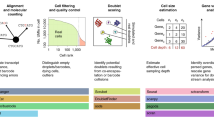
Abstract
Single-cell technologies, particularly single-cell RNA sequencing (scRNA-seq) methods, together with associated computational tools and the growing availability of public data resources, are transforming drug discovery and development. New opportunities are emerging in target identification owing to improved disease understanding through cell subtyping, and highly multiplexed functional genomics screens incorporating scRNA-seq are enhancing target credentialling and prioritization. ScRNA-seq is also aiding the selection of relevant preclinical disease models and providing new insights into drug mechanisms of action. In clinical development, scRNA-seq can inform decision-making via improved biomarker identification for patient stratification and more precise monitoring of drug response and disease progression. Here, we illustrate how scRNA-seq methods are being applied in key steps in drug discovery and development, and discuss ongoing challenges for their implementation in the pharmaceutical industry.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
DiMasi, J. A., Grabowski, H. G. & Hansen, R. W. Innovation in the pharmaceutical industry: new estimates of R&D costs. J. Health Econ. 47, 20–33 (2016).
Wouters, O. J., McKee, M. & Luyten, J. Estimated research and development investment needed to bring a new medicine to market, 2009–2018. JAMA 323, 844–853 (2020).
Paul, S. M. et al. How to improve R&D productivity: the pharmaceutical industry’s grand challenge. Nat. Rev. Drug Discov. 9, 203–214 (2010).
Nelson, M. R. et al. The support of human genetic evidence for approved drug indications. Nat. Genet. 47, 856–860 (2015).
1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012).
Sernoskie, S. C., Jee, A. & Uetrecht, J. P. The emerging role of the innate immune response in idiosyncratic drug reactions. Pharmacol. Rev. 73, 861–896 (2021).
Heid, C. A., Stevens, J., Livak, K. J. & Williams, P. M. Real time quantitative PCR. Genome Res. 6, 986–994 (1996).
Cheung, R. K. & Utz, P. J. CyTOF — the next generation of cell detection. Nat. Rev. Rheumatol. 7, 502–503 (2011).
Bendall, S. C. et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332, 687–696 (2011).
Nassar, A. F., Ogura, H. & Wisnewski, A. V. Impact of recent innovations in the use of mass cytometry in support of drug development. Drug. Discov. Today 20, 1169–1175 (2015).
Wen, L. & Tang, F. Recent advances in single-cell sequencing technologies. Precis. Clin. Med. 5, pbac002 (2022).
Jovic, D. et al. Single‐cell RNA sequencing technologies and applications: a brief overview. Clin. Transl. Med. 12, e694 (2022).
Kashima, Y. et al. Single-cell sequencing techniques from individual to multiomics analyses. Exp. Mol. Med. 52, 1419–1427 (2020).
Svensson, V., Vento-Tormo, R. & Teichmann, S. A. Exponential scaling of single-cell RNA-seq in the past decade. Nat. Protoc. 13, 599–604 (2018).
Aldridge, S. & Teichmann, S. A. Single cell transcriptomics comes of age. Nat. Commun. 11, 4307 (2020).
Tang, F. et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 6, 377–382 (2009). Successful attempt to sequence the full transcriptome of a single cell in an unbiased way.
Navin, N. E., Rozenblatt-Rosen, O. & Zhang, N. R. New frontiers in single-cell genomics. Genome Res. 31, ix–x (2021).
Zilionis, R. et al. Single-cell transcriptomics of human and mouse lung cancers reveals conserved myeloid populations across individuals and species. Immunity 50, 1317–1334.e10 (2019). A detailed study correlating immune cell populations in mouse and human lung cancer.
Sade-Feldman, M. et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell 175, 998–1013.e20 (2018). Illustration of how scRNA-seq approaches can be used to identify new predictive biomarkers for the response or resistance to ICI therapies in cancer.
Jang, J. S. et al. Molecular signatures of multiple myeloma progression through single cell RNA-Seq. Blood Cancer J. 9, 2 (2019).
Tanaka, N. et al. Single-cell RNA-seq analysis reveals the platinum resistance gene COX7B and the surrogate marker CD63. Cancer Med. 7, 6193–6204 (2018).
Jerby-Arnon, L. et al. A cancer cell program promotes T cell exclusion and resistance to checkpoint blockade. Cell 175, 984–997.e24 (2018). This work demonstrates the utility of scRNA-seq for the identification of an immune resistance programme associated with T cell exclusion and immune evasion. It also provides new therapeutic approaches to overcome resistance to ICI.
Cohen, Y. C. et al. Identification of resistance pathways and therapeutic targets in relapsed multiple myeloma patients through single-cell sequencing. Nat. Med. 27, 491–503 (2021).
Villani, A.-C. et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356, eaah4573 (2017).
Park, J.-E. et al. A cell atlas of human thymic development defines T cell repertoire formation. Science 367, eaay3224 (2020).
GTEx Consortium.Landscape of X chromosome inactivation across human tissues. Nature 550, 244–248 (2017).
Ramachandran, P. et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature 575, 512–518 (2019).
Song, H. et al. Single-cell analysis of human primary prostate cancer reveals the heterogeneity of tumor-associated epithelial cell states. Nat. Commun. 13, 141 (2022).
Wang, Q. et al. Single-cell chromatin accessibility landscape in kidney identifies additional cell-of-origin in heterogenous papillary renal cell carcinoma. Nat. Commun. 13, 31 (2022).
Nowicki-Osuch, K. et al. Molecular phenotyping reveals the identity of Barrett’s esophagus and its malignant transition. Science 373, 760–767 (2021). Illustrative example of how SC studies can help to understand tumorigenesis.
Steen, C. B. et al. The landscape of tumor cell states and ecosystems in diffuse large B cell lymphoma. Cancer Cell 39, 1422–1437.e10 (2021).
Wu, S. Z. et al. A single-cell and spatially resolved atlas of human breast cancers. Nat. Genet. 53, 1334–1347 (2021).
Zhang, X. et al. Dissecting esophageal squamous-cell carcinoma ecosystem by single-cell transcriptomic analysis. Nat. Commun. 12, 5291 (2021).
Pu, W. et al. Single-cell transcriptomic analysis of the tumor ecosystems underlying initiation and progression of papillary thyroid carcinoma. Nat. Commun. 12, 6058 (2021).
Ursu, O. et al. Massively parallel phenotyping of coding variants in cancer with Perturb-seq. Nat. Biotechnol. 40, 896–905 (2022). High-throughput analysis of oncogene and tumour suppressor variant phenotypes at single-cell level.
Chaligne, R. et al. Epigenetic encoding, heritability and plasticity of glioma transcriptional cell states. Nat. Genet. 53, 1469–1479 (2021).
Johnson, K. C. et al. Single-cell multimodal glioma analyses identify epigenetic regulators of cellular plasticity and environmental stress response. Nat. Genet. 53, 1456–1468 (2021).
Croucher, D. C. et al. Longitudinal single-cell analysis of a myeloma mouse model identifies subclonal molecular programs associated with progression. Nat. Commun. 12, 6322 (2021).
Salehi, S. et al. Clonal fitness inferred from time-series modelling of single-cell cancer genomes. Nature 595, 585–590 (2021). SC-based study showing how TP53 mutations alter tumour clonal fitness in TNBC and the impact on resistance to cisplatin chemotherapy.
