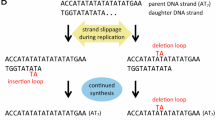
Abstract
DNA mismatch repair (MMR) is one of many evolutionarily conserved processes that act as guardians of genomic integrity. MMR proteins recognize errors that occur during DNA replication and initiate countermeasures to rectify those mistakes. MMR deficiency (MMRd) therefore leads to a dramatic accumulation of mutations. The MMRd genomic signature is characterized by a high frequency of single-base substitutions as well as insertions and/or deletions that preferentially occur in short nucleotide repeat sequences known as microsatellites. This accumulation leads to a phenomenon termed microsatellite instability, which accordingly serves as a marker of underlying MMRd. MMRd is associated with hereditary cancer syndromes such as Lynch syndrome and constitutional MMRd as well as with sporadic tumour development across a variety of tissues. High baseline immune cell infiltration is a characteristic feature of MMRd/microsatellite instability-high tumours, as is the upregulation of immune checkpoints. Importantly, the molecular profile of MMRd tumours confers remarkable sensitivity to immune-checkpoint inhibitors (ICIs). Many patients with MMRd disease derive durable clinical benefit when treated with these agents regardless of the primary tumour site. Nevertheless, a substantial subset of these patients will fail to respond to ICI, and increasing research is focused on identifying the factors that confer resistance. In this Review, we begin by discussing the biological function of the MMR machinery as well as the genomic sequelae of MMRd before then examining the clinical implications of MMRd with a specific focus on cancer predisposition, diagnostic approaches, therapeutic strategies and potential mechanisms of resistance to ICIs.
Key points
-
Immune-checkpoint inhibitors (ICIs) confer remarkably durable clinical benefit in many patients with DNA mismatch repair-deficient (MMRd) tumours.
-
MMRd tumours are thought to be responsive to ICIs because they harbour many single-base substitutions and frameshift mutations, which, if expressed, have the potential to encode tumour-specific immunogenic neoantigens.
-
Immune-mediated killing of MMRd cancer cells can be orchestrated by various effector cells, enabling MMRd tumours to respond to ICIs despite major histocompatibility complex (MHC) class I loss.
-
Most patients with MMRd tumours derive benefit from ICIs, although a substantial number have primary resistance and many more develop acquired resistance.
-
Many potential predictors of response and resistance to ICIs are under active investigation, but none are currently ready for clinical implementation.
-
The accurate diagnosis of MMRd status is an important determinant of ICI response. This is best achieved through a multimodal approach that involves immunohistochemical analysis of mismatch repair protein expression and microsatellite profiling.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Lynch, H. T., Snyder, C. L., Shaw, T. G., Heinen, C. D. & Hitchins, M. P. Milestones of Lynch syndrome: 1895–2015. Nat. Rev. Cancer 15, 181–194 (2015).
Holliday, R. A mechanism for gene conversion in fungi. Genet. Res. 89, 285–307 (2007).
Kunkel, T. A. & Erie, D. A. Eukaryotic mismatch repair in relation to DNA replication. Annu. Rev. Genet. 49, 291–313 (2015).
Le, D. T. et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357, 409–413 (2017).
Mandal, R. et al. Genetic diversity of tumors with mismatch repair deficiency influences anti-PD-1 immunotherapy response. Science 364, 485–491 (2019).
Germano, G. et al. Inactivation of DNA repair triggers neoantigen generation and impairs tumour growth. Nature 552, 116–120 (2017).
Le, D. T. et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 372, 2509–2520 (2015).
Hause, R. J., Pritchard, C. C., Shendure, J. & Salipante, S. J. Classification and characterization of microsatellite instability across 18 cancer types. Nat. Med. 22, 1342–1350 (2016).
Kim, T. M., Laird, P. W. & Park, P. J. The landscape of microsatellite instability in colorectal and endometrial cancer genomes. Cell 155, 858–868 (2013).
Cortes-Ciriano, I., Lee, S., Park, W. Y., Kim, T. M. & Park, P. J. A molecular portrait of microsatellite instability across multiple cancers. Nat. Commun. 8, 15180 (2017).
Ercan, A. B. et al. Clinical and biological landscape of constitutional mismatch-repair deficiency syndrome: an international replication repair deficiency consortium cohort study. Lancet Oncol. 25, 668–682 (2024).
Veigl, M. L. et al. Biallelic inactivation of hMLH1 by epigenetic gene silencing, a novel mechanism causing human MSI cancers. Proc. Natl Acad. Sci. USA 95, 8698–8702 (1998).
Llosa, N. J. et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 5, 43–51 (2015).
Marabelle, A. et al. Efficacy of pembrolizumab in patients with noncolorectal high microsatellite instability/mismatch repair-deficient cancer: results from the phase II KEYNOTE-158 study. J. Clin. Oncol. 38, 1–10 (2020).
André, T. et al. Pembrolizumab in microsatellite-instability-high advanced colorectal cancer. N. Engl. J. Med. 383, 2207–2218 (2020).
Mirza, M. R. et al. Dostarlimab for primary advanced or recurrent endometrial cancer. N. Engl. J. Med. 388, 2145–2158 (2023).
Lenz, H. J. et al. First-line nivolumab plus low-dose ipilimumab for microsatellite instability-high/mismatch repair-deficient metastatic colorectal cancer: the phase II CheckMate 142 study. J. Clin. Oncol. 40, 161–170 (2022).
André, T. et al. Antitumor activity and safety of dostarlimab monotherapy in patients with mismatch repair deficient solid tumors: a nonrandomized controlled trial. JAMA Netw. Open. 6, e2341165 (2023).
Andre, T. et al. Nivolumab plus ipilimumab in microsatellite-instability-high metastatic colorectal cancer. N. Engl. J. Med. 391, 2014–2026 (2024).
André, T. et al. Nivolumab plus ipilimumab versus nivolumab in microsatellite instability-high metastatic colorectal cancer (CheckMate 8HW): a randomised, open-label, phase 3 trial. Lancet Lond. Engl. 405, 383–395 (2025).
Iyer, R. R., Pluciennik, A., Burdett, V. & Modrich, P. L. DNA mismatch repair: functions and mechanisms. Chem. Rev. 106, 302–323 (2006).
Kunkel, T. A. DNA replication fidelity. J. Biol. Chem. 279, 16895–16898 (2004).
Kunkel, T. A. & Bebenek, K. DNA replication fidelity. Annu. Rev. Biochem. 69, 497–529 (2000).
Schofield, M. J. & Hsieh, P. DNA mismatch repair: molecular mechanisms and biological function. Annu. Rev. Microbiol. 57, 579–608 (2003).
Kolodner, R. D. A personal historical view of DNA mismatch repair with an emphasis on eukaryotic DNA mismatch repair. DNA Repair. 38, 3–13 (2016).
Wildenberg, J. & Meselson, M. Mismatch repair in heteroduplex DNA. Proc. Natl Acad. Sci. USA 72, 2202–2206 (1975).
Wagner, R. & Meselson, M. Repair tracts in mismatched DNA heteroduplexes. Proc. Natl Acad. Sci. USA 73, 4135–4139 (1976).
Modrich, P. Mechanisms and biological effects of mismatch repair. Annu. Rev. Genet. 25, 229–253 (1991).
Fishel, R. Mismatch repair. J. Biol. Chem. 290, 26395–26403 (2015).
Miller, J. H. Mutators in Escherichia coli. Mutat. Res. 409, 99–106 (1998).
Kunkel, T. A. DNA-mismatch repair. The intricacies of eukaryotic spell-checking. Curr. Biol. CB 5, 1091–1094 (1995).
Kolodner, R. Biochemistry and genetics of eukaryotic mismatch repair. Genes. Dev. 10, 1433–1442 (1996).
Acharya, S., Foster, P. L., Brooks, P. & Fishel, R. The coordinated functions of the E. coli MutS and MutL proteins in mismatch repair. Mol. Cell 12, 233–246 (2003).
Wang, H. et al. DNA bending and unbending by MutS govern mismatch recognition and specificity. Proc. Natl Acad. Sci. USA 100, 14822–14827 (2003).
Cho, W. K. et al. ATP alters the diffusion mechanics of MutS on mismatched DNA. Struct. 20, 1264–1274 (2012).
Liu, J. et al. Cascading MutS and MutL sliding clamps control DNA diffusion to activate mismatch repair. Nature 539, 583–587 (2016).
Yang, X. W. et al. MutS functions as a clamp loader by positioning MutL on the DNA during mismatch repair. Nat. Commun. 13, 5808 (2022).
Hall, M. C. & Matson, S. W. The Escherichia coli MutL protein physically interacts with MutH and stimulates the MutH-associated endonuclease activity. J. Biol. Chem. 274, 1306–1312 (1999).
Au, K. G., Welsh, K. & Modrich, P. Initiation of methyl-directed mismatch repair. J. Biol. Chem. 267, 12142–12148 (1992).
Kunkel, T. A. & Erie, D. A. DNA mismatch repair. Annu. Rev. Biochem. 74, 681–710 (2005).
Modrich, P. Mechanisms in E. coli and human mismatch repair (Nobel lecture). Angew. Chem. Int. Ed. Engl. 55, 8490–8501 (2016).
Dai, J. et al. Molecular basis of the dual role of the Mlh1–Mlh3 endonuclease in MMR and in meiotic crossover formation. Proc. Natl Acad. Sci. USA 118, e2022704118 (2021).
Flores-Rozas, H. & Kolodner, R. D. The Saccharomyces cerevisiae MLH3 gene functions in MSH3-dependent suppression of frameshift mutations. Proc. Natl Acad. Sci. USA 95, 12404–12409 (1998).
