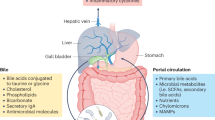
Abstract
Metabolic dysfunction-associated steatotic liver disease (MASLD) is the most prevalent chronic liver disease worldwide, affecting billions of the global population. It can gradually progress to more severe diseases, including steatohepatitis, cirrhosis and hepatocellular carcinoma. Studies have highlighted the importance of the gut microbiome in the pathogenesis and progression of MASLD. On the other hand, increasing evidence has revealed the clinical potential of targeting the gut microbiome to treat MASLD. In this Review, we summarize gut microbial alterations in MASLD, metabolic dysfunction-associated steatohepatitis and hepatocellular carcinoma. The mechanisms by which a dysregulated gut–liver axis contributes to disease progression are also described, including intestinal barrier dysfunction, chronic inflammation, and altered metabolic pathways (for example, bile acids) and microbial-derived metabolites (for example, short-chain fatty acids, tryptophan derivatives and endogenous ethanol). In addition, we discuss the clinical implications of utilizing the gut microbiome as a diagnostic biomarker and the therapeutic approaches to treat MASLD and related diseases such as faecal microbiota transplantation, probiotics and engineered bacteria, prebiotics and postbiotics, microbial-derived metabolites, antimicrobials and bacteriophages. Finally, we discuss current challenges in basic and translational research on the microbiome in MASLD and propose future directions to drive progress in this field.
Key points
-
Progression from metabolic dysfunction-associated steatotic liver disease (MASLD) to steatohepatitis, fibrosis, cirrhosis and, eventually, hepatocellular carcinoma is closely associated with gut microbial dysbiosis and a dysfunctional gut–liver axis.
-
As MASLD progresses, the gut microbiome exhibits enriched Proteobacteria, Clostridium and Streptococcus as well as depleted beneficial bacteria including Eubacterium and Faecalibacterium.
-
A dysregulated gut–liver axis as well as microbial dysbiosis promote MASLD progression by impairing the intestinal barrier, altering microbial-mediated metabolic pathways and metabolites, and contributing to chronic inflammation in the liver.
-
Gut microorganisms and metabolites can be utilized as diagnostic biomarkers of MASLD, which have been shown to stratify individuals predisposed to an increased risk of disease progression with high accuracy.
-
Owing to its pivotal role, emerging studies have developed various therapeutic approaches that target the gut microbiome to treat MASLD, including faecal microbiota transplantation, probiotics, prebiotics and postbiotics, antimicrobials, microbial-derived metabolites and bacteriophages.
-
Research into the microbiome in MASLD is challenged by the lack of mechanistic understanding and consistency across studies, and unclear duration, dosage and efficacy have limited the clinical application of microbiome-targeting interventions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Tripathi, A. et al. The gut–liver axis and the intersection with the microbiome. Nat. Rev. Gastroenterol. Hepatol. 15, 397–411 (2018).
Albillos, A., de Gottardi, A. & Rescigno, M. The gut-liver axis in liver disease: pathophysiological basis for therapy. J. Hepatol. 72, 558–577 (2020).
Hsu, C. L. & Schnabl, B. The gut–liver axis and gut microbiota in health and liver disease. Nat. Rev. Microbiol. 21, 719–733 (2023).
Powell, E. E., Wong, V. W. & Rinella, M. Non-alcoholic fatty liver disease. Lancet 397, 2212–2224 (2021).
Singh, S. et al. Fibrosis progression in nonalcoholic fatty liver vs nonalcoholic steatohepatitis: a systematic review and meta-analysis of paired-biopsy studies. Clin. Gastroenterol. Hepatol. 13, 643–654.e9 (2015).
Chalasani, N. et al. The diagnosis and management of nonalcoholic fatty liver disease: practice guidance from the American Association for the Study of Liver Diseases. Hepatology 67, 328–357 (2018).
Raman, M. et al. Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clin. Gastroenterol. Hepatol. 11, 868–875.e3 (2013).
Wong, V. W. et al. Molecular characterization of the fecal microbiota in patients with nonalcoholic steatohepatitis — a longitudinal study. PLoS ONE 8, e62885 (2013).