Quinn, J. J. et al. Single-cell lineages reveal the rates, routes, and drivers of metastasis in cancer xenografts. Science 371, eabc1944 (2021).
Yaddanapudi, K. et al. Single-cell immune mapping of melanoma sentinel lymph nodes reveals an actionable immunotolerant microenvironment. Clin. Cancer Res. 28, 2069–2081 (2022).
Lund, A. W. Standing watch: immune activation and failure in melanoma sentinel lymph nodes. Clin. Cancer Res. 28, 1996–1998 (2022).
Li, J. et al. Single-cell characterization of the cellular landscape of acral melanoma identifies novel targets for immunotherapy. Clin. Cancer Res. 28, 2131–2146 (2022).
Sun, Y.-F. et al. Dissecting spatial heterogeneity and the immune-evasion mechanism of CTCs by single-cell RNA-seq in hepatocellular carcinoma. Nat. Commun. 12, 4091 (2021).
Diamantopoulou, Z. et al. The metastatic spread of breast cancer accelerates during sleep. Nature 607, 156–162 (2022).
Rozenblatt-Rosen, O. et al. The Human Tumor Atlas Network: charting tumor transitions across space and time at single-cell resolution. Cell 181, 236–249 (2020). Description of the goals of the Human Tumor Atlas Network project — building a SC and spatially resolved pan-cancer atlas also covering the dynamics from cancer initiation to metastasis.
Becker, W. R. et al. Single-cell analyses define a continuum of cell state and composition changes in the malignant transformation of polyps to colorectal cancer. Nat. Genet. 54, 985–995 (2022).
Arenas, E. Parkinson’s disease in the single-cell era. Nat. Neurosci. 25, 536–538 (2022).
Kamath, T. et al. Single-cell genomic profiling of human dopamine neurons identifies a population that selectively degenerates in Parkinson’s disease. Nat. Neurosci. 25, 588–595 (2022). Identification and characterization of a dopamine neuron subpopulation that selectively degenerates in Parkinson disease.
Miller, M. B. et al. Somatic genomic changes in single Alzheimer’s disease neurons. Nature 604, 714–722 (2022).
Keren-Shaul, H. et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell 169, 1276–1290.e17 (2017). Identification and characterization of a disease-associated microglia population in Alzheimer disease.
Wang, P. et al. Single-cell transcriptome and TCR profiling reveal activated and expanded T cell populations in Parkinson’s disease. Cell Discov. 7, 52 (2021).
Cadwell, C. R. et al. Electrophysiological, transcriptomic and morphologic profiling of single neurons using Patch-seq. Nat. Biotechnol. 34, 199–203 (2016).
Fuzik, J. et al. Integration of electrophysiological recordings with single-cell RNA-seq data identifies neuronal subtypes. Nat. Biotechnol. 34, 175–183 (2016).
Yang, A. C. et al. A human brain vascular atlas reveals diverse mediators of Alzheimer’s risk. Nature 603, 885–892 (2022).
Berg, J. et al. Human neocortical expansion involves glutamatergic neuron diversification. Nature 598, 151–158 (2021).
Simone, D. et al. Single cell analysis of spondyloarthritis regulatory T cells identifies distinct synovial gene expression patterns and clonal fates. Commun. Biol. 4, 1395 (2021).
Penkava, F. et al. Single-cell sequencing reveals clonal expansions of pro-inflammatory synovial CD8 T cells expressing tissue-homing receptors in psoriatic arthritis. Nat. Commun. 11, 4767 (2020).
Wu, X. et al. Single-cell sequencing of immune cells from anticitrullinated peptide antibody positive and negative rheumatoid arthritis. Nat. Commun. 12, 4977 (2021).
Liu, Y. et al. Classification of human chronic inflammatory skin disease based on single-cell immune profiling. Sci. Immunol. 7, eabl9165 (2022).
Ingelfinger, F. et al. Twin study reveals non-heritable immune perturbations in multiple sclerosis. Nature 603, 152–158 (2022).
Bjornevik, K. et al. Longitudinal analysis reveals high prevalence of Epstein-Barr virus associated with multiple sclerosis. Science 375, 296–301 (2022).
Lanz, T. V. et al. Clonally expanded B cells in multiple sclerosis bind EBV EBNA1 and GlialCAM. Nature 603, 321–327 (2022).
Nathan, A. et al. Single-cell eQTL models reveal dynamic T cell state dependence of disease loci. Nature 606, 120–128 (2022). Describes the discovery of cell-state-specific and dynamic eQTL patterns in human memory T cells revealing new eQTL associations for non-coding variants linked to disease.
Ma, K.-Y. et al. High-throughput and high-dimensional single-cell analysis of antigen-specific CD8+ T cells. Nat. Immunol. 22, 1590–1598 (2021).
Wauters, E. et al. Discriminating mild from critical COVID-19 by innate and adaptive immune single-cell profiling of bronchoalveolar lavages. Cell Res. 31, 272–290 (2021).
Stephenson, E. et al. Single-cell multi-omics analysis of the immune response in COVID-19. Nat. Med. 27, 904–916 (2021).
Lee, J. W. et al. Integrated analysis of plasma and single immune cells uncovers metabolic changes in individuals with COVID-19. Nat. Biotechnol. 40, 110–120 (2022).
Georg, P. et al. Complement activation induces excessive T cell cytotoxicity in severe COVID-19. Cell 185, 493–512.e25 (2022).
Wang, S. et al. A single-cell transcriptomic landscape of the lungs of patients with COVID-19. Nat. Cell Biol. 23, 1314–1328 (2021). Study using SC sequencing to better understand severe COVID-19.
Delorey, T. M. et al. COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature 595, 107–113 (2021). Study using SC sequencing to better understand severe COVID-19.
Tian, Y. et al. Single-cell immunology of SARS-CoV-2 infection. Nat. Biotechnol. 40, 30–41 (2022).
Dar, D., Dar, N., Cai, L. & Newman, D. K. Spatial transcriptomics of planktonic and sessile bacterial populations at single-cell resolution. Science 373, eabi4882 (2021).
Gideon, H. P. et al. Multimodal profiling of lung granulomas in macaques reveals cellular correlates of tuberculosis control. Immunity 55, 827–846.e10 (2022).
Abdelfattah, N. et al. Single-cell analysis of human glioma and immune cells identifies S100A4 as an immunotherapy target. Nat. Commun. 13, 767 (2022).
Lareau, C. A., Parker, K. R. & Satpathy, A. T. Charting the tumor antigen maps drawn by single-cell genomics. Cancer Cell 39, 1553–1557 (2021).
Gladka, M. M. et al. Single-cell sequencing of the healthy and diseased heart reveals cytoskeleton-associated protein 4 as a new modulator of fibroblasts activation. Circulation 138, 166–180 (2018). Illustrative example of how SC approaches can help to identify candidate targets. Here, CKAP4 for cardiac fibrosis.
Kuppe, C. et al. Decoding myofibroblast origins in human kidney fibrosis. Nature 589, 281–286 (2021).
Li, Z. et al. Chromatin-accessibility estimation from single-cell ATAC-seq data with scOpen. Nat. Commun. 12, 6386 (2021).
Cano-Gamez, E. & Trynka, G. From GWAS to function: using functional genomics to identify the mechanisms underlying complex diseases. Front. Genet. 11, 424 (2020).