Chen, P. C. et al. Contributions by MutL homologues Mlh3 and Pms2 to DNA mismatch repair and tumor suppression in the mouse. Cancer Res. 65, 8662–8670 (2005).
Kadyrova, L. Y. & Kadyrov, F. A. Endonuclease activities of MutLα and its homologs in DNA mismatch repair. DNA Repair 38, 42–49 (2016).
Putnam, C. D. Strand discrimination in DNA mismatch repair. DNA Repair 105, 103161 (2021).
Drummond, J. T., Li, G. M., Longley, M. J. & Modrich, P. Isolation of an hMSH2-p160 heterodimer that restores DNA mismatch repair to tumor cells. Science 268, 1909–1912 (1995).
Genschel, J., Littman, S. J., Drummond, J. T. & Modrich, P. Isolation of MutSbeta from human cells and comparison of the mismatch repair specificities of MutSbeta and MutSalpha. J. Biol. Chem. 273, 19895–19901 (1998).
Hombauer, H., Campbell, C. S., Smith, C. E., Desai, A. & Kolodner, R. D. Visualization of eukaryotic DNA mismatch repair reveals distinct recognition and repair intermediates. Cell 147, 1040–1053 (2011).
Warren, J. J. et al. Structure of the human MutSalpha DNA lesion recognition complex. Mol. Cell 26, 579–592 (2007).
Lamers, M. H. et al. The crystal structure of DNA mismatch repair protein MutS binding to a G × T mismatch. Nature 407, 711–717 (2000).
Edelbrock, M. A., Kaliyaperumal, S. & Williams, K. J. Structural, molecular and cellular functions of MSH2 and MSH6 during DNA mismatch repair, damage signaling and other noncanonical activities. Mutat. Res. 743-744, 53–66 (2013).
Bradford, K. C. et al. Dynamic human MutSα-MutLα complexes compact mismatched DNA. Proc. Natl Acad. Sci. USA 117, 16302–16312 (2020).
Bowers, J., Sokolsky, T., Quach, T. & Alani, E. A mutation in the MSH6 subunit of the Saccharomyces cerevisiae MSH2-MSH6 complex disrupts mismatch recognition. J. Biol. Chem. 274, 16115–16125 (1999).
Dufner, P., Marra, G., Räschle, M. & Jiricny, J. Mismatch recognition and DNA-dependent stimulation of the ATPase activity of hMutSalpha is abolished by a single mutation in the hMSH6 subunit. J. Biol. Chem. 275, 36550–36555 (2000).
Schofield, M. J. et al. The Phe–X–Glu DNA binding motif of MutS. The role of hydrogen bonding in mismatch recognition. J. Biol. Chem. 276, 45505–45508 (2001).
Drotschmann, K., Yang, W., Brownewell, F. E., Kool, E. T. & Kunkel, T. A. Asymmetric recognition of DNA local distortion. Structure-based functional studies of eukaryotic Msh2–Msh6. J. Biol. Chem. 276, 46225–46229 (2001).
Gupta, S., Gellert, M. & Yang, W. Mechanism of mismatch recognition revealed by human MutSβ bound to unpaired DNA loops. Nat. Struct. Mol. Biol. 19, 72–78 (2011).
Gradia, S., Acharya, S. & Fishel, R. The human mismatch recognition complex hMSH2–hMSH6 functions as a novel molecular switch. Cell 91, 995–1005 (1997).
Geng, H. et al. Biochemical analysis of the human mismatch repair proteins hMutSα MSH2(G674A)–MSH6 and MSH2–MSH6(T1219D). J. Biol. Chem. 287, 9777–9791 (2012).
Iaccarino, I., Marra, G., Palombo, F. & Jiricny, J. hMSH2 and hMSH6 play distinct roles in mismatch binding and contribute differently to the ATPase activity of hMutSalpha. EMBO J. 17, 2677–2686 (1998).
Fishel, R. Mismatch repair, molecular switches, and signal transduction. Genes. Dev. 12, 2096–2101 (1998).
Antony, E. & Hingorani, M. M. Mismatch recognition-coupled stabilization of Msh2–Msh6 in an ATP-bound state at the initiation of DNA repair. Biochemistry 42, 7682–7693 (2003).
Gradia, S. et al. hMSH2–hMSH6 forms a hydrolysis-independent sliding clamp on mismatched DNA. Mol. Cell 3, 255–261 (1999).
Plotz, G., Raedle, J., Brieger, A., Trojan, J. & Zeuzem, S. N-terminus of hMLH1 confers interaction of hMutLalpha and hMutLbeta with hMutSalpha. Nucleic Acids Res. 31, 3217–3226 (2003).
Plotz, G. et al. Mutations in the MutSalpha interaction interface of MLH1 can abolish DNA mismatch repair. Nucleic Acids Res. 34, 6574–6586 (2006).
Kadyrov, F. A., Dzantiev, L., Constantin, N. & Modrich, P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell 126, 297–308 (2006).
Sacho, E. J., Kadyrov, F. A., Modrich, P., Kunkel, T. A. & Erie, D. A. Direct visualization of asymmetric adenine-nucleotide-induced conformational changes in MutL alpha. Mol. Cell 29, 112–121 (2008).
Genschel, J. et al. Interaction of proliferating cell nuclear antigen with PMS2 is required for MutLα activation and function in mismatch repair. Proc. Natl Acad. Sci. USA 114, 4930–4935 (2017).
Pluciennik, A. et al. PCNA function in the activation and strand direction of MutLα endonuclease in mismatch repair. Proc. Natl Acad. Sci. USA 107, 16066–16071 (2010).
Umar, A. et al. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell 87, 65–73 (1996).
Schmutte, C. et al. Human exonuclease I interacts with the mismatch repair protein hMSH2. Cancer Res. 58, 4537–4542 (1998).
Goellner, E. M., Putnam, C. D. & Kolodner, R. D. Exonuclease 1-dependent and independent mismatch repair. DNA Repair. 32, 24–32 (2015).
Amin, N. S., Nguyen, M. N., Oh, S. & Kolodner, R. D. exo1-Dependent mutator mutations: model system for studying functional interactions in mismatch repair. Mol. Cell Biol. 21, 5142–5155 (2001).
Calil, F. A. et al. Rad27 and Exo1 function in different excision pathways for mismatch repair in Saccharomyces cerevisiae. Nat. Commun. 12, 5568 (2021).
Zhang, Y. et al. Reconstitution of 5′-directed human mismatch repair in a purified system. Cell 122, 693–705 (2005).
Genschel, J. & Modrich, P. Mechanism of 5′-directed excision in human mismatch repair. Mol. Cell 12, 1077–1086 (2003).
Goellner, E. M. et al. Identification of Exo1-Msh2 interaction motifs in DNA mismatch repair and new Msh2-binding partners. Nat. Struct. Mol. Biol. 25, 650–659 (2018).
Budczies, J. et al. Tumour mutational burden: clinical utility, challenges and emerging improvements. Nat. Rev. Clin. Oncol. 21, 725–742 (2024).
Chow, R. D. et al. Distinct mechanisms of mismatch-repair deficiency delineate two modes of response to anti-PD-1 immunotherapy in endometrial carcinoma. Cancer Discov. 13, 312–331 (2023).
Kwon, M. et al. Determinants of response and intrinsic resistance to PD-1 blockade in microsatellite instability-high gastric cancer. Cancer Discov. 11, 2168–2185 (2021).
Schrock, A. B. et al. Tumor mutational burden is predictive of response to immune checkpoint inhibitors in MSI-high metastatic colorectal cancer. Ann. Oncol. 30, 1096–1103 (2019).
Campbell, B. B. et al. Comprehensive analysis of hypermutation in human cancer. Cell 171, 1042–1056.e10 (2017).
Rousseau, B. et al. The spectrum of benefit from checkpoint blockade in hypermutated tumors. N. Engl. J. Med. 384, 1168–1170 (2021).
Rousseau, B. et al. PD-1 blockade in solid tumors with defects in polymerase epsilon. Cancer Discov. 12, 1435–1448 (2022).
Chung, J. et al. DNA polymerase and mismatch repair exert distinct microsatellite instability signatures in normal and malignant human cells. Cancer Discov. 11, 1176–1191 (2021).
Westcott, P. M. K. et al. Mismatch repair deficiency is not sufficient to elicit tumor immunogenicity. Nat. Genet. 55, 1686–1695 (2023).
Lander, E. S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Ellegren, H. Microsatellites: simple sequences with complex evolution. Nat. Rev. Genet. 5, 435–445 (2004).
Kelkar, Y. D. et al. What is a microsatellite: a computational and experimental definition based upon repeat mutational behavior at A/T and GT/AC repeats. Genome Biol. Evol. 2, 620–635 (2010).
Viguera, E., Canceill, D. & Ehrlich, S. D. Replication slippage involves DNA polymerase pausing and dissociation. EMBO J. 20, 2587–2595 (2001).
Hile, S. E. & Eckert, K. A. Positive correlation between DNA polymerase alpha-primase pausing and mutagenesis within polypyrimidine/polypurine microsatellite sequences. J. Mol. Biol. 335, 745–759 (2004).
Brinkmann, B., Klintschar, M., Neuhuber, F., Hühne, J. & Rolf, B. Mutation rate in human microsatellites: influence of the structure and length of the tandem repeat. Am. J. Hum. Genet. 62, 1408–1415 (1998).
Brohede, J., Primmer, C. R., Møller, A. & Ellegren, H. Heterogeneity in the rate and pattern of germline mutation at individual microsatellite loci. Nucleic Acids Res. 30, 1997–2003 (2002).