Shen, F. et al. Gut microbiota dysbiosis in patients with non-alcoholic fatty liver disease. Hepatobiliary Pancreat. Dis. Int. 16, 375–381 (2017).
Zhu, L. et al. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology 57, 601–609 (2013).
Aron-Wisnewsky, J. et al. Gut microbiota and human NAFLD: disentangling microbial signatures from metabolic disorders. Nat. Rev. Gastroenterol. Hepatol. 17, 279–297 (2020).
Lee, G. et al. Distinct signatures of gut microbiome and metabolites associated with significant fibrosis in non-obese NAFLD. Nat. Commun. 11, 4982 (2020).
Loomba, R. et al. Gut microbiome-based metagenomic signature for non-invasive detection of advanced fibrosis in human nonalcoholic fatty liver disease. Cell Metab. 25, 1054–1062.e5 (2017).
Hoyles, L. et al. Molecular phenomics and metagenomics of hepatic steatosis in non-diabetic obese women. Nat. Med. 24, 1070–1080 (2018).
Kwan, S. Y. et al. Gut microbiome features associated with liver fibrosis in Hispanics, a population at high risk for fatty liver disease. Hepatology 75, 955–967 (2022).
Carr, R. M. et al. An integrated analysis of fecal microbiome and metabolomic features distinguish non-cirrhotic NASH from healthy control populations. Hepatology 78, 1843–1857 (2023).
Schwimmer, J. B. et al. Microbiome signatures associated with steatohepatitis and moderate to severe fibrosis in children with nonalcoholic fatty liver disease. Gastroenterology 157, 1109–1122 (2019).
Yang, R. et al. Effects of probiotics on nonalcoholic fatty liver disease: a systematic review and meta-analysis. Expert Rev. Gastroenterol. Hepatol. 15, 1401–1409 (2021).
Zeng, X. et al. Pien Tze Huang protects against non-alcoholic steatohepatitis by modulating the gut microbiota and metabolites in mice. Engineering 35, 257–269 (2024).
Kajikawa, A. et al. Characterization of flagellins isolated from a highly motile strain of Lactobacillus agilis. BMC Microbiol. 16, 49 (2016).
Kim, D. & Kim, W. R. Nonobese fatty liver disease. Clin. Gastroenterol. Hepatol. 15, 474–485 (2017).
Leung, J. C. et al. Histological severity and clinical outcomes of nonalcoholic fatty liver disease in nonobese patients. Hepatology 65, 54–64 (2017).
Duarte, S. M. B. et al. Gut microbiome composition in lean patients with NASH is associated with liver damage independent of caloric intake: a prospective pilot study. Nutr. Metab. Cardiovasc. Dis. 28, 369–384 (2018).
Iwaki, M. et al. Gut microbiota composition associated with hepatic fibrosis in non-obese patients with non-alcoholic fatty liver disease. J. Gastroenterol. Hepatol. 36, 2275–2284 (2021).
Kuchay, M. S., Martinez-Montoro, J. I., Choudhary, N. S., Fernandez-Garcia, J. C. & Ramos-Molina, B. Non-alcoholic fatty liver disease in lean and non-obese individuals: current and future challenges. Biomedicines 9, 1346 (2021).
Ha, Y. et al. Intimate association of visceral obesity with non-alcoholic fatty liver disease in healthy Asians: a case-control study. J. Gastroenterol. Hepatol. 30, 1666–1672 (2015).
Feng, R. N. et al. Lean-non-alcoholic fatty liver disease increases risk for metabolic disorders in a normal weight Chinese population. World J. Gastroenterol. 20, 17932–17940 (2014).
Qin, N. et al. Alterations of the human gut microbiome in liver cirrhosis. Nature 513, 59–64 (2014).
Ponziani, F. R. et al. Hepatocellular carcinoma is associated with gut microbiota profile and inflammation in nonalcoholic fatty liver disease. Hepatology 69, 107–120 (2019).
Caussy, C. et al. A gut microbiome signature for cirrhosis due to nonalcoholic fatty liver disease. Nat. Commun. 10, 1406 (2019).