Jagadeesh, K. A. et al. Identifying disease-critical cell types and cellular processes by integrating single-cell RNA-sequencing and human genetics. Nat. Genet. 54, 1479–1492 (2022). The method scLinker combines GWAS summary statistics with scRNA-seq data sets and thereby enables the discovery of cell types (and biological processes) linked to disease.
Muslu, O., Hoyt, C. T., Lacerda, M., Hofmann-Apitius, M. & Frohlich, H. Guiltytargets: prioritization of novel therapeutic targets with network representation learning. IEEE/ACM Trans. Comput. Biol. Bioinform. 19, 491–500 (2022).
Gawel, D. R. et al. A validated single-cell-based strategy to identify diagnostic and therapeutic targets in complex diseases. Genome Med. 11, 47 (2019).
Dixit, A. et al. Perturb-seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell 167, 1853–1866.e17 (2016). Technique for pooled CRISPR screening with scRNA-seq readouts.
Adamson, B. et al. A multiplexed single-cell CRISPR screening platform enables systematic dissection of the unfolded protein response. Cell 167, 1867–1882.e21 (2016).
Datlinger, P. et al. Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods 14, 297–301 (2017).
Shifrut, E. et al. Genome-wide CRISPR screens in primary human T cells reveal key regulators of immune function. Cell 175, 1958–1971.e15 (2018).
Jin, X. et al. In vivo Perturb-Seq reveals neuronal and glial abnormalities associated with autism risk genes. Science 370, eaaz6063 (2020).
Lazo, J. S. et al. Credentialing and pharmacologically targeting PTP4A3 phosphatase as a molecular target for ovarian cancer. Biomolecules 11, 969 (2021).
Wang, W. et al. MAPK4 promotes triple negative breast cancer growth and reduces tumor sensitivity to PI3K blockade. Nat. Commun. 13, 245 (2022).
Wang, P.-X. et al. Targeting CASP8 and FADD-like apoptosis regulator ameliorates nonalcoholic steatohepatitis in mice and nonhuman primates. Nat. Med. 23, 439–449 (2017).
Bertin, S. et al. Dual-specificity phosphatase 6 regulates CD4+ T-cell functions and restrains spontaneous colitis in IL-10-deficient mice. Mucosal Immunol. 8, 505–515 (2015).
Ruan, J.-W. et al. Dual-specificity phosphatase 6 deficiency regulates gut microbiome and transcriptome response against diet-induced obesity in mice. Nat. Microbiol. 2, 16220 (2016).
Chang, C.-S. et al. Single-cell RNA sequencing uncovers the individual alteration of intestinal mucosal immunocytes in Dusp6 knockout mice. iScience 25, 103738 (2022).
Llewellyn, H. P. et al. T cells and monocyte-derived myeloid cells mediate immunotherapy-related hepatitis in a mouse model. J. Hepatol. 75, 1083–1095 (2021).
Chen, S.-H. et al. Dual checkpoint blockade of CD47 and PD-L1 using an affinity-tuned bispecific antibody maximizes antitumor immunity. J. Immunother. Cancer 9, e003464 (2021).
Mimitou, E. P. et al. Multiplexed detection of proteins, transcriptomes, clonotypes and CRISPR perturbations in single cells. Nat. Methods 16, 409–412 (2019).
Katzenelenbogen, Y. et al. Coupled scRNA-Seq and intracellular protein activity reveal an immunosuppressive role of TREM2 in cancer. Cell 182, 872–885.e19 (2020).
Frangieh, C. J. et al. Multimodal pooled Perturb-CITE-seq screens in patient models define mechanisms of cancer immune evasion. Nat. Genet. 53, 332–341 (2021).
Schütte, M. et al. Molecular dissection of colorectal cancer in pre-clinical models identifies biomarkers predicting sensitivity to EGFR inhibitors. Nat. Commun. 8, 14262 (2017).
Kinker, G. S. et al. Pan-cancer single-cell RNA-seq identifies recurring programs of cellular heterogeneity. Nat. Genet. 52, 1208–1218 (2020).
Mead, B. E. et al. Screening for modulators of the cellular composition of gut epithelia via organoid models of intestinal stem cell differentiation. Nat. Biomed. Eng. 6, 476–494 (2022).
Bock, C. et al. The organoid cell atlas. Nat. Biotechnol. 39, 13–17 (2021).
Shinozawa, T. et al. High-fidelity drug-induced liver injury screen using human pluripotent stem cell-derived organoids. Gastroenterology 160, 831–846.e10 (2021). Characterization of organoid preclinical models for liver injury drug screening using scRNA-seq.
Krieger, T. G. et al. Single-cell analysis of patient-derived PDAC organoids reveals cell state heterogeneity and a conserved developmental hierarchy. Nat. Commun. 12, 5826 (2021).
Bondoc, A. et al. Identification of distinct tumor cell populations and key genetic mechanisms through single cell sequencing in hepatoblastoma. Commun. Biol. 4, 1049 (2021).
Kim, K.-T. et al. Single-cell mRNA sequencing identifies subclonal heterogeneity in anti-cancer drug responses of lung adenocarcinoma cells. Genome Biol. 16, 127 (2015).
Hosein, A. N. et al. Cellular heterogeneity during mouse pancreatic ductal adenocarcinoma progression at single-cell resolution. JCI Insight 5, 129212 (2019).
Tabula Muris Consortium. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367–372 (2018). The Tabula Muris project generated a SC multi-tissue atlas at SC resolution for the frequently used Mus musculus animal model in preclinical research.
Kumar, M. P. et al. Analysis of single-cell RNA-Seq identifies cell-cell communication associated with tumor characteristics. Cell Rep. 25, 1458–1468.e4 (2018).
Taukulis, I. A. et al. Single-cell RNA-Seq of cisplatin-treated adult stria vascularis identifies cell type-specific regulatory networks and novel therapeutic gene targets. Front. Mol. Neurosci. 14, 718241 (2021). Illustrative example of how SC approaches can be used to explain toxic undesirable effects of therapies.
Yofe, I., Dahan, R. & Amit, I. Single-cell genomic approaches for developing the next generation of immunotherapies. Nat. Med. 26, 171–177 (2020).
McFarland, J. M. et al. Multiplexed single-cell transcriptional response profiling to define cancer vulnerabilities and therapeutic mechanism of action. Nat. Commun. 11, 4296 (2020).
Shin, D., Lee, W., Lee, J. H. & Bang, D. Multiplexed single-cell RNA-seq via transient barcoding for simultaneous expression profiling of various drug perturbations. Sci. Adv. 5, eaav2249 (2019).
Srivatsan, S. R. et al. Massively multiplex chemical transcriptomics at single-cell resolution. Science 367, 45–51 (2020). Illustration of how a high-content screening method that uses scRNA-seq as readout can provide new hints on HDAC inhibitor MoA in cancer.
Ji, Y., Lotfollahi, M., Wolf, F. A. & Theis, F. J. Machine learning for perturbational single-cell omics. Cell Syst. 12, 522–537 (2021).
Lotfollahi, M. J., Wolf, F. A. & Theis, F.J. scGen predicts single-cell perturbation responses. Nat. Methods 16, 715–721 (2019).
Lotfollahi, M. et al. Learning interpretable cellular responses to complex perturbations in high-throughput screens. Preprint at bioRxiv https://doi.org/10.1101/2021.04.14.439903 (2021).
Brewer, R. C. et al. BNT162b2 vaccine induces divergent B cell responses to SARS-CoV-2 S1 and S2. Nat. Immunol. 23, 33–39 (2022).