Chakraborty, R., Kimmel, M., Stivers, D. N., Davison, L. J. & Deka, R. Relative mutation rates at di-, tri-, and tetranucleotide microsatellite loci. Proc. Natl Acad. Sci. USA 94, 1041–1046 (1997).
Bachtrog, D., Agis, M., Imhof, M. & Schlötterer, C. Microsatellite variability differs between dinucleotide repeat motifs-evidence from Drosophila melanogaster. Mol. Biol. Evol. 17, 1277–1285 (2000).
Strand, M., Prolla, T. A., Liskay, R. M. & Petes, T. D. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature 365, 274–276 (1993).
Mizutani, T. et al. Recapitulating the adenoma-carcinoma sequence by selection of four spontaneous oncogenic mutations in mismatch-repair-deficient human colon organoids. Nat. Cancer 5, 1852–1867 (2024).
Markowitz, S. et al. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science 268, 1336–1338 (1995).
Myeroff, L. L. et al. A transforming growth factor beta receptor type II gene mutation common in colon and gastric but rare in endometrial cancers with microsatellite instability. Cancer Res. 55, 5545–5547 (1995).
Wang, J. et al. Demonstration that mutation of the type II transforming growth factor beta receptor inactivates its tumor suppressor activity in replication error-positive colon carcinoma cells. J. Biol. Chem. 270, 22044–22049 (1995).
Gurin, C. C., Federici, M. G., Kang, L. & Boyd, J. Causes and consequences of microsatellite instability in endometrial carcinoma. Cancer Res. 59, 462–466 (1999).
Giannakis, M. et al. RNF43 is frequently mutated in colorectal and endometrial cancers. Nat. Genet. 46, 1264–1266 (2014).
Perucho, M. Microsatellite instability: the mutator that mutates the other mutator. Nat. Med. 2, 630–631 (1996).
Kayhanian, H. et al. Homopolymer switches mediate adaptive mutability in mismatch repair-deficient colorectal cancer. Nat. Genet. 56, 1420–1433 (2024).
Cunningham, J. M. et al. Hypermethylation of the hMLH1 promoter in colon cancer with microsatellite instability. Cancer Res. 58, 3455–3460 (1998).
Herman, J. G. et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc. Natl Acad. Sci. USA 95, 6870–6875 (1998).
Fang, M., Ou, J., Hutchinson, L. & Green, M. R. The BRAF oncoprotein functions through the transcriptional repressor MAFG to mediate the CpG island methylator phenotype. Mol. Cell 55, 904–915 (2014).
Metcalf, A. M. & Spurdle, A. B. Endometrial tumour BRAF mutations and MLH1 promoter methylation as predictors of germline mismatch repair gene mutation status: a literature review. Fam. Cancer 13, 1–12 (2014).
Drews, R. M. et al. A pan-cancer compendium of chromosomal instability. Nature 606, 976–983 (2022).
Taylor, A. M. et al. Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell 33, 676–689.e3 (2018).
Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513, 202–209 (2014).
Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012).
Cancer Genome Atlas Research Network, Kandoth, C. et al. Integrated genomic characterization of endometrial carcinoma. Nature 497, 67–73 (2013).
Trautmann, K. et al. Chromosomal instability in microsatellite-unstable and stable colon cancer. Clin. Cancer Res. 12, 6379–6385 (2006).
Shlien, A. et al. Combined hereditary and somatic mutations of replication error repair genes result in rapid onset of ultra-hypermutated cancers. Nat. Genet. 47, 257–262 (2015).
Cocco, E. et al. Colorectal carcinomas containing hypermethylated MLH1 promoter and wild-type BRAF/KRAS are enriched for targetable kinase fusions. Cancer Res. 79, 1047–1053 (2019).
Spies, M. & Fishel, R. Mismatch repair during homologous and homeologous recombination. Cold Spring Harb. Perspect. Biol. 7, a022657 (2015).
Li, L. S. et al. Chromosomal imbalances in the colorectal carcinomas with microsatellite instability. Am. J. Pathol. 163, 1429–1436 (2003).
Spurr, L. F., Weichselbaum, R. R. & Pitroda, S. P. Tumor aneuploidy predicts survival following immunotherapy across multiple cancers. Nat. Genet. 54, 1782–1785 (2022).
Roudko, V. et al. Shared immunogenic poly-epitope frameshift mutations in microsatellite unstable tumors. Cell 183, 1634–1649.e17 (2020).
Ballhausen, A. et al. The shared frameshift mutation landscape of microsatellite-unstable cancers suggests immunoediting during tumor evolution. Nat. Commun. 11, 4740 (2020).
Saeterdal, I. et al. Frameshift-mutation-derived peptides as tumor-specific antigens in inherited and spontaneous colorectal cancer. Proc. Natl Acad. Sci. USA 98, 13255–13260 (2001).
Schwitalle, Y. et al. Immune response against frameshift-induced neopeptides in HNPCC patients and healthy HNPCC mutation carriers. Gastroenterology 134, 988–997 (2008).
Kang, Y. J. et al. A scoping review and meta-analysis on the prevalence of pan-tumour biomarkers (dMMR, MSI, high TMB) in different solid tumours. Sci. Rep. 12, 20495 (2022).
Caris Life Sciences. Caris molecular database. Caris Life Sciences https://www.carislifesciences.com/ (2024).
Bonneville, R. et al. Landscape of microsatellite instability across 39 cancer types. JCO Precis. Oncol. 2017, PO.17.00073 (2017).
Latham, A. et al. Microsatellite instability is associated with the presence of Lynch syndrome pan-cancer. J. Clin. Oncol. 37, 286–295 (2019).
Papke, D. J. et al. Prevalence of mismatch-repair deficiency in rectal adenocarcinomas. N. Engl. J. Med. 387, 1714–1716 (2022).
Gordhandas, S. et al. Comprehensive analysis of germline drivers in endometrial cancer. J. Natl Cancer Inst. 115, 560–569 (2023).
Ryan, Na. J. et al. The proportion of endometrial cancers associated with Lynch syndrome: a systematic review of the literature and meta-analysis. Genet. Med. 21, 2167–2180 (2019).
Post, C. C. B. et al. Prevalence and prognosis of Lynch syndrome and sporadic mismatch repair deficiency in endometrial cancer. J. Natl Cancer Inst. 113, 1212–1220 (2021).
Koopman, M. et al. Deficient mismatch repair system in patients with sporadic advanced colorectal cancer. Br. J. Cancer 100, 266–273 (2009).
Gutierrez, C., Ogino, S., Meyerhardt, J. A. & Iorgulescu, J. B. The prevalence and prognosis of microsatellite instability-high/mismatch repair-deficient colorectal adenocarcinomas in the United States. JCO Precis. Oncol. 7, e2200179 (2023).
Germano, G., Amirouchene-Angelozzi, N., Rospo, G. & Bardelli, A. The clinical impact of the genomic landscape of mismatch repair-deficient cancers. Cancer Discov. 8, 1518–1528 (2018).
Cristescu, R. et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat. Med. 21, 449–456 (2015).
Popat, S., Hubner, R. & Houlston, R. S. Systematic review of microsatellite instability and colorectal cancer prognosis. J. Clin. Oncol. 23, 609–618 (2005).
Goldstein, J. et al. Multicenter retrospective analysis of metastatic colorectal cancer (CRC) with high-level microsatellite instability (MSI-H). Ann. Oncol. 25, 1032–1038 (2014).
Merok, M. A. et al. Microsatellite instability has a positive prognostic impact on stage II colorectal cancer after complete resection: results from a large, consecutive Norwegian series. Ann. Oncol. 24, 1274–1282 (2013).
Klingbiel, D. et al. Prognosis of stage II and III colon cancer treated with adjuvant 5-fluorouracil or FOLFIRI in relation to microsatellite status: results of the PETACC-3 trial. Ann. Oncol. 26, 126–132 (2015).
Cohen, R. et al. Microsatellite instability in patients with stage III colon cancer receiving fluoropyrimidine with or without oxaliplatin: an ACCENT pooled analysis of 12 adjuvant trials. J. Clin. Oncol. 39, 642–651 (2021).
Sargent, D. J. et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J. Clin. Oncol. 28, 3219–3226 (2010).
Uhlig, J. et al. Microsatellite instability and KRAS mutation in stage IV colorectal cancer: prevalence, geographic discrepancies, and outcomes from the national cancer database. J. Natl Compr. Cancer Netw. 19, 307–318 (2021).
Polom, K. et al. Meta-analysis of microsatellite instability in relation to clinicopathological characteristics and overall survival in gastric cancer. Br. J. Surg. 105, 159–167 (2018).
Anglesio, M. S. et al. Cancer-associated mutations in endometriosis without cancer. N. Engl. J. Med. 376, 1835–1848 (2017).
Leskela, S. et al. Mismatch repair deficiency in ovarian carcinoma: frequency, causes, and consequences. Am. J. Surg. Pathol. 44, 649–656 (2020).
Sinicrope, F. A. et al. Prognostic impact of deficient DNA mismatch repair in patients with stage III colon cancer from a randomized trial of FOLFOX-based adjuvant chemotherapy. J. Clin. Oncol. 31, 3664–3672 (2013).
Cercek, A. et al. Mismatch repair-deficient rectal cancer and resistance to neoadjuvant chemotherapy. Clin. Cancer Res. 26, 3271–3279 (2020).
Jover, R. et al. The efficacy of adjuvant chemotherapy with 5-fluorouracil in colorectal cancer depends on the mismatch repair status. Eur. J. Cancer Oxf. Engl. 45, 365–373 (2009).
Tajima, A., Hess, M. T., Cabrera, B. L., Kolodner, R. D. & Carethers, J. M. The mismatch repair complex hMutS alpha recognizes 5-fluorouracil-modified DNA: implications for chemosensitivity and resistance. Gastroenterology 127, 1678–1684 (2004).