Oh, T. G. et al. A universal gut-microbiome-derived signature predicts cirrhosis. Cell Metab. 32, 878–888.e6 (2020).
Behary, J. et al. Gut microbiota impact on the peripheral immune response in non-alcoholic fatty liver disease related hepatocellular carcinoma. Nat. Commun. 12, 187 (2021).
Quevrain, E. et al. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn’s disease. Gut 65, 415–425 (2016).
Lu, H. et al. Butyrate-producing Eubacterium rectale suppresses lymphomagenesis by alleviating the TNF-induced TLR4/MyD88/NF-κB axis. Cell Host Microbe 30, 1139–1150.e7 (2022).
Atarashi, K. et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500, 232–236 (2013).
Narushima, S. et al. Characterization of the 17 strains of regulatory T cell-inducing human-derived Clostridia. Gut Microbes 5, 333–339 (2014).
Scheiman, J. et al. Meta-omics analysis of elite athletes identifies a performance-enhancing microbe that functions via lactate metabolism. Nat. Med. 25, 1104–1109 (2019).
Leung, H. et al. Risk assessment with gut microbiome and metabolite markers in NAFLD development. Sci. Transl Med. 14, eabk0855 (2022).
Rothschild, D. et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 555, 210–215 (2018).
Yatsunenko, T. et al. Human gut microbiome viewed across age and geography. Nature 486, 222–227 (2012).
Qin, J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010).
Arumugam, M. et al. Enterotypes of the human gut microbiome. Nature 473, 174–180 (2011).
Underhill, D. M. & Iliev, I. D. The mycobiota: interactions between commensal fungi and the host immune system. Nat. Rev. Immunol. 14, 405–416 (2014).
You, N. et al. Fecal fungi dysbiosis in nonalcoholic fatty liver disease. Obesity 29, 350–358 (2021).
Demir, M. et al. The fecal mycobiome in non-alcoholic fatty liver disease. J. Hepatol. 76, 788–799 (2022).
Niu, C. et al. Mapping the human oral and gut fungal microbiota in patients with metabolic dysfunction-associated fatty liver disease. Front. Cell Infect. Microbiol. 13, 1157368 (2023).
Viebahn, G. et al. Fungal signature differentiates alcohol-associated liver disease from nonalcoholic fatty liver disease. Gut Microbes 16, 2307586 (2024).
Pak, J., Tucci, V. T., Vincent, A. L., Sandin, R. L. & Greene, J. N. Mucormycosis in immunochallenged patients. J. Emerg. Trauma Shock 1, 106–113 (2008).
Lopes, J. P. & Lionakis, M. S. Pathogenesis and virulence of Candida albicans. Virulence 13, 89–121 (2022).
Guerin, E. et al. Biology and taxonomy of crass-like bacteriophages, the most abundant virus in the human gut. Cell Host Microbe 24, 653–664.e6 (2018).
Lang, S. et al. Intestinal virome signature associated with severity of nonalcoholic fatty liver disease. Gastroenterology 159, 1839–1852 (2020).
Strathdee, S. A., Hatfull, G. F., Mutalik, V. K. & Schooley, R. T. Phage therapy: from biological mechanisms to future directions. Cell 186, 17–31 (2023).
Manrique, P. et al. Healthy human gut phageome. Proc. Natl Acad. Sci. USA 113, 10400–10405 (2016).
Shkoporov, A. N. et al. The human gut virome is highly diverse, stable, and individual specific. Cell Host Microbe 26, 527–541.e5 (2019).
Shkoporov, A. N. & Hill, C. Bacteriophages of the human gut: the “Known Unknown” of the microbiome. Cell Host Microbe 25, 195–209 (2019).
Aggarwala, V., Liang, G. & Bushman, F. D. Viral communities of the human gut: metagenomic analysis of composition and dynamics. Mob. DNA 8, 12 (2017).
Leinwand, J. C. et al. Intrahepatic microbes govern liver immunity by programming NKT cells. J. Clin. Invest. 132, e151725 (2022).
Sookoian, S. et al. Intrahepatic bacterial metataxonomic signature in non-alcoholic fatty liver disease. Gut 69, 1483–1491 (2020).