Andreano, E. et al. Hybrid immunity improves B cells and antibodies against SARS-CoV-2 variants. Nature 600, 530–535 (2021).
Hall, V. et al. Protection against SARS-CoV-2 after Covid-19 vaccination and previous infection. N. Engl. J. Med. 386, 1207–1220 (2022).
RECOVERY Collaborative Group. Dexamethasone in hospitalized patients with Covid-19. N. Engl. J. Med. 384, 693–704 (2021).
Sinha, S. et al. Dexamethasone modulates immature neutrophils and interferon programming in severe COVID-19. Nat. Med. 28, 201–211 (2022).
Aissa, A. F. et al. Single-cell transcriptional changes associated with drug tolerance and response to combination therapies in cancer. Nat. Commun. 12, 1628 (2021).
Guinney, J. et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 21, 1350–1356 (2015).
Mehrvarz Sarshekeh, A. et al. Consensus molecular subtype (CMS) as a novel integral biomarker in colorectal cancer: a phase II trial of bintrafusp alfa in CMS4 metastatic CRC. JCO 38, 4084–4084 (2020).
Khaliq, A. M. et al. Refining colorectal cancer classification and clinical stratification through a single-cell atlas. Genome Biol. 23, 113 (2022).
Joanito, I. et al. Single-cell and bulk transcriptome sequencing identifies two epithelial tumor cell states and refines the consensus molecular classification of colorectal cancer. Nat. Genet. 54, 963–975 (2022). Novel classification of CRC for biomarker prognosis proposed by using SC approaches and the tumour environment.
Litchfield, K. et al. Meta-analysis of tumor- and T cell-intrinsic mechanisms of sensitization to checkpoint inhibition. Cell 184, 596–614.e14 (2021).
Li, H., van der Merwe, P. A. & Sivakumar, S. Biomarkers of response to PD-1 pathway blockade. Br. J. Cancer 126, 1663–1675 (2022).
Leader, A. M. et al. Single-cell analysis of human non-small cell lung cancer lesions refines tumor classification and patient stratification. Cancer Cell 39, 1594–1609.e12 (2021).
Xiong, D., Wang, Y. & You, M. A gene expression signature of TREM2hi macrophages and γδ T cells predicts immunotherapy response. Nat. Commun. 11, 5084 (2020).
Kieffer, Y. et al. Single-cell analysis reveals fibroblast clusters linked to immunotherapy resistance in cancer. Cancer Discov. 10, 1330–1351 (2020).
Dominguez, C. X. et al. Single-cell RNA sequencing reveals stromal evolution into LRRC15+ myofibroblasts as a determinant of patient response to cancer immunotherapy. Cancer Discov. 10, 232–253 (2020).
Guo, X. et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat. Med. 24, 978–985 (2018).
Zheng, C. et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell 169, 1342–1356.e16 (2017).
Pittet, M. J., Michielin, O. & Migliorini, D. Clinical relevance of tumour-associated macrophages. Nat. Rev. Clin. Oncol. 19, 402–421 (2022).
Färkkilä, A. et al. Immunogenomic profiling determines responses to combined PARP and PD-1 inhibition in ovarian cancer. Nat. Commun. 11, 1459 (2020).
Jansen, C. S. et al. An intra-tumoral niche maintains and differentiates stem-like CD8 T cells. Nature 576, 465–470 (2019).
Vanhersecke, L. et al. Mature tertiary lymphoid structures predict immune checkpoint inhibitor efficacy in solid tumors independently of PD-L1 expression. Nat. Cancer 2, 794–802 (2021).
Zhang, K. et al. Longitudinal single-cell RNA-seq analysis reveals stress-promoted chemoresistance in metastatic ovarian cancer. Sci. Adv. 8, eabm1831 (2022).
Candelli, T. et al. Identification and characterization of relapse-initiating cells in MLL-rearranged infant ALL by single-cell transcriptomics. Leukemia 36, 58–67 (2022).
Pieters, R. et al. A treatment protocol for infants younger than 1 year with acute lymphoblastic leukaemia (Interfant-99): an observational study and a multicentre randomised trial. Lancet 370, 240–250 (2007).
Martin, J. C. et al. Single-cell analysis of Crohn’s disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy. Cell 178, 1493–1508.e20 (2019).
Smillie, C. S. et al. Intra- and inter-cellular rewiring of the human colon during ulcerative colitis. Cell 178, 714–730.e22 (2019).
Wang, Z. et al. Single-cell RNA sequencing of peripheral blood mononuclear cells from acute Kawasaki disease patients. Nat. Commun. 12, 5444 (2021).
Zhang, Y. et al. Single-cell analyses of renal cell cancers reveal insights into tumor microenvironment, cell of origin, and therapy response. Proc. Natl Acad. Sci. USA 118, e2103240118 (2021).
Tyner, J. W. et al. Functional genomic landscape of acute myeloid leukaemia. Nature 562, 526–531 (2018).
Schuurhuis, G. J. et al. Minimal/measurable residual disease in AML: a consensus document from the European LeukemiaNet MRD Working Party. Blood 131, 1275–1291 (2018).
Ediriwickrema, A. et al. Single-cell mutational profiling enhances the clinical evaluation of AML MRD. Blood Adv. 4, 943–952 (2020). Minimal residual disease in acute myeloid leukaemia can be better assessed by using SC mutational profiling.
Oren, Y. et al. Cycling cancer persister cells arise from lineages with distinct programs. Nature 596, 576–582 (2021). Shows that SC approaches are key for the identification of cancer persister cells induced in response to treatment.
Kim, C. et al. Chemoresistance evolution in triple-negative breast cancer delineated by single-cell sequencing. Cell 173, 879–893.e13 (2018).
Yost, K. E. et al. Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat. Med. 25, 1251–1259 (2019).
Zhang, Y. et al. Single-cell analyses reveal key immune cell subsets associated with response to PD-L1 blockade in triple-negative breast cancer. Cancer Cell 39, 1578–1593.e8 (2021).
Bassez, A. et al. A single-cell map of intratumoral changes during anti-PD1 treatment of patients with breast cancer. Nat. Med. 27, 820–832 (2021).
Wu, T. D. et al. Peripheral T cell expansion predicts tumour infiltration and clinical response. Nature 579, 274–278 (2020).
Stoeckius, M. et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017). Explains the CITE-seq technique, which enables researchers to simultaneously assess the full transcriptome at SC resolution with the protein expression of selected cell surface markers.
Peterson, V. M. et al. Multiplexed quantification of proteins and transcripts in single cells. Nat. Biotechnol. 35, 936–939 (2017).
Chen, S., Lake, B. B. & Zhang, K. High-throughput sequencing of the transcriptome and chromatin accessibility in the same cell. Nat. Biotechnol. 37, 1452–1457 (2019).
Ma, S. et al. Chromatin potential identified by shared single-cell profiling of RNA and chromatin. Cell 183, 1103–1116.e20 (2020).
Clark, S. J. et al. scNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells. Nat. Commun. 9, 781 (2018).
Ren, X. et al. COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas. Cell 184, 1895–1913.e19 (2021).
Mathys, H. et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 570, 332–337 (2019).
Melms, J. C. et al. A molecular single-cell lung atlas of lethal COVID-19. Nature 595, 114–119 (2021).
Ding, J. et al. Systematic comparison of single-cell and single-nucleus RNA-sequencing methods. Nat. Biotechnol. 38, 737–746 (2020).