Dolcetti, R. et al. High prevalence of activated intraepithelial cytotoxic T lymphocytes and increased neoplastic cell apoptosis in colorectal carcinomas with microsatellite instability. Am. J. Pathol. 154, 1805–1813 (1999).
Phillips, S. M. et al. Tumour-infiltrating lymphocytes in colorectal cancer with microsatellite instability are activated and cytotoxic. Br. J. Surg. 91, 469–475 (2004).
Kim, H., Jen, J., Vogelstein, B. & Hamilton, S. R. Clinical and pathological characteristics of sporadic colorectal carcinomas with DNA replication errors in microsatellite sequences. Am. J. Pathol. 145, 148–156 (1994).
Alexander, J. et al. Histopathological identification of colon cancer with microsatellite instability. Am. J. Pathol. 158, 527–535 (2001).
Howitt, B. E. et al. Association of polymerase e-mutated and microsatellite-instable endometrial cancers with neoantigen load, number of tumor-infiltrating lymphocytes, and expression of PD-1 and PD-L1. JAMA Oncol. 1, 1319–1323 (2015).
Shin, S. J. et al. Mismatch repair status of gastric cancer and its association with the local and systemic immune response. Oncol 24, e835–e844 (2019).
Tougeron, D. et al. Tumor-infiltrating lymphocytes in colorectal cancers with microsatellite instability are correlated with the number and spectrum of frameshift mutations. Mod. Pathol. 22, 1186–1195 (2009).
Maby, P. et al. Correlation between density of CD8+ T-cell infiltrate in microsatellite unstable colorectal cancers and frameshift mutations: a rationale for personalized immunotherapy. Cancer Res. 75, 3446–3455 (2015).
Mlecnik, B. et al. Integrative analyses of colorectal cancer show immunoscore is a stronger predictor of patient survival than microsatellite instability. Immunity 44, 698–711 (2016).
Espenschied, C. R. et al. Multigene panel testing provides a new perspective on Lynch syndrome. J. Clin. Oncol. 35, 2568–2575 (2017).
Lynch, H. T. et al. Phenotypic and genotypic heterogeneity of Lynch syndrome: a complex diagnostic challenge. Fam. Cancer 17, 403–414 (2018).
Ligtenberg, M. J. L. et al. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3′ exons of TACSTD1. Nat. Genet. 41, 112–117 (2009).
Dámaso, E. et al. Primary constitutional MLH1 epimutations: a focal epigenetic event. Br. J. Cancer 119, 978–987 (2018).
Win, A. K. et al. Prevalence and penetrance of major genes and polygenes for colorectal cancer. Cancer Epidemiol. Biomark. Prev. 26, 404–412 (2017).
Thompson, B. A. et al. Application of a 5-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants in the InSiGHT locus-specific database. Nat. Genet. 46, 107–115 (2014).
de la Chapelle, A. Genetic predisposition to colorectal cancer. Nat. Rev. Cancer 4, 769–780 (2004).
Fishel, R. & Kolodner, R. D. Identification of mismatch repair genes and their role in the development of cancer. Curr. Opin. Genet. Dev. 5, 382–395 (1995).
Hemminki, A. et al. Loss of the wild type MLH1 gene is a feature of hereditary nonpolyposis colorectal cancer. Nat. Genet. 8, 405–410 (1994).
Liu, B. et al. Mismatch repair gene defects in sporadic colorectal cancers with microsatellite instability. Nat. Genet. 9, 48–55 (1995).
Kane, M. F. et al. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 57, 808–811 (1997).
Pande, M. et al. Cancer spectrum in DNA mismatch repair gene mutation carriers: results from a hospital based Lynch syndrome registry. Fam. Cancer 11, 441–447 (2012).
Dominguez-Valentin, M. et al. Cancer risks by gene, age, and gender in 6350 carriers of pathogenic mismatch repair variants: findings from the prospective Lynch syndrome database. Genet. Med. 22, 15–25 (2020).
Ponti, G. & Ponz de Leon, M. Muir–Torre syndrome. Lancet Oncol. 6, 980–987 (2005).
Kempers, M. J. E. et al. Risk of colorectal and endometrial cancers in EPCAM deletion-positive Lynch syndrome: a cohort study. Lancet Oncol. 12, 49–55 (2011).
Baglietto, L. et al. Risks of Lynch syndrome cancers for MSH6 mutation carriers. J. Natl Cancer Inst. 102, 193–201 (2010).
Ten Broeke, S. W. et al. Cancer risks for PMS2-associated Lynch syndrome. J. Clin. Oncol. 36, 2961–2968 (2018).
Salem, M. E. et al. Relationship between MLH1, PMS2, MSH2 and MSH6 gene-specific alterations and tumor mutational burden in 1057 microsatellite instability-high solid tumors. Int. J. Cancer 147, 2948–2956 (2020).
Sekine, S. et al. Mismatch repair deficiency commonly precedes adenoma formation in Lynch syndrome-associated colorectal tumorigenesis. Mod. Pathol. 30, 1144–1151 (2017).
De Jong, A. E. et al. The role of mismatch repair gene defects in the development of adenomas in patients with HNPCC. Gastroenterology 126, 42–48 (2004).
Ahadova, A. et al. Three molecular pathways model colorectal carcinogenesis in Lynch syndrome. Int. J. Cancer 143, 139–150 (2018).
Yurgelun, M. B. et al. Microsatellite instability and DNA mismatch repair protein deficiency in Lynch syndrome colorectal polyps. Cancer Prev. Res. 5, 574–582 (2012).
Engel, C. et al. Associations of pathogenic variants in MLH1, MSH2, and MSH6 with risk of colorectal adenomas and tumors and with somatic mutations in patients with Lynch syndrome. Gastroenterology 158, 1326–1333 (2020).
Ahadova, A., von Knebel Doeberitz, M., Bläker, H. & Kloor, M. CTNNB1-mutant colorectal carcinomas with immediate invasive growth: a model of interval cancers in Lynch syndrome. Fam. Cancer 15, 579–586 (2016).
Ahadova, A. et al. A ‘Two-in-One Hit’ model of shortcut carcinogenesis in MLH1 Lynch syndrome carriers. Gastroenterology 165, 267–270.e4 (2023).
Pussila, M. et al. Mitotic abnormalities precede microsatellite instability in lynch syndrome-associated colorectal tumourigenesis. eBioMedicine 103, 105111 (2024).
Dominguez-Valentin, M. et al. No difference in penetrance between truncating and missense/aberrant splicing pathogenic variants in MLH1 and MSH2: a prospective Lynch syndrome database study. J. Clin. Med. 10, 2856 (2021).
Mecklin, J. P. et al. Development of colorectal tumors in colonoscopic surveillance in Lynch syndrome. Gastroenterology 133, 1093–1098 (2007).
Kloor, M. et al. Prevalence of mismatch repair-deficient crypt foci in Lynch syndrome: a pathological study. Lancet Oncol. 13, 598–606 (2012).
Møller, P. et al. Incidences of colorectal adenomas and cancers under colonoscopy surveillance suggest an accelerated “Big Bang” pathway to CRC in three of the four Lynch syndromes. Hered. Cancer Clin. Pract. 22, 6 (2024).
Staffa, L. et al. Mismatch repair-deficient crypt foci in Lynch syndrome-molecular alterations and association with clinical parameters. PLoS ONE 10, e0121980 (2015).
Brand, R. E. et al. Detection of DNA mismatch repair deficient crypts in random colonoscopic biopsies identifies Lynch syndrome patients. Fam. Cancer 19, 169–175 (2020).
Pai, R. K. et al. DNA mismatch repair protein deficient non-neoplastic colonic crypts: a novel indicator of Lynch syndrome. Mod. Pathol. 31, 1608–1618 (2018).
Ricciardone, M. D. et al. Human MLH1 deficiency predisposes to hematological malignancy and neurofibromatosis type 1. Cancer Res. 59, 290–293 (1999).
Wang, Q. et al. Neurofibromatosis and early onset of cancers in hMLH1-deficient children. Cancer Res. 59, 294–297 (1999).
Wimmer, K. et al. Diagnostic criteria for constitutional mismatch repair deficiency syndrome: suggestions of the European consortium ‘care for CMMRD’ (C4CMMRD). J. Med. Genet. 51, 355–365 (2014).
Baris, H. N. et al. Constitutional mismatch repair deficiency in Israel: high proportion of founder mutations in MMR genes and consanguinity. Pediatr. Blood Cancer 63, 418–427 (2016).
Lavoine, N. et al. Constitutional mismatch repair deficiency syndrome: clinical description in a French cohort. J. Med. Genet. 52, 770–778 (2015).
Durno, C. et al. Survival benefit for individuals with constitutional mismatch repair deficiency undergoing surveillance. J. Clin. Oncol. 39, 2779–2790 (2021).
Wimmer, K. & Kratz, C. P. Constitutional mismatch repair-deficiency syndrome. Haematologica 95, 699–701 (2010).
Aronson, M. et al. Diagnostic criteria for constitutional mismatch repair deficiency (CMMRD): recommendations from the international consensus working group. J. Med. Genet. 59, 318–327 (2022).
Bouffet, E. et al. Immune checkpoint inhibition for hypermutant glioblastoma multiforme resulting from germline biallelic mismatch repair deficiency. J. Clin. Oncol. 34, 2206–2211 (2016).
Gallon, R. et al. Constitutional microsatellite instability, genotype, and phenotype correlations in constitutional mismatch repair deficiency. Gastroenterology 164, 579–592.e8 (2023).
