin-based molecular clutches, see Fig. 5 in the main text for additional details.
Extended Data Fig. 6 Time evolution of cell adhesion complexes in MEF WT cells on elastic-like substrates (Kelvin-Voigt model).
(a, b) Time evolution of cell traction (P) and retrograde actin flow (v) calculated by solving the master-equation [Eq. (B10), SI] using the finite-difference method, starting from an initial configuration with zero molecular clutches between the cell and the substrate. Three different values of the substrate Young’s modulus (E) and viscosity coefficient (ξs) were tested: 100 Pa, 1 kPa, 10 kPa and 0 pN ⋅ s/nm, 1 pN ⋅ s/nm, 1000 pN ⋅ s/nm, respectively. The values of the other model parameters were the same as in the linear WT model, see Table T1, SI. In all cases, cell traction and retrograde actin flow rapidly reach steady-state values within 10-20 s, regardless of the substrate viscosity. (c) Probability distribution (pon) of molecular clutch extension (l) after 20 s of time evolution of the molecular clutch system, calculated for three different values of the Young’s modulus (E) and viscosity coefficient (ξs) of the substrate. The graph shows that the molecular clutch system reaches the steady-state probability distribution given by Eq. (9) in the main text within 20 s (yellow dashed lines), regardless of the substrate viscosity. (d) Time evolution of substrate deformation (xs). The graph shows that the characteristic relaxation time of the substrate strongly depends on its viscosity, but it has little effect on the rapid convergence of the probability distribution of molecular clutch extension, cell traction, and retrograde actin flow to their steady-state values for elastic-like substrates described by the Kelvin-Voigt model, see panels (a-c). (e) The characteristic relaxation time of retrograde actin flow obtained by fitting the curves shown in panel (b) to an exponential decay function as a function of the Young’s modulus and viscosity coefficient of the substrate. It can be seen that the characteristic relaxation time decreases with increasing Young’s modulus of the substrate. However, it is practically independent of the viscous properties of elastic-like substrates.
Extended Data Fig. 7 Time evolution of cell adhesion complexes in MEF Talin 1 KO, Talin 2 shRNA cells on elastic-like substrates (Kelvin-Voigt model).
(a, b) Time evolution of cell traction (P) and retrograde actin flow (v) calculated by solving the master-equation [Eq. (B10), SI] using the finite-difference method, starting from an initial configuration with zero molecular clutches between the cell and the substrate. Three different values of the substrate Young’s modulus (E) and viscosity coefficient (ξs) were tested: 100 Pa, 1 kPa, 10 kPa and 0 pN ⋅ s/nm, 1 pN ⋅ s/nm, 1000 pN ⋅ s/nm, respectively. The values of the other model parameters were the same as in the linear KD model, see Table T1, SI. In all cases, cell traction and retrograde actin flow rapidly reach steady-state values within 10-20 s, regardless of the substrate viscosity. (c) Probability distribution (pon) of molecular clutch extension (l) after 20 s of time evolution of the molecular clutch system, calculated for three different values of the Young’s modulus (E) and viscosity coefficient (ξs) of the substrate. The graph shows that the molecular clutch system reaches the steady-state probability distribution given by Eq. (9) in the main text within 20 s (yellow dashed lines), regardless of the substrate viscosity. (d) Time evolution of substrate deformation (xs). The graph shows that the characteristic relaxation time of the substrate strongly depends on its viscosity, but it has little effect on the rapid convergence of the probability distribution of molecular clutch extension, cell traction, and retrograde actin flow to their steady-state values for elastic-like substrates described by the Kelvin-Voigt model, see panels (a-c). (e) The characteristic relaxation time of retrograde actin flow obtained by fitting the curves shown in panel (b) to an exponential decay function as a function of the Young’s modulus and viscosity coefficient of the substrate. It can be seen that the characteristic relaxation time decreases with increasing Young’s modulus of the substrate. However, it is practically independent of the viscous properties of elastic-like substrates.
Extended Data Fig. 8 Segmentation of FAs and measurement of their area and average intensities of talin and vinculin in them.
(a) Segmentation and tracking of FAs was carried out using the FAA server [71]. The tracked FAs were then filtered to remove tracks with lifetimes of less than 5 minutes and tracks in which the size of FAs exceeded 0.5 μm2 in more than 3 of the first 5 frames in order to remove tracks in which FAs were already mature when detected and leave only tracks corresponding to NAs. (b) After segmentation and tracking, all NAs were divided into two main groups - peripheral NAs and central NAs - using an eroded cell mask as described in Methods in the main text. Only peripheral NAs were used for comparison with the model. (c) The mean area of peripheral NAs measured in 4 different NIH-3T3 cells. (d, e) The mean normalized signal intensities of talin and vinculin measured in peripheral NAs in 4 different NIH-3T3 cells. The signal measured in each NA was normalized to the first frame, after which the mean normalized intensity was calculated by averaging over all peripheral NAs for each cell. In panels (c-e), the total number of tracked peripheral NAs in each cell was N = 133 in cell 1, N = 100 in cell 2, N = 101 in cell 3 and N = 103 in cell 4. The error bars in panels (c-e) represent SEM.
Supplementary information
Supplementary Information
Supplementary Appendices A–I, Figs. 1–6 and Tables 1–5.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, P., Wang, Q., Dai, X. et al. Elastic properties of force-transmitting linkages determine multistable mechanosensitive behaviour of cell adhesion. Nat. Phys. 21, 1431–1443 (2025). https://doi.org/10.1038/s41567-025-02964-z
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41567-025-02964-z
This article is cited by
-
Ca2+ transmembrane transport enhances oscillatory growth of cancer cell invadopodia
Communications Physics (2025)