Thrupp, N. et al. Single-nucleus RNA-Seq is not suitable for detection of microglial activation genes in humans. Cell Rep. 32, 108189 (2020).
Der, E. et al. Tubular cell and keratinocyte single-cell transcriptomics applied to lupus nephritis reveal type I IFN and fibrosis relevant pathways. Nat. Immunol. 20, 915–927 (2019).
Haque, A., Engel, J., Teichmann, S. A. & Lönnberg, T. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med. 9, 75 (2017).
Ding, J., Sharon, N. & Bar-Joseph, Z. Temporal modelling using single-cell transcriptomics. Nat. Rev. Genet. 23, 355–368 (2022). An excellent review on how to design and analyse SC time-series experiments.
Guillaumet-Adkins, A. et al. Single-cell transcriptome conservation in cryopreserved cells and tissues. Genome Biol. 18, 45 (2017).
Wilkinson, M. D. et al. The FAIR guiding principles for scientific data management and stewardship. Sci. Data 3, 160018 (2016).
Svensson, V., da Veiga Beltrame, E. & Pachter, L. A curated database reveals trends in single-cell transcriptomics. Database 2020, baaa073 (2020).
Han, X. et al. Construction of a human cell landscape at single-cell level. Nature 581, 303–309 (2020).
Tabula Sapiens Consortium. The Tabula Sapiens: a multiple-organ, single-cell transcriptomic atlas of humans. Science 376, eabl4896 (2022). The Tabula Sapiens consortium created and publicly released a multi-tissue transcriptome SC atlas covering 15 human donors.
Füllgrabe, A. et al. Guidelines for reporting single-cell RNA-seq experiments. Nat. Biotechnol. 38, 1384–1386 (2020).
Meghill, C. et al. Cellxgene: a performant, scalable exploration platform for high dimensional sparse matrices. Preprint at bioRxiv https://doi.org/10.1101/2021.04.05.438318 (2021).
Li, B. et al. Cumulus provides cloud-based data analysis for large-scale single-cell and single-nucleus RNA-seq. Nat. Methods 17, 793–798 (2020).
Papatheodorou, I. et al. Expression Atlas update: from tissues to single cells. Nucleic Acids Res. 48, D77–D83 (2020). EMBL-EBI SCEA is a valuable public SC resource used by industry.
Moreno, P. et al. User-friendly, scalable tools and workflows for single-cell RNA-seq analysis. Nat. Methods 18, 327–328 (2021).
Lähnemann, D. et al. Eleven grand challenges in single-cell data science. Genome Biol. 21, 31 (2020).
Angerer, P. et al. Single cells make big data: new challenges and opportunities in transcriptomics. Curr. Opin. Syst. Biol. 4, 85–91 (2017).
Zhang, M. J. et al. Polygenic enrichment distinguishes disease associations of individual cells in single-cell RNA-seq data. Nat. Genet. 54, 1572–1580 (2022).
Efremova, M., Vento-Tormo, M., Teichmann, S. A. & Vento-Tormo, R. CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nat. Protoc. 15, 1484–1506 (2020).
Fu, Y. et al. Pan-cancer computational histopathology reveals mutations, tumor composition and prognosis. Nat. Cancer 1, 800–810 (2020).
Warnat-Herresthal, S. et al. Swarm Learning as a privacy-preserving machine learning approach for disease classification. Preprint at bioRxiv https://doi.org/10.1101/2020.06.25.171009 (2020).
Regev, A. et al. The human cell atlas. eLife 6, e27041 (2017). Clearly explains the idea and goals of the HCA project.
Han, L. et al. Cell transcriptomic atlas of the non-human primate Macaca fascicularis. Nature 604, 723–731 (2022).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021).
Domínguez Conde, C. et al. Cross-tissue immune cell analysis reveals tissue-specific features in humans. Science 376, eabl5197 (2022).
Qian, J. et al. A pan-cancer blueprint of the heterogeneous tumor microenvironment revealed by single-cell profiling. Cell Res. 30, 745–762 (2020).
Zheng, L. et al. Pan-cancer single-cell landscape of tumor-infiltrating T cells. Science 374, abe6474 (2021).
Sun, D. et al. TISCH: a comprehensive web resource enabling interactive single-cell transcriptome visualization of tumor microenvironment. Nucleic Acids Res. 49, D1420–D1430 (2021).
Nieto, P. et al. A single-cell tumor immune atlas for precision oncology. Genome Res. 31, 1913–1926 (2021).
Zhang, F. et al. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat. Immunol. 20, 928–942 (2019).
Cao, J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019).
Cusanovich, D. A. et al. A single-cell atlas of in vivo mammalian chromatin accessibility. Cell 174, 1309–1324.e18 (2018).
Cao, J. et al. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357, 661–667 (2017).
Cusanovich, D. A. et al. The cis-regulatory dynamics of embryonic development at single-cell resolution. Nature 555, 538–542 (2018).
Zhang, K. et al. A single-cell atlas of chromatin accessibility in the human genome. Cell 184, 5985–6001.e19 (2021).
Cheng, J., Liao, J., Shao, X., Lu, X. & Fan, X. Multiplexing methods for simultaneous large‐scale transcriptomic profiling of samples at single‐cell resolution. Adv. Sci. 8, 2101229 (2021).
Picelli, S. et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 10, 1096–1098 (2013).
Hwang, B., Lee, J. H. & Bang, D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp. Mol. Med. 50, 1–14 (2018).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Kaminow, B., Yunusov, D. & Dobin, A. STARsolo: accurate, fast and versatile mapping/quantification of single-cell and single-nucleus RNA-seq data. Preprint at bioRxiv https://doi.org/10.1101/2021.05.05.442755 (2021).
Srivastava, A., Malik, L., Smith, T., Sudbery, I. & Patro, R. Alevin efficiently estimates accurate gene abundances from dscRNA-seq data. Genome Biol. 20, 65 (2019).
Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016).
Melsted, P., Ntranos, V. & Pachter, L. The barcode, UMI, set format and BUStools. Bioinformatics 35, 4472–4473 (2019).
Lun, A. T. L. et al. EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol. 20, 63 (2019).
Muskovic, W. & Powell, J. E. DropletQC: improved identification of empty droplets and damaged cells in single-cell RNA-seq data. Genome Biol. 22, 329 (2021).
Yang, S. et al. Decontamination of ambient RNA in single-cell RNA-seq with DecontX. Genome Biol. 21, 57 (2020).
Young, M. D. & Behjati, S. SoupX removes ambient RNA contamination from droplet-based single-cell RNA sequencing data. GigaScience 9, giaa151 (2020).
Wolock, S. L., Lopez, R. & Klein, A. M. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 8, 281–291.e9 (2019).
McGinnis, C. S., Murrow, L. M. & Gartner, Z. J. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 8, 329–337.e4 (2019).
DePasquale, E. A. K. et al. DoubletDecon: deconvoluting doublets from single-cell RNA-sequencing data. Cell Rep. 29, 1718–1727.e8 (2019).
Lun, A. T. L., McCarthy, D. J. & Marioni, J. C. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res 5, 2122 (2016).
Hafemeister, C. & Satija, R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296 (2019).
Bacher, R. et al. SCnorm: robust normalization of single-cell RNA-seq data. Nat. Methods 14, 584–586 (2017).