Pearlman, R. et al. Clinical characteristics of patients with colorectal cancer with double somatic mismatch repair mutations compared with Lynch syndrome. J. Med. Genet. 56, 462–470 (2019).
Cohen, R. et al. Clinical and molecular characterisation of hereditary and sporadic metastatic colorectal cancers harbouring microsatellite instability/DNA mismatch repair deficiency. Eur. J. Cancer 86, 266–274 (2017).
Parsons, M. T., Buchanan, D. D., Thompson, B., Young, J. P. & Spurdle, A. B. Correlation of tumour BRAF mutations and MLH1 methylation with germline mismatch repair (MMR) gene mutation status: a literature review assessing utility of tumour features for MMR variant classification. J. Med. Genet. 49, 151–157 (2012).
Weisenberger, D. J. et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat. Genet. 38, 787–793 (2006).
Diaz, L. A. et al. Pembrolizumab versus chemotherapy for microsatellite instability-high or mismatch repair-deficient metastatic colorectal cancer (KEYNOTE-177): final analysis of a randomised, open-label, phase 3 study. Lancet Oncol. 23, 659–670 (2022).
Ambrosini, M. et al. BRAF + EGFR +/− MEK inhibitors after immune checkpoint inhibitors in BRAF V600E mutated and deficient mismatch repair or microsatellite instability high metastatic colorectal cancer. Eur. J. Cancer 210, 114290 (2024).
Luchini, C. et al. ESMO recommendations on microsatellite instability testing for immunotherapy in cancer, and its relationship with PD-1/PD-L1 expression and tumour mutational burden: a systematic review-based approach. Ann. Oncol. 30, 1232–1243 (2019).
Wang, C., Zhang, L., Vakiani, E. & Shia, J. Detecting mismatch repair deficiency in solid neoplasms: immunohistochemistry, microsatellite instability, or both? Mod. Pathol. 35, 1515–1528 (2022).
Graham, R. P. et al. Heterogenous MSH6 loss is a result of microsatellite instability within MSH6 and occurs in sporadic and hereditary colorectal and endometrial carcinomas. Am. J. Surg. Pathol. 39, 1370–1376 (2015).
Bartley, A. N., Luthra, R., Saraiya, D. S., Urbauer, D. L. & Broaddus, R. R. Identification of cancer patients with Lynch syndrome: clinically significant discordances and problems in tissue-based mismatch repair testing. Cancer Prev. Res. 5, 320–327 (2012).
Hampel, H. et al. Feasibility of screening for Lynch syndrome among patients with colorectal cancer. J. Clin. Oncol. 26, 5783–5788 (2008).
Hechtman, J. F. et al. Retained mismatch repair protein expression occurs in approximately 6% of microsatellite instability-high cancers and is associated with missense mutations in mismatch repair genes. Mod. Pathol. 33, 871–879 (2020).
Kato, A. et al. Isolated loss of PMS2 immunohistochemical expression is frequently caused by heterogenous MLH1 promoter hypermethylation in Lynch syndrome screening for endometrial cancer patients. Am. J. Surg. Pathol. 40, 770–776 (2016).
Goel, A., Nagasaka, T., Hamelin, R. & Boland, C. R. An optimized pentaplex PCR for detecting DNA mismatch repair-deficient colorectal cancers. PLoS ONE 5, e9393 (2010).
Suraweera, N. et al. Evaluation of tumor microsatellite instability using five quasimonomorphic mononucleotide repeats and pentaplex PCR. Gastroenterology 123, 1804–1811 (2002).
Wang, Y., Shi, C., Eisenberg, R. & Vnencak-Jones, C. L. Differences in microsatellite instability profiles between endometrioid and colorectal cancers: a potential cause for false-negative results? J. Mol. Diagn. JMD 19, 57–64 (2017).
Kuismanen, S. A. et al. Endometrial and colorectal tumors from patients with hereditary nonpolyposis colon cancer display different patterns of microsatellite instability. Am. J. Pathol. 160, 1953–1958 (2002).
Middha, S. et al. Reliable pan-cancer microsatellite instability assessment by using targeted next-generation sequencing data. JCO Precis. Oncol. 2017, PO.17.00084 (2017).
Rios-Doria, E. et al. Integration of clinical sequencing and immunohistochemistry for the molecular classification of endometrial carcinoma. Gynecol. Oncol. 174, 262–272 (2023).
Dedeurwaerdere, F. et al. Comparison of microsatellite instability detection by immunohistochemistry and molecular techniques in colorectal and endometrial cancer. Sci. Rep. 11, 12880 (2021).
Willis, J. et al. Validation of microsatellite instability detection using a comprehensive plasma-based genotyping panel. Clin. Cancer Res. 25, 7035–7045 (2019).
Woodhouse, R. et al. Clinical and analytical validation of FoundationOne liquid CDx, a novel 324-gene cfDNA-based comprehensive genomic profiling assay for cancers of solid tumor origin. PLoS ONE 15, e0237802 (2020).
Silveira, A. B. et al. High-accuracy determination of microsatellite instability compatible with liquid biopsies. Clin. Chem. 66, 606–613 (2020).
Ali-Fehmi, R. et al. Analysis of concordance between next-generation sequencing assessment of microsatellite instability and immunohistochemistry-mismatch repair from solid tumors. JCO Precis. Oncol. 8, e2300648 (2024).
Bartley, A. N. et al. Mismatch repair and microsatellite instability testing for immune checkpoint inhibitor therapy: guideline from the college of American pathologists in collaboration with the association for molecular pathology and fight colorectal cancer. Arch. Pathol. Lab. Med. 146, 1194–1210 (2022).
Niu, B. et al. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinforma. Oxf. Engl. 30, 1015–1016 (2014).
Koh, W. J. et al. Uterine neoplasms, version 1.2018, NCCN clinical practice guidelines in oncology. J. Natl Compr. Cancer Netw. 16, 170–199 (2018).
Ajani, J. A. et al. Gastric cancer, version 2.2022, NCCN clinical practice guidelines in oncology. J. Natl Compr. Cancer Netw. 20, 167–192 (2022).
Benson, A. B. et al. Colon cancer, version 1.2017, NCCN clinical practice guidelines in oncology. J. Natl Compr. Cancer Netw. 15, 370–398 (2017).
Heald, B. et al. Implementation of universal microsatellite instability and immunohistochemistry screening for diagnosing lynch syndrome in a large academic medical center. J. Clin. Oncol. 31, 1336–1340 (2013).
Ladabaum, U. et al. Strategies to identify the Lynch syndrome among patients with colorectal cancer: a cost-effectiveness analysis. Ann. Intern. Med. 155, 69–79 (2011).
Dhimal T. et al. Mismatch repair and microsatellite testing for individuals with colorectal cancer. JAMA Oncol. https://doi.org/10.1001/jamaoncol.2024.4342 (2024).
Hampel, H. et al. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer). N. Engl. J. Med. 352, 1851–1860 (2005).
Beamer, L. C. et al. Reflex immunohistochemistry and microsatellite instability testing of colorectal tumors for Lynch syndrome among US cancer programs and follow-up of abnormal results. J. Clin. Oncol. 30, 1058–1063 (2012).
Giardiello, F. M. et al. Guidelines on genetic evaluation and management of Lynch syndrome: a consensus statement by the US multi-society task force on colorectal cancer. Gastroenterology 147, 502–526 (2014).
Carnevali, I. W. et al. MLH1 promoter methylation could be the second hit in lynch syndrome carcinogenesis. Genes 14, 2060 (2023).
Brahmer, J. R. et al. Phase I study of single-agent anti-programmed death-1 (MDX-1106) in refractory solid tumors: safety, clinical activity, pharmacodynamics, and immunologic correlates. J. Clin. Oncol. 28, 3167–3175 (2010).
Topalian, S. L. et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 366, 2443–2454 (2012).
Lipson, E. J. et al. Durable cancer regression off-treatment and effective reinduction therapy with an anti-PD-1 antibody. Clin. Cancer Res. 19, 462–468 (2013).
Overman, M. J. et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol. 18, 1182–1191 (2017).
André, T. et al. Nivolumab plus low-dose ipilimumab in previously treated patients with microsatellite instability-high/mismatch repair-deficient metastatic colorectal cancer: 4-year follow-up from CheckMate 142. Ann. Oncol. 33, 1052–1060 (2022).
Overman, M. J. et al. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair-deficient/microsatellite instability-high metastatic colorectal cancer. J. Clin. Oncol. 36, 773–779 (2018).
Le, D. T. et al. Phase II open-label study of pembrolizumab in treatment-refractory, microsatellite instability-high/mismatch repair-deficient metastatic colorectal cancer: KEYNOTE-164. J. Clin. Oncol. 38, 11–19 (2020).
O’Malley, D. M. et al. Pembrolizumab in patients with microsatellite instability-high advanced endometrial cancer: results from the KEYNOTE-158 study. J. Clin. Oncol. 40, 752–761 (2022).
Oaknin, A. et al. Clinical activity and safety of the anti-programmed death 1 monoclonal antibody dostarlimab for patients with recurrent or advanced mismatch repair-deficient endometrial cancer: a nonrandomized phase 1 clinical trial. JAMA Oncol. 6, 1766–1772 (2020).
Friedman, C. F. et al. Nivolumab for mismatch-repair-deficient or hypermutated gynecologic cancers: a phase 2 trial with biomarker analyses. Nat. Med. 30, 1330–1338 (2024).
Eskander, R. N. et al. Pembrolizumab plus chemotherapy in advanced endometrial cancer. N. Engl. J. Med. 388, 2159–2170 (2023).
Eskander, R. N. et al. Pembrolizumab plus chemotherapy in advanced or recurrent endometrial cancer: overall survival and exploratory analyses of the NRG GY018 phase 3 randomized trial. Nat. Med. 31, 1539–1546 (2025).