Liu, B. et al. Hepatic stellate cell activation and senescence induced by intrahepatic microbiota disturbances drive progression of liver cirrhosis toward hepatocellular carcinoma. J. Immunother. Cancer 10, e003069 (2022).
Sun, L. et al. Intratumoural microbiome can predict the prognosis of hepatocellular carcinoma after surgery. Clin. Transl Med. 13, e1331 (2023).
Wang, X. et al. Gut-liver translocation of pathogen Klebsiella pneumoniae promotes hepatocellular carcinoma in mice. Nat. Microbiol. 10, 169–184 (2025).
Huang, J. H. et al. The intratumoral bacterial metataxonomic signature of hepatocellular carcinoma. Microbiol. Spectr. 10, e0098322 (2022).
He, Y., Zhang, Q., Yu, X., Zhang, S. & Guo, W. Overview of microbial profiles in human hepatocellular carcinoma and adjacent nontumor tissues. J. Transl Med. 21, 68 (2023).
Li, S. et al. Intratumoral microbial heterogeneity affected tumor immune microenvironment and determined clinical outcome of HBV-related HCC. Hepatology 78, 1079–1091 (2023).
Effenberger, M. et al. A gut bacterial signature in blood and liver tissue characterizes cirrhosis and hepatocellular carcinoma. Hepatol. Commun. 7, e00182 (2023).
Guo, W. et al. Diet and risk of non-alcoholic fatty liver disease, cirrhosis, and liver cancer: a large prospective cohort study in UK Biobank. Nutrients 14, 5335 (2022).
Petermann-Rocha, F. et al. Associations between an inflammatory diet index and severe non-alcoholic fatty liver disease: a prospective study of 171,544 UK Biobank participants. BMC Med. 21, 123 (2023).
Loomba, R., Friedman, S. L. & Shulman, G. I. Mechanisms and disease consequences of nonalcoholic fatty liver disease. Cell 184, 2537–2564 (2021).
European Association for the Study of the Liver; European Association for the Study of Diabetes; European Association for the Study of Obesity. EASL-EASD-EASO clinical practice guidelines on the management of metabolic dysfunction-associated steatotic liver disease (MASLD). J. Hepatol. 81, 492–542 (2024).
George, E. S. et al. The association between diet and hepatocellular carcinoma: a systematic review. Nutrients 13, 172 (2021).
Yaskolka Meir, A. et al. Effect of green-Mediterranean diet on intrahepatic fat: the DIRECT PLUS randomised controlled trial. Gut 70, 2085–2095 (2021).
Vilar-Gomez, E. et al. High-quality diet, physical activity, and college education are associated with low risk of NAFLD among the US population. Hepatology 75, 1491–1506 (2022).
Zhang, X. et al. Dietary cholesterol drives fatty liver-associated liver cancer by modulating gut microbiota and metabolites. Gut 70, 761–774 (2021).
Wei, W. et al. Parabacteroides distasonis uses dietary inulin to suppress NASH via its metabolite pentadecanoic acid. Nat. Microbiol. 8, 1534–1548 (2023).
Lange, N. F., Radu, P. & Dufour, J. F. Prevention of NAFLD-associated HCC: role of lifestyle and chemoprevention. J. Hepatol. 75, 1217–1227 (2021).
Chen, B. et al. Gut bacteria alleviate smoking-related NASH by degrading gut nicotine. Nature 610, 562–568 (2022).
Zhang, X., Chang, K. M., Yu, J. & Loomba, R. Unraveling mechanisms of genetic risks in metabolic dysfunction-associated steatotic liver diseases: a pathway to precision medicine. Annu. Rev. Pathol. 20, 375–403 (2025).
Ha, S., Wong, V. W., Zhang, X. & Yu, J. Interplay between gut microbiome, host genetic and epigenetic modifications in MASLD and MASLD-related hepatocellular carcinoma. Gut 74, 141–152 (2025).
Zhang, X. et al. Intestinal TM6SF2 protects against metabolic dysfunction-associated steatohepatitis through the gut-liver axis. Nat. Metab. 7, 102–119 (2025).
Harte, A. L. et al. Elevated endotoxin levels in non-alcoholic fatty liver disease. J. Inflamm. 7, 15 (2010).