Duò, A., Robinson, M. D. & Soneson, C. A systematic performance evaluation of clustering methods for single-cell RNA-seq data. F1000Res 7, 1141 (2020).
Kobak, D. & Berens, P. The art of using t-SNE for single-cell transcriptomics. Nat. Commun. 10, 5416 (2019). Best practices on applying tSNE non-linear projections on scRNA-seq data sets.
Becht, E. et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38–44 (2019). Comparison of UMAP with respect to other non-linear projection methods when applied to scRNA-seq data sets.
Jaitin, D. A. et al. Dissecting immune circuits by linking CRISPR-pooled screens with single-cell RNA-Seq. Cell 167, 1883–1896.e15 (2016).
Papalexi, E. et al. Characterizing the molecular regulation of inhibitory immune checkpoints with multimodal single-cell screens. Nat. Genet. 53, 322–331 (2021).
Yang, L. et al. scMAGeCK links genotypes with multiple phenotypes in single-cell CRISPR screens. Genome Biol. 21, 19 (2020).
Duan, B. et al. Model-based understanding of single-cell CRISPR screening. Nat. Commun. 10, 2233 (2019).
Wang, R., Lin, D.-Y. & Jiang, Y. SCOPE: a normalization and copy-number estimation method for single-cell DNA sequencing. Cell Syst. 10, 445–452.e6 (2020).
Zaccaria, S. & Raphael, B. J. Characterizing allele- and haplotype-specific copy numbers in single cells with CHISEL. Nat. Biotechnol. 39, 207–214 (2021).
Zafar, H., Wang, Y., Nakhleh, L., Navin, N. & Chen, K. Monovar: single-nucleotide variant detection in single cells. Nat. Methods 13, 505–507 (2016).
Dong, X. et al. Accurate identification of single-nucleotide variants in whole-genome-amplified single cells. Nat. Methods 14, 491–493 (2017).
Luquette, L. J., Bohrson, C. L., Sherman, M. A. & Park, P. J. Identification of somatic mutations in single cell DNA-seq using a spatial model of allelic imbalance. Nat. Commun. 10, 3908 (2019).
Singer, J., Kuipers, J., Jahn, K. & Beerenwinkel, N. Single-cell mutation identification via phylogenetic inference. Nat. Commun. 9, 5144 (2018).
Mallory, X. F., Edrisi, M., Navin, N. & Nakhleh, L. Methods for copy number aberration detection from single-cell DNA-sequencing data. Genome Biol. 21, 208 (2020).
Gao, R. et al. Delineating copy number and clonal substructure in human tumors from single-cell transcriptomes. Nat. Biotechnol. 39, 599–608 (2021).
Patel, A. P. et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344, 1396–1401 (2014).
Petti, A. A. et al. A general approach for detecting expressed mutations in AML cells using single cell RNA-sequencing. Nat. Commun. 10, 3660 (2019).
Vu, T. N. et al. Cell-level somatic mutation detection from single-cell RNA sequencing. Bioinformatics 35, 4679–4687 (2019).
Cuomo, A. S. E. et al. Optimizing expression quantitative trait locus mapping workflows for single-cell studies. Genome Biol. 22, 188 (2021).
Stubbington, M. J. T. et al. T cell fate and clonality inference from single-cell transcriptomes. Nat. Methods 13, 329–332 (2016).
Lindeman, I. et al. BraCeR: B-cell-receptor reconstruction and clonality inference from single-cell RNA-seq. Nat. Methods 15, 563–565 (2018).
Song, L. et al. TRUST4: immune repertoire reconstruction from bulk and single-cell RNA-seq data. Nat. Methods 18, 627–630 (2021).
Upadhyay, A. A. et al. BALDR: a computational pipeline for paired heavy and light chain immunoglobulin reconstruction in single-cell RNA-seq data. Genome Med. 10, 20 (2018).
Rizzetto, S. et al. B-cell receptor reconstruction from single-cell RNA-seq with VDJPuzzle. Bioinformatics 34, 2846–2847 (2018).
Borcherding, N., Bormann, N. L. & Kraus, G. scRepertoire: an R-based toolkit for single-cell immune receptor analysis. F1000Res 9, 47 (2020).
McDavid, A., Gu, Y. & VonKaenel, E. CellaRepertorium: data structures, clustering and testing for single cell immune receptor repertoires (scRNAseq RepSeq/AIRR-seq). https://rdrr.io/bioc/CellaRepertorium (2021).
Zhang, Z., Xiong, D., Wang, X., Liu, H. & Wang, T. Mapping the functional landscape of T cell receptor repertoires by single-T cell transcriptomics. Nat. Methods 18, 92–99 (2021).
Buenrostro, J. D. et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490 (2015).
Wu, S. J. et al. Single-cell CUT&Tag analysis of chromatin modifications in differentiation and tumor progression. Nat. Biotechnol. 39, 819–824 (2021).
Grosselin, K. et al. High-throughput single-cell ChIP-seq identifies heterogeneity of chromatin states in breast cancer. Nat. Genet. 51, 1060–1066 (2019).
Clark, S. J. et al. Genome-wide base-resolution mapping of DNA methylation in single cells using single-cell bisulfite sequencing (scBS-seq). Nat. Protoc. 12, 534–547 (2017).
Slavov, N. Learning from natural variation across the proteomes of single cells. PLoS Biol. 20, e3001512 (2022).
Vistain, L. F. & Tay, S. Single-cell proteomics. Trends Biochem. Sci. 46, 661–672 (2021).
Perkel, J. M. Single-cell proteomics takes centre stage. Nature 597, 580–582 (2021).
Brinkerhoff, H., Kang, A. S. W., Liu, J., Aksimentiev, A. & Dekker, C. Multiple rereads of single proteins at single–amino acid resolution using nanopores. Science 374, 1509–1513 (2021).
Mimitou, E. P. et al. Scalable, multimodal profiling of chromatin accessibility, gene expression and protein levels in single cells. Nat. Biotechnol. 10, 1246–1258 (2021).
Hücker, S. M. et al. Single-cell microRNA sequencing method comparison and application to cell lines and circulating lung tumor cells. Nat. Commun. 12, 4316 (2021).
Gawronski, K. A. B. & Kim, J. Single cell transcriptomics of noncoding RNAs and their cell-specificity: Single cell transcriptomics of noncoding RNAs. WIREs RNA 8, e1433 (2017).
Seydel, C. Single-cell metabolomics hits its stride. Nat. Methods 18, 1452–1456 (2021).
VanInsberghe, M., van den Berg, J., Andersson-Rolf, A., Clevers, H. & van Oudenaarden, A. Single-cell Ribo-seq reveals cell cycle-dependent translational pausing. Nature 597, 561–565 (2021).
Arrastia, M. V. et al. Single-cell measurement of higher-order 3D genome organization with scSPRITE. Nat. Biotechnol. 40, 64–73 (2022).
Zhang, R., Zhou, T. & Ma, J. Multiscale and integrative single-cell Hi-C analysis with Higashi. Nat. Biotechnol. 40, 254–261 (2022).
Chen, K. H., Boettiger, A. N., Moffitt, J. R., Wang, S. & Zhuang, X. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015).
Rodriques, S. G. et al. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science 363, 1463–1467 (2019).
Vickovic, S. et al. High-definition spatial transcriptomics for in situ tissue profiling. Nat. Methods 16, 987–990 (2019).