Powell, M. A. et al. Overall survival in patients with endometrial cancer treated with dostarlimab plus carboplatin-paclitaxel in the randomized ENGOT-EN6/GOG-3031/RUBY trial. Ann. Oncol. 35, 728–738 (2024).
Westin, S. N. et al. Durvalumab plus carboplatin/paclitaxel followed by maintenance durvalumab with or without olaparib as first-line treatment for advanced endometrial cancer: the phase III DUO-E trial. J. Clin. Oncol. 42, 283–299 (2024).
Subbiah, V., Gouda, M. A., Ryll, B., Burris, H. A. & Kurzrock, R. The evolving landscape of tissue-agnostic therapies in precision oncology. CA Cancer J. Clin. 74, 433–452 (2024).
Lemery, S., Keegan, P. & Pazdur, R. First FDA approval agnostic of cancer site — when a biomarker defines the indication. N. Engl. J. Med. 377, 1409–1412 (2017).
Das, A. et al. Genomic predictors of response to PD-1 inhibition in children with germline DNA replication repair deficiency. Nat. Med. 28, 125–135 (2022).
Margalit, O. et al. Duration of immunotherapy in dMMR/MSI-H metastatic colorectal cancer patients. Eur. J. Cancer 212, 114336 (2024).
Chalabi, M. et al. Neoadjuvant immunotherapy leads to pathological responses in MMR-proficient and MMR-deficient early-stage colon cancers. Nat. Med. 26, 566–576 (2020).
Chalabi, M. et al. Neoadjuvant immunotherapy in locally advanced mismatch repair-deficient colon cancer. N. Engl. J. Med. 390, 1949–1958 (2024).
de Gooyer, P. G. M. et al. Neoadjuvant nivolumab and relatlimab in locally advanced MMR-deficient colon cancer: a phase 2 trial. Nat. Med. 30, 3284–3290 (2024).
André, T. et al. Neoadjuvant nivolumab plus ipilimumab and adjuvant nivolumab in localized deficient mismatch repair/microsatellite instability-high gastric or esophagogastric junction adenocarcinoma: the GERCOR NEONIPIGA phase II study. J. Clin. Oncol. 41, 255–265 (2023).
Ludford, K. et al. Neoadjuvant pembrolizumab in localized microsatellite instability high/deficient mismatch repair solid tumors. J. Clin. Oncol. 41, 2181–2190 (2023).
Cercek, A. et al. PD-1 blockade in mismatch repair-deficient, locally advanced rectal cancer. N. Engl. J. Med. 386, 2363–2376 (2022).
Cercek, A. et al. Nonoperative management of mismatch repair-deficient tumors. N. Engl. J. Med. 392, 2297–2308 (2025).
Li, Y. et al. Efficacy and safety of neoadjuvant subcutaneous envafolimab in dMMR/MSI-H locally advanced colon cancer. Target. Oncol. 19, 601–610 (2024).
Li, J. et al. Biomarkers of pathologic complete response to neoadjuvant immunotherapy in mismatch repair-deficient colorectal cancer. Clin. Cancer Res. 30, 368–378 (2024).
Xie, Y. et al. Prevalent pseudoprogression and pseudoresidue in patients with rectal cancer treated with neoadjuvant immune checkpoint inhibitors. J. Natl Compr. Cancer Netw. 21, 133–142.e3 (2023).
Zhang, X. et al. Efficacy and safety of neoadjuvant monoimmunotherapy with PD-1 Inhibitor for dMMR/MSI-H locally advanced colorectal cancer: a single-center real-world study. Front. Immunol. 13, 913483 (2022).
Pan, T. et al. Neoadjuvant immunotherapy with ipilimumab plus nivolumab in mismatch repair deficient/microsatellite instability-high colorectal cancer: a preliminary report of case series. Clin. Colorectal Cancer 23, 104–110 (2024).
Yu, J. H. et al. Neoadjuvant camrelizumab plus apatinib for locally advanced microsatellite instability-high or mismatch repair-deficient colorectal cancer (NEOCAP): a single-arm, open-label, phase 2 study. Lancet Oncol. 25, 843–852 (2024).
Kothari, A. et al. Pathological response following neoadjuvant immunotherapy in mismatch repair-deficient/microsatellite instability-high locally advanced, non-metastatic colorectal cancer. Br. J. Surg. 109, 489–492 (2022).
Raimondi, A. et al. Multicentre, multi-cohort, single-arm phase II trial of tremelimumab and durvalumab as neoadjuvant or definitive treatment of patients (pts) with microsatellite instability-high (MSI) resectable gastric or gastroesophageal junction adenocarcinoma (GAC/GEJAC): the INFINITY study. Ann. Oncol. 35, S172 (2024).
Pei, F. et al. Single-agent neoadjuvant immunotherapy with a PD-1 antibody in locally advanced mismatch repair-deficient or microsatellite instability-high colorectal cancer. Clin. Colorectal Cancer 22, 85–91 (2023).
Shiu, K. K. et al. NEOPRISM-CRC: neoadjuvant pembrolizumab stratified to tumour mutation burden for high risk stage 2 or stage 3 deficient-MMR/MSI-high colorectal cancer. J. Clin. Oncol. 42, LBA3504 (2024).
Chen, G. et al. Neoadjuvant PD-1 blockade with sintilimab in mismatch-repair deficient, locally advanced rectal cancer: an open-label, single-centre phase 2 study. Lancet Gastroenterol. Hepatol. 8, 422–431 (2023).
De La Fouchardiere, C. et al. IMHOTEP Phase II trial of neoadjuvant pembrolizumab in dMMR/MSI tumors: results of the colorectal cancer cohort. Ann. Oncol. 35, S429 (2024).
De La Fouchardiere, C. et al. IMHOTEP phase II trial of neoadjuvant pembrolizumab in dMMR/MSI localized cancers: results of the digestive non-colorectal cancer cohorts. Ann. Oncol. 35, S899–S900 (2024).
Eerkens, A. L. et al. Neoadjuvant immune checkpoint blockade in women with mismatch repair deficient endometrial cancer: a phase I study. Nat. Commun. 15, 7695 (2024).
Xu, R. H. et al. Neoadjuvant treatment of IBI310 (anti-CTLA-4 antibody) plus sintilimab (anti-PD-1 antibody) in patients with microsatellite instability-high/mismatch repair-deficient colorectal cancer: results from a randomized, open-labeled, phase Ib study. J. Clin. Oncol. 42, 3505 (2024).
Avallone, A. et al. Neoadjuvant nivolumab in early stage colorectal cancer. Ann. Oncol. 31, S449 (2020).
Rousseau, B., White, J. R., Cercek, A. & Diaz, L. A. The duration of immunotherapy for mismatch repair-deficient cancers. N. Engl. J. Med. 392, 824–826 (2025).
Cercek, A. et al. Durable complete responses to PD-1 blockade alone in mismatch repair deficient locally advanced rectal cancer. J. Clin. Oncol. 42, LBA3512 (2024).
Van Gorp T. et al. ENGOT-en11/GOG-3053/KEYNOTE-B21: a randomised, double-blind, phase III study of pembrolizumab or placebo plus adjuvant chemotherapy with or without radiotherapy in patients with newly diagnosed, high-risk endometrial cancer. Ann. Oncol. https://doi.org/10.1016/j.annonc.2024.08.2242 (2024).
Sinicrope, F. et al. Randomized trial of standard chemotherapy alone or combined with atezolizumab as adjuvant therapy for patients with stage III deficient DNA mismatch repair (dMMR) colon cancer (Alliance A021502; ATOMIC). J. Clin. Oncol. 43, LBA1 (2025).
Janjigian, Y. Y. et al. Circulating tumor DNA status to direct adjuvant immunotherapy for mismatch repair deficient tumors [abstract]. Cancer Res. 85, CT002 (2025).
Manning-Geist, B. L. et al. Microsatellite instability-high endometrial cancers with MLH1 promoter hypermethylation have distinct molecular and clinical profiles. Clin. Cancer Res. 28, 4302–4311 (2022).
Ramchander, N. C. et al. Distinct immunological landscapes characterize inherited and sporadic mismatch repair deficient endometrial cancer. Front. Immunol. 10, 3023 (2019).
Bellone, S. et al. A phase 2 evaluation of pembrolizumab for recurrent Lynch-like versus sporadic endometrial cancers with microsatellite instability. Cancer 128, 1206–1218 (2022).
Eskander, R. N. et al. LBA43 Updated response data and analysis of progression free survival by mechanism of mismatch repair loss in endometrial cancer (EC) patients (pts) treated with pembrolizumab plus carboplatin/paclitaxel (CP) as compared to CP plus placebo (PBO) in the NRG GY018 trial. J. Clin. Oncol. 34, S1284 (2024).
Mirza, M. R. et al. Post hoc analysis of progression-free survival (PFS) and overall survival (OS) by mechanism of mismatch repair (MMR) protein loss in patients with endometrial cancer (EC) treated with dostarlimab plus chemotherapy in the RUBY trial. J. Clin. Oncol. 42, 5606 (2024).
Liu, G. C. et al. The heterogeneity between Lynch-associated and sporadic MMR deficiency in colorectal cancers. J. Natl Cancer Inst. 110, 975–984 (2018).
Khushman, M. M. et al. Differential responses to immune checkpoint inhibitors are governed by diverse mismatch repair gene alterations. Clin. Cancer Res. 30, 1906–1915 (2024).
Ratovomanana, T. et al. Prediction of response to immune checkpoint blockade in patients with metastatic colorectal cancer with microsatellite instability. Ann. Oncol. 34, 703–713 (2023).
Hwang, H. S., Kim, D. & Choi, J. Distinct mutational profile and immune microenvironment in microsatellite-unstable and POLE-mutated tumors. J. Immunother. Cancer 9, e002797 (2021).
Sena, L. A. et al. Tumor frameshift mutation proportion predicts response to immunotherapy in mismatch repair-deficient prostate cancer. Oncologist 26, e270–e278 (2021).
von Loga, K. et al. Extreme intratumour heterogeneity and driver evolution in mismatch repair deficient gastro-oesophageal cancer. Nat. Commun. 11, 139 (2020).
Clendenning, M. et al. Somatic mutations of the coding microsatellites within the beta-2-microglobulin gene in mismatch repair-deficient colorectal cancers and adenomas. Fam. Cancer 17, 91–100 (2018).
Ozcan, M., Janikovits, J., von Knebel Doeberitz, M. & Kloor, M. Complex pattern of immune evasion in MSI colorectal cancer. Oncoimmunology 7, e1445453 (2018).
Germano, G. et al. CD4 T cell-dependent rejection of beta-2 microglobulin null mismatch repair-deficient tumors. Cancer Discov. 11, 1844–1859 (2021).
de Vries, N. L. et al. γδ T cells are effectors of immunotherapy in cancers with HLA class I defects. Nature 613, 743–750 (2023).
Middha, S. et al. Majority of B2M-mutant and -deficient colorectal carcinomas achieve clinical benefit from immune checkpoint inhibitor therapy and are microsatellite instability-high. JCO Precis. Oncol. 3, PO.18.00321, https://doi.org/10.1200/PO.18.00321 (2019).
Talhouk, A. et al. Molecular subtype not immune response drives outcomes in endometrial carcinoma. Clin. Cancer Res. 25, 2537–2548 (2019).
Narayanan, S. et al. Cytolytic activity score to assess anticancer immunity in colorectal cancer. Ann. Surg. Oncol. 25, 2323–2331 (2018).
Giannakis, M. et al. Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep. 17, 1206 (2016).
Chang, K. et al. Immune profiling of premalignant lesions in patients with Lynch syndrome. JAMA Oncol. 4, 1085–1092 (2018).
Liu, S. et al. Cellular localization of PD-L1 expression in mismatch-repair-deficient and proficient colorectal carcinomas. Mod. Pathol. 32, 110–121 (2019).
Andre, T. et al. Analysis of tumor PD-L1 expression and biomarkers in relation to clinical activity in patients (pts) with deficient DNA mismatch repair (dMMR)/hgih microsatellite instability (MSI-H) metastatic colorectal cancer (mCRC) treated with nivolumab (NIVO) + ipilimumab (IPI): CheckMate 142. Ann. Oncol. 28, V163 (2017).
Angell, H. K. et al. PD-L1 and immune infiltrates are differentially expressed in distinct subgroups of gastric cancer. Oncoimmunology 8, e1544442 (2019).
Pelka, K. et al. Spatially organized multicellular immune hubs in human colorectal cancer. Cell 184, 4734–4752.e20 (2021).
Oaknin, A. et al. Safety, efficacy, and biomarker analyses of dostarlimab in patients with endometrial cancer: interim results of the phase I GARNET study. Clin. Cancer Res. 29, 4564–4574 (2023).
Lee, L. H. et al. Patterns and prognostic relevance of PD-1 and PD-L1 expression in colorectal carcinoma. Mod. Pathol. 29, 1433–1442 (2016).
Rousseau, B., Cercek, A. & Diaz, L. A. From neoadjuvant to organ-sparing immunotherapy for colorectal cancer. Nat. Med. 30, 2407–2408 (2024).
Gallois, C. et al. Transcriptomic signatures of MSI-high metastatic colorectal cancer predict efficacy of immune checkpoint inhibitors. Clin. Cancer Res. 29, 3771–3778 (2023).
Tauriello, D. V. F. et al. TGFβ drives immune evasion in genetically reconstituted colon cancer metastasis. Nature 554, 538–543 (2018).
Lu, C. et al. DNA sensing in mismatch repair-deficient tumor cells is essential for anti-tumor immunity. Cancer Cell 39, 96–108.e6 (2021).
Decout, A., Katz, J. D., Venkatraman, S. & Ablasser, A. The cGAS–STING pathway as a therapeutic target in inflammatory diseases. Nat. Rev. Immunol. 21, 548–569 (2021).
Dvorkin, S., Cambier, S., Volkman, H. E. & Stetson, D. B. New frontiers in the cGAS–STING intracellular DNA-sensing pathway. Immunity 57, 718–730 (2024).
Guan, J. et al. MLH1 deficiency-triggered DNA hyperexcision by exonuclease 1 activates the cGAS–STING pathway. Cancer Cell 39, 109–121.e5 (2021).
Pietrantonio, F. et al. Nomogram to predict the outcomes of patients with microsatellite instability-high metastatic colorectal cancer receiving immune checkpoint inhibitors. J. Immunother. Cancer 9, e003370 (2021).
Maio, M. et al. Pembrolizumab in microsatellite instability high or mismatch repair deficient cancers: updated analysis from the phase II KEYNOTE-158 study. Ann. Oncol. 33, 929–938 (2022).
Alouani, E. et al. Efficacy of immunotherapy in mismatch repair-deficient advanced colorectal cancer in routine clinical practice. An AGEO study. ESMO Open. 8, 101574 (2023).
Belkouchi, Y. et al. Predicting immunotherapy outcomes in patients with MSI tumors using NLR and CT global tumor volume. Front. Oncol. 12, 982790 (2022).
Cohen, R. et al. Association of primary resistance to immune checkpoint inhibitors in metastatic colorectal cancer with misdiagnosis of microsatellite instability or mismatch repair deficiency status. JAMA Oncol. 5, 551–555 (2019).
Tian, J. et al. Combined PD-1, BRAF and MEK inhibition in BRAFV600E colorectal cancer: a phase 2 trial. Nat. Med. 29, 458–466 (2023).
Kopetz, S. et al. Encorafenib, binimetinib, and cetuximab in BRAF V600E-mutated colorectal cancer. N. Engl. J. Med. 381, 1632–1643 (2019).
Gray, M. D. et al. The Werner syndrome protein is a DNA helicase. Nat. Genet. 17, 100–103 (1997).
Orren, D. K., Theodore, S. & Machwe, A. The Werner syndrome helicase/exonuclease (WRN) disrupts and degrades D-loops in vitro. Biochemistry 41, 13483–13488 (2002).
Mohaghegh, P., Karow, J. K., Brosh, R. M., Bohr, V. A. & Hickson, I. D. The Bloom’s and Werner’s syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 29, 2843–2849 (2001).
van Wietmarschen, N. et al. Repeat expansions confer WRN dependence in microsatellite-unstable cancers. Nature 586, 292–298 (2020).
Lieb, S. et al. Werner syndrome helicase is a selective vulnerability of microsatellite instability-high tumor cells. eLife 8, e43333 (2019).
Kategaya, L., Perumal, S. K., Hager, J. H. & Belmont, L. D. Werner syndrome helicase is required for the survival of cancer cells with microsatellite instability. iScience 13, 488–497 (2019).
Chan, E. M. et al. WRN helicase is a synthetic lethal target in microsatellite unstable cancers. Nature 568, 551–556 (2019).
Picco, G. et al. Novel WRN helicase inhibitors selectively target microsatellite-unstable cancer cells. Cancer Discov. 14, 1457–1475 (2024).
Baltgalvis, K. A. et al. Chemoproteomic discovery of a covalent allosteric inhibitor of WRN helicase. Nature 629, 435–442 (2024).
Ferretti, S. et al. Discovery of WRN inhibitor HRO761 with synthetic lethality in MSI cancers. Nature 629, 443–449 (2024).
Rubenstein, J. H., Enns, R., Heidelbaugh, J. & Barkun, A. Clinical guidelines committee. american gastroenterological association institute guideline on the diagnosis and management of Lynch syndrome. Gastroenterology 149, 777–782 (2015).
Seppälä, T. T. et al. European guidelines from the EHTG and ESCP for Lynch syndrome: an updated third edition of the Mallorca guidelines based on gene and gender. Br. J. Surg. 108, 484–498 (2021).
Stjepanovic, N. et al. Hereditary gastrointestinal cancers: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up†. Ann. Oncol. 30, 1558–1571 (2019).
National Comprehensive Cancer Network. Genetic/familial high-risk assessment: colorectal, endometrial, and gastric (version 2.2024). J. Natl Compr. Cancer Netw. https://www.nccn.org/professionals/physician_gls/pdf/genetics_ceg.pdf (2024).
Schmeler, K. M. et al. Prophylactic surgery to reduce the risk of gynecologic cancers in the Lynch syndrome. N. Engl. J. Med. 354, 261–269 (2006).
Syngal, S. et al. ACG clinical guideline: genetic testing and management of hereditary gastrointestinal cancer syndromes. Am. J. Gastroenterol. 110, 223–262 (2015).
Burn, J. et al. Cancer prevention with aspirin in hereditary colorectal cancer (Lynch syndrome), 10-year follow-up and registry-based 20-year data in the CAPP2 study: a double-blind, randomised, placebo-controlled trial. Lancet Lond. Engl. 395, 1855–1863 (2020).
Kloor, M. et al. A frameshift peptide neoantigen-based vaccine for mismatch repair-deficient cancers: a phase I/IIa clinical trial. Clin. Cancer Res. 26, 4503–4510 (2020).
Leoni, G. et al. A genetic vaccine encoding shared cancer neoantigens to treat tumors with microsatellite instability. Cancer Res. 80, 3972–3982 (2020).