Liu, B., Li, Y. & Zhang, L. Analysis and visualization of spatial transcriptomic data. Front. Genet. 12, 785290 (2022).
Hu, J. et al. Statistical and machine learning methods for spatially resolved transcriptomics with histology. Comput. Struct. Biotechnol. J. 19, 3829–3841 (2021).
Zeng, Z., Li, Y., Li, Y. & Luo, Y. Statistical and machine learning methods for spatially resolved transcriptomics data analysis. Genome Biol. 23, 83 (2022).
Palla, G., Fischer, D. S., Regev, A. & Theis, F. J. Spatial components of molecular tissue biology. Nat. Biotechnol. 40, 308–318 (2022).
Jiang, R., Sun, T., Song, D. & Li, J. J. Statistics or biology: the zero-inflation controversy about scRNA-seq data. Genome Biol. 23, 31 (2022).
Luecken, M. D. et al. Benchmarking atlas-level data integration in single-cell genomics. Nat. Methods 19, 41–50 (2022). Benchmark of data integration methods of scRNA-seq data sets.
Tran, H. T. N. et al. A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome Biol. 21, 12 (2020).
Song, F., Chan, G. M. A. & Wei, Y. Flexible experimental designs for valid single-cell RNA-sequencing experiments allowing batch effects correction. Nat. Commun. 11, 3274 (2020).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019).
Pliner, H. A., Shendure, J. & Trapnell, C. Supervised classification enables rapid annotation of cell atlases. Nat. Methods 16, 983–986 (2019).
Kiselev, V. Y., Yiu, A. & Hemberg, M. scmap: projection of single-cell RNA-seq data across data sets. Nat. Methods 15, 359–362 (2018).
Aran, D. et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 20, 163–172 (2019).
Cortal, A., Martignetti, L., Six, E. & Rausell, A. Gene signature extraction and cell identity recognition at the single-cell level with Cell-ID. Nat. Biotechnol. 39, 1095–1102 (2021).
Qiu, X. et al. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982 (2017).
Wolf, F. A. et al. PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol. 20, 59 (2019).
Street, K. et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19, 477 (2018).
Velten, L. et al. Human haematopoietic stem cell lineage commitment is a continuous process. Nat. Cell Biol. 19, 271–281 (2017).
Schlitzer, A. et al. Identification of cDC1- and cDC2-committed DC progenitors reveals early lineage priming at the common DC progenitor stage in the bone marrow. Nat. Immunol. 16, 718–728 (2015).
La Manno, G. et al. RNA velocity of single cells. Nature 560, 494–498 (2018).
Bergen, V., Lange, M., Peidli, S., Wolf, F. A. & Theis, F. J. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol. 38, 1408–1414 (2020).
Lange, M. et al. CellRank for directed single-cell fate mapping. Nat. Methods 19, 159–170 (2022).
Hänzelmann, S., Castelo, R. & Guinney, J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinforma. 14, 7 (2013).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Fan, J. et al. Characterizing transcriptional heterogeneity through pathway and gene set overdispersion analysis. Nat. Methods 13, 241–244 (2016).
DeTomaso, D. et al. Functional interpretation of single cell similarity maps. Nat. Commun. 10, 4376 (2019).
Wei, C.-J., Xu, X. & Lo, C. W. Connexins and cell signaling in development and disease. Annu. Rev. Cell Dev. Biol. 20, 811–838 (2004).
Noël, F. et al. Dissection of intercellular communication using the transcriptome-based framework ICELLNET. Nat. Commun. 12, 1089 (2021).
Browaeys, R., Saelens, W. & Saeys, Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat. Methods 17, 159–162 (2020).
Jin, S. et al. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 12, 1088 (2021).
Cabello-Aguilar, S. et al. SingleCellSignalR: inference of intercellular networks from single-cell transcriptomics. Nucleic Acids Res. 48, e55–e55 (2020).
Wang, S., Karikomi, M., MacLean, A. L. & Nie, Q. Cell lineage and communication network inference via optimization for single-cell transcriptomics. Nucleic Acids Res. 47, e66 (2019).
Dimitrov, D. et al. Comparison of methods and resources for cell-cell communication inference from single-cell RNA-Seq data. Nat. Commun. 13, 3224 (2022). A review of methods for inferring intercellular interactions from SC transcriptomics data sets.
Zhang, Q. et al. Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell 179, 829–845.e20 (2019).
Wang, X., Park, J., Susztak, K., Zhang, N. R. & Li, M. Bulk tissue cell type deconvolution with multi-subject single-cell expression reference. Nat. Commun. 10, 380 (2019).
Erdmann-Pham, D. D., Fischer, J., Hong, J. & Song, Y. S. Likelihood-based deconvolution of bulk gene expression data using single-cell references. Genome Res. 31, 1794–1806 (2021).
Wang, J., Roeder, K. & Devlin, B. Bayesian estimation of cell type-specific gene expression with prior derived from single-cell data. Genome Res. 31, 1807–1818 (2021).
Sokolowski, D. J. et al. Single-cell mapper (scMappR): using scRNA-seq to infer the cell-type specificities of differentially expressed genes. NAR Genom. Bioinform. 3, lqab011 (2021).
Newman, A. M. et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 37, 773–782 (2019). This paper presents CIBERSORTx — a method to computationally infer cell-type-specific gene expression profiles and their relative proportions from bulk RNA-seq samples relying on scRNA-seq data sets as the reference for relevant cell types and their markers.
Luca, B. A. et al. Atlas of clinically distinct cell states and ecosystems across human solid tumors. Cell 184, 5482–5496.e28 (2021).
Goldstein, L. D. et al. Massively parallel single-cell B-cell receptor sequencing enables rapid discovery of diverse antigen-reactive antibodies. Commun. Biol. 2, 304 (2019).
Marks, C. & Deane, C. M. How repertoire data are changing antibody science. J. Biol. Chem. 295, 9823–9837 (2020).
Setliff, I. et al. High-throughput mapping of B cell receptor sequences to antigen specificity. Cell 179, 1636–1646.e15 (2019).
Peng, L. et al. Monospecific and bispecific monoclonal SARS-CoV-2 neutralizing antibodies that maintain potency against B.1.617. Nat. Commun. 13, 1638 (2022).
Castellanos-Rueda, R., Di Roberto, R. B., Schlatter, F. S. & Reddy, S. T. Leveraging single-cell sequencing for chimeric antigen receptor T cell therapies. Trends Biotechnol. 39, 1308–1320 (2021). Review paper on how SC sequencing is helping to characterize and identify CAR-T cells.
Li, X. et al. Single-cell transcriptomic analysis reveals BCMA CAR-T cell dynamics in a patient with refractory primary plasma cell leukemia. Mol. Ther. 29, 645–657 (2021). Illustrative example of how scRNA-seq can be used to analyse the dynamics of CAR-T cells in a clinically successful case of relapsed or refractory primary plasma cell leukaemia.
Deng, Q. et al. Characteristics of anti-CD19 CAR T cell infusion products associated with efficacy and toxicity in patients with large B cell lymphomas. Nat. Med. 26, 1878–1887 (2020).
Chen, G. M. et al. Integrative bulk and single-cell profiling of premanufacture T-cell populations reveals factors mediating long-term persistence of CAR T-cell therapy. Cancer Discov. 11, 2186–2199 (2021).
Parker, K. R. et al. Single-cell analyses identify brain mural cells expressing CD19 as potential off-tumor targets for CAR-T immunotherapies. Cell 183, 126–142.e17 (2020).