Overman, M. et al. Results of phase I–II bridging study for Nous-209, a neoantigen cancer immunotherapy, in combination with pembrolizumab as first line treatment in patients with advanced dMMR/MSI-h colorectal cancer. J. Clin. Oncol. 41, e14665 (2023).
Fakih, M. et al. First clinical and immunogenicity results including all subjects enrolled in a phase I study of Nous-209, an off-the-shelf immunotherapy, with pembrolizumab, for the treatment of tumors with a deficiency in mismatch repair/microsatellite instability (dMMR/MSI). J. Clin. Oncol. 40, 2515 (2022).
D’Alise, A. M. et al. Adenoviral-based vaccine promotes neoantigen-specific CD8+ T cell stemness and tumor rejection. Sci. Transl. Med. 14, eabo7604 (2022).
Harrold, E. C. et al. Neoplasia risk in patients with Lynch syndrome treated with immune checkpoint blockade. Nat. Med. 29, 2458–2463 (2023).
Feng, Y. et al. Spatially organized tumor-stroma boundary determines the efficacy of immunotherapy in colorectal cancer patients. Nat. Commun. 15, 10259 (2024).
Boland, C. R. & Lynch, H. T. The history of Lynch syndrome. Fam. Cancer 12, 145–157 (2013).
Classics in oncology. Heredity with reference to carcinoma as shown by the study of the cases examined in the pathological laboratory of the University of Michigan, 1895–1913. By Aldred Scott Warthin. 1913. CA Cancer J. Clin. 35, 348–359 (1985).
Warthin, A. S. The further study of a cancer family. J. Cancer Res. 9, 279–286 (1925).
Hauser, I. & Weller, C. A further report on the cancer family of Warthin. J. Cancer Res. 27, 434–449 (1936).
Douglas, J. A. et al. History and molecular genetics of Lynch syndrome in family G: a century later. JAMA 294, 2195–2202 (2005).
Lynch, H. T., Shaw, M. W., Magnuson, C. W., Larsen, A. L. & Krush, A. J. Hereditary factors in cancer. Study of two large midwestern kindreds. Arch. Intern. Med. 117, 206–212 (1966).
Lynch, H. T. & Krush, A. J. Heredity and adenocarcinoma of the colon. Gastroenterology 53, 517–527 (1967).
Lynch, H. T., Krush, A. J. & Larsen, A. L. Heredity and multiple primary malignant neoplasms: six cancer families. Am. J. Med. Sci. 254, 322–329 (1967).
Lynch, H. T. & Krush, A. J. Cancer family ‘G’ revisited: 1895–1970. Cancer 27, 1505–1511 (1971).
Boland, C. Cancer family syndrome. Am. J. Dig. Dis. 23, S25–S27 (1978).
Boland, C. R. Evolution of the nomenclature for the hereditary colorectal cancer syndromes. Fam. Cancer 4, 211–218 (2005).
Kalady, M. F., Kravochuck, S. E., Heald, B., Burke, C. A. & Church, J. M. Defining the adenoma burden in lynch syndrome. Dis. Colon. Rectum 58, 388–392 (2015).
Vasen, H. F., Mecklin, J. P., Khan, P. M. & Lynch, H. T. The international collaborative group on hereditary non-polyposis colorectal cancer (ICG-HNPCC). Dis. Colon. Rectum 34, 424–425 (1991).
Vasen, H. F., Watson, P., Mecklin, J. P. & Lynch, H. T. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology 116, 1453–1456 (1999).
Lynch, H. T. et al. History of the international collaborative group on hereditary non polyposis colorectal cancer. Fam. Cancer 2, 3–5 (2003).
Farabaugh, P. J., Schmeissner, U., Hofer, M. & Miller, J. H. Genetic studies of the lac repressor. VII. On the molecular nature of spontaneous hotspots in the lacI gene of Escherichia coli. J. Mol. Biol. 126, 847–857 (1978).
Streisinger, G. & Owen, J. Mechanisms of spontaneous and induced frameshift mutation in bacteriophage T4. Genetics 109, 633–659 (1985).
Levinson, G. & Gutman, G. A. High frequencies of short frameshifts in poly-CA/TG tandem repeats borne by bacteriophage M13 in Escherichia coli K-12. Nucleic Acids Res. 15, 5323–5338 (1987).
Schaaper, R. M. & Dunn, R. L. Spectra of spontaneous mutations in Escherichia coli strains defective in mismatch correction: the nature of in vivo DNA replication errors. Proc. Natl Acad. Sci. USA 84, 6220–6224 (1987).
Peinado, M. A., Malkhosyan, S., Velazquez, A. & Perucho, M. Isolation and characterization of allelic losses and gains in colorectal tumors by arbitrarily primed polymerase chain reaction. Proc. Natl Acad. Sci. USA 89, 10065–10069 (1992).
Ionov, Y., Peinado, M. A., Malkhosyan, S., Shibata, D. & Perucho, M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature 363, 558–561 (1993).
Thibodeau, S. N., Bren, G. & Schaid, D. Microsatellite instability in cancer of the proximal colon. Science 260, 816–819 (1993).
Peltomäki, P. et al. Genetic mapping of a locus predisposing to human colorectal cancer. Science 260, 810–812 (1993).
Aaltonen LA et al. Clues to the pathogenesis of familial colorectal cancer. Science 260, 812–816 (1993).
Lindblom, A., Tannergård, P., Werelius, B. & Nordenskjöld, M. Genetic mapping of a second locus predisposing to hereditary non-polyposis colon cancer. Nat. Genet. 5, 279–282 (1993).
Leach, F. S. et al. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell 75, 1215–1225 (1993).
Papadopoulos, N. et al. Mutation of a mutL homolog in hereditary colon cancer. Science 263, 1625–1629 (1994).
Fishel, R. et al. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell 75, 1027–1038 (1993).
Bronner, C. E. et al. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature 368, 258–261 (1994).
Liu, B. et al. hMSH2 mutations in hereditary nonpolyposis colorectal cancer kindreds. Cancer Res. 54, 4590–4594 (1994).
Nicolaides, N. C. et al. Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature 371, 75–80 (1994).
Miyaki, M. et al. Germline mutation of MSH6 as the cause of hereditary nonpolyposis colorectal cancer. Nat. Genet. 17, 271–272 (1997).
Strand, M., Earley, M. C., Crouse, G. F. & Petes, T. D. Mutations in the MSH3 gene preferentially lead to deletions within tracts of simple repetitive DNA in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA 92, 10418–10421 (1995).
Tran, H. T., Keen, J. D., Kricker, M., Resnick, M. A. & Gordenin, D. A. Hypermutability of homonucleotide runs in mismatch repair and DNA polymerase proofreading yeast mutants. Mol. Cell Biol. 17, 2859–2865 (1997).
Risinger, J. I., Umar, A., Barrett, J. C. & Kunkel, T. A. A hPMS2 mutant cell line is defective in strand-specific mismatch repair. J. Biol. Chem. 270, 18183–18186 (1995).
Parsons, R. et al. Hypermutability and mismatch repair deficiency in RER+ tumor cells. Cell 75, 1227–1236 (1993).
Umar, A. et al. Defective mismatch repair in extracts of colorectal and endometrial cancer cell lines exhibiting microsatellite instability. J. Biol. Chem. 269, 14367–14370 (1994).
Author information
Authors and Affiliations
Contributions
All authors made a substantial contribution to all aspects of the preparation of this manuscript.
Corresponding author
Ethics declarations
Competing interests
B.R. has acted as a consultant and/or adviser for Neophore and Artios Pharma and is an inventor of a patent related to MMRd and immunotherapy. C.A. has received grants or contracts from Abbvie, Artios, AstraZeneca, Clovis and Genentech/Roche; has participated on a Data Safety Monitoring Board or Advisory Board for AstraZeneca, Merck and WIRB-Copernicus Group (WCG); and has served in a leadership or fiduciary role for the GOG Foundation and NRG Oncology. M.B.F. has acted as a consultant and/or adviser to Abbott Laboratories, Bristol Myers Squibb and Genzyme. L.A.D.J. is a member of the board of directors of Epitope and Quest Diagnostics and is a compensated consultant to Absci, Blackstone, Delfi, GSK, Innovatus Capital Partners, Seer and Neophore. L.A.D.J. is also an inventor of multiple licenced patents related to technology for circulating tumour DNA analyses and MMRd for diagnosis and therapy; some of these licences and relationships are associated with equity or royalty payments to the inventors. He holds equity in Absci, Delfi, Epitope, Neophore, Quest Diagnostics and Seer. He divested his equity in Personal Genome Diagnostics to LabCorp in February 2022 and divested his equity in Thrive Earlier Detection to Exact Biosciences in January 2021. His spouse holds equity in Amgen. The terms of all these arrangements are being managed by Memorial Sloan Kettering in accordance with their conflict-of-interest policy. P.J. declares no competing interests.
Peer review
Peer review information
Nature Reviews Clinical Oncology thanks Toni Seppälä, who co-reviewed with Joni Panula; Julien Taieb; and the other, anonymous, reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Johannet, P., Rousseau, B., Aghajanian, C. et al. Therapeutic targeting of mismatch repair-deficient cancers. Nat Rev Clin Oncol 22, 734–759 (2025). https://doi.org/10.1038/s41571-025-01054-6
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41571-025-01054-6
This article is cited by
-
Molecular Testing for Intrahepatic Cholangiocarcinoma: What, When, How?
Journal of Gastrointestinal Cancer (2026)
-
Immune checkpoint inhibitors for the treatment of solid tumors and lymphoma in the past 26 years (2000–2025)
Journal of Hematology & Oncology (2025)