Jing, Y. et al. Expression of chimeric antigen receptor therapy targets detected by single-cell sequencing of normal cells may contribute to off-tumor toxicity. Cancer Cell 39, 1558–1559 (2021).
Wang, D. et al. CRISPR screening of CAR T cells and cancer stem cells reveals critical dependencies for cell-based therapies. Cancer Discov. 11, 1192–1211 (2021).
Legut, M. et al. A genome-scale screen for synthetic drivers of T cell proliferation. Nature 603, 728–735 (2022).
Kumar, N. et al. Rapid single cell evaluation of human disease and disorder targets using REVEAL: SingleCellTM. BMC Genomics 22, 5 (2021). Illustrative example of how the pharmaceutical industry is using publicly available SC resources internally.
Lachmann, A. et al. Massive mining of publicly available RNA-seq data from human and mouse. Nat. Commun. 9, 1366 (2018).
Collado-Torres, L. et al. Reproducible RNA-seq analysis using recount2. Nat. Biotechnol. 35, 319–321 (2017).
Vivian, J. et al. Toil enables reproducible, open source, big biomedical data analyses. Nat. Biotechnol. 35, 314–316 (2017).
Soneson, C. & Robinson, M. D. Bias, robustness and scalability in single-cell differential expression analysis. Nat. Methods 15, 255–261 (2018).
Stuart, T. & Satija, R. Integrative single-cell analysis. Nat. Rev. Genet. 20, 257–272 (2019).
Luecken, M. D. & Theis, F. J. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol. Syst. Biol. 15, e8746 (2019). Provides best practices on analysing SC transcriptomics data sets.
Cannoodt, R., Saelens, W., Deconinck, L. & Saeys, Y. Spearheading future omics analyses using dyngen, a multi-modal simulator of single cells. Nat. Commun. 12, 3942 (2021).
Treppner, M. et al. Synthetic single cell RNA sequencing data from small pilot studies using deep generative models. Sci. Rep. 11, 9403 (2021).
Zappia, L., Phipson, B. & Oshlack, A. Splatter: simulation of single-cell RNA sequencing data. Genome Biol. 18, 174 (2017).
Saelens, W., Cannoodt, R., Todorov, H. & Saeys, Y. A comparison of single-cell trajectory inference methods. Nat. Biotechnol. 37, 547–554 (2019). A comprehensive review that compares trajectory inference methods for SC data sets and provides guidance on their limitations and usage.
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Mayr, C. H. et al. Integrative analysis of cell state changes in lung fibrosis with peripheral protein biomarkers. EMBO Mol. Med. 13, e12871 (2021).
Nguyen, Q. H., Pervolarakis, N., Nee, K. & Kessenbrock, K. Experimental considerations for single-cell RNA sequencing approaches. Front. Cell Dev. Biol. 6, 108 (2018).
Dal Molin, A. & Di Camillo, B. How to design a single-cell RNA-sequencing experiment: pitfalls, challenges and perspectives. Brief. Bioinform. 20, 1384–1394 (2019).
Acknowledgements
The authors thank I. Papatheodorou (Research Group Leader, EMBL-EBI), B. Kidd (Director, Bristol Myers Squibb (BMS)), R. Loos (Director, BMS) and M. Hall (Senior Scientific Officer, EMBL-EBI) for constructive criticism and proofreading of the original article before this revision.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
N.K. is an employee and shareholder of BMS. M.M. is an employee and shareholder of GSK. B.V.d.S. is an employee and shareholder of UCB Pharma. M.K. is an employee and shareholder of GSK. J.H. is an employee of Boehringer Ingelheim Pharmaceuticals, Inc. B.N. is an employee of Eisai, Inc. J.S.L. is an employee and shareholder of Sanofi. Y.W. was previously a shareholder of BMS. J.P. was previously an employee and shareholder of Sanofi. J.W. is an employee of Pfizer. E.F. is a shareholder of Sanofi and Board Director of Pulmobiotics. A.L. is a GSK shareholder, has consulted for Astex Therapeutics, LifeArc and Syncona and has received research funding from Novo Nordisk and AstraZeneca. X.C. is a former employee and shareholder of AbbVie. E.M.-G., W.B. and J.M. declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Glossary
- Barcode
-
A short DNA sequence ‘tag’ to identify reads that originate from the same cell.
- Biomarkers
-
Readouts used to classify biological states, often in the context of patient stratification.
- Cell-type deconvolution
-
Estimation of the proportion of particular cell types in a bulk RNA sequencing sample, based on cell markers or a labelled single-cell expression matrix.
- CRISPR screening
-
A pooled or arrayed screen of cells harbouring CRISPR-mediated gene edits.
- Doublets
-
Sets of two (or more) cells mistakenly considered as single cells, owing to being captured and processed in the same droplet and thus with the same barcode in data.
- Hashing
-
A labelling technique that attaches barcoded antibodies to cell surface proteins, allowing multiplexing of samples for single-cell sequencing, and subsequent disambiguation of sample of origin during analysis.
- Metadata
-
A set of data that describe and give information about other data (Oxford dictionary). For example, patient or sample characteristics in an RNA sequencing experiment.
- Seurat
-
A popular R package for the quality control, analysis and exploration of single-cell RNA sequencing data.
- Target credentialling
-
Also called target qualification. Exploration of target quality more expansively than a straightforward target validation. May include contextually informed enquiries into biological characteristics such as network, pathway or interactome mapping, regulatory landscape or other investigations intended to either help rank target quality or inform on-target biology.
- t-Distributed stochastic neighbour embedding
-
(t-SNE). A popular dimensionality reduction technique for the visualization of single-cell experiments.
- Trajectory inference
-
Inference from single-cell data of the order of cells along a dynamic biological process (for example, developmental trajectory). Relies on the fact that a heterogeneous sample provides a snapshot view on a mixture of cells in different phases along the developmental or dynamic biological process. Also called ‘pseudo-time analysis’.
- Uniform manifold approximation and projection
-
(UMAP). A popular dimensionality reduction technique for the visualization of single-cell experiments, with some advantages in preservation of global data structure and performance compared with t-distributed stochastic neighbour embedding.
- Unique molecular identifier
-
(UMI). Reads with the same UMI are from the same mRNA molecule. UMIs help in the assessment of sequencing accuracy and precision.
- Unsupervised clustering
-
Analysis grouping of similar samples together that does not require labelling or prior knowledge.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Van de Sande, B., Lee, J.S., Mutasa-Gottgens, E. et al. Applications of single-cell RNA sequencing in drug discovery and development. Nat Rev Drug Discov 22, 496–520 (2023). https://doi.org/10.1038/s41573-023-00688-4
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41573-023-00688-4
This article is cited by
-
Navigating single-cell RNA-sequencing: protocols, tools, databases, and applications
Genomics & Informatics (2025)
-
A dynamic molecular landscape in colorectal cancer progression at single-cell resolution
Journal of Translational Medicine (2025)
-
Transmembrane emp24 domain-containing protein 3 promotes the malignant progression of glioma by regulating the ZBTB7A signaling axis
Molecular Biomedicine (2025)
-
Old drugs, new challenges: reassigning drugs for cancer therapies
Cellular & Molecular Biology Letters (2025)
-
Advancing omics technologies in acute respiratory distress syndrome: paving the way for personalized medicine
Intensive Care Medicine Experimental (2025)