Verdam, F. J., Rensen, S. S., Driessen, A., Greve, J. W. & Buurman, W. A. Novel evidence for chronic exposure to endotoxin in human nonalcoholic steatohepatitis. J. Clin. Gastroenterol. 45, 149–152 (2011).
Giorgio, V. et al. Intestinal permeability is increased in children with non-alcoholic fatty liver disease, and correlates with liver disease severity. Dig. Liver Dis. 46, 556–560 (2014).
Ruiz, A. G. et al. Lipopolysaccharide-binding protein plasma levels and liver TNF-alpha gene expression in obese patients: evidence for the potential role of endotoxin in the pathogenesis of non-alcoholic steatohepatitis. Obes. Surg. 17, 1374–1380 (2007).
Miele, L. et al. Increased intestinal permeability and tight junction alterations in nonalcoholic fatty liver disease. Hepatology 49, 1877–1887 (2009).
Xin, D., Zong-Shun, L., Bang-Mao, W. & Lu, Z. Expression of intestinal tight junction proteins in patients with non-alcoholic fatty liver disease. Hepatogastroenterology 61, 136–140 (2014).
Rahman, K. et al. Loss of junctional adhesion molecule a promotes severe steatohepatitis in mice on a diet high in saturated fat, fructose, and cholesterol. Gastroenterology 151, 733–746.e12 (2016).
Mouries, J. et al. Microbiota-driven gut vascular barrier disruption is a prerequisite for non-alcoholic steatohepatitis development. J. Hepatol. 71, 1216–1228 (2019).
Feng, Y., Wang, Y., Wang, P., Huang, Y. & Wang, F. Short-chain fatty acids manifest stimulative and protective effects on intestinal barrier function through the inhibition of NLRP3 inflammasome and autophagy. Cell Physiol. Biochem. 49, 190–205 (2018).
Singh, R. et al. Enhancement of the gut barrier integrity by a microbial metabolite through the Nrf2 pathway. Nat. Commun. 10, 89 (2019).
Ghosh, S., Whitley, C. S., Haribabu, B. & Jala, V. R. Regulation of intestinal barrier function by microbial metabolites. Cell Mol. Gastroenterol. Hepatol. 11, 1463–1482 (2021).
Ravcheev, D. A. & Thiele, I. Systematic genomic analysis reveals the complementary aerobic and anaerobic respiration capacities of the human gut microbiota. Front. Microbiol. 5, 674 (2014).
Patel, D. & Witt, S. N. Ethanolamine and phosphatidylethanolamine: partners in health and disease. Oxid. Med. Cell Longev. 2017, 4829180 (2017).
Mishra, S. P. et al. A mechanism by which gut microbiota elevates permeability and inflammation in obese/diabetic mice and human gut. Gut 72, 1848–1865 (2023).
Jiao, N. et al. Suppressed hepatic bile acid signalling despite elevated production of primary and secondary bile acids in NAFLD. Gut 67, 1881–1891 (2018).
Smirnova, E. et al. Metabolic reprogramming of the intestinal microbiome with functional bile acid changes underlie the development of NAFLD. Hepatology 76, 1811–1824 (2022).
Yoshimoto, S. et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 499, 97–101 (2013).
Xie, G. et al. Dysregulated hepatic bile acids collaboratively promote liver carcinogenesis. Int. J. Cancer 139, 1764–1775 (2016).
Jia, W., Xie, G. & Jia, W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat. Rev. Gastroenterol. Hepatol. 15, 111–128 (2018).
Loo, T. M. et al. Gut microbiota promotes obesity-associated liver cancer through PGE2-mediated suppression of antitumor immunity. Cancer Discov. 7, 522–538 (2017).
Liu, L. et al. Deoxycholic acid disrupts the intestinal mucosal barrier and promotes intestinal tumorigenesis. Food Funct. 9, 5588–5597 (2018).
Zhou, H. et al. Bile acid toxicity in Paneth cells contributes to gut dysbiosis induced by high-fat feeding. JCI Insight 5, e138881 (2020).
Ma, C. et al. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science 360, eaan5931 (2018).
Kuang, J. et al. Hyodeoxycholic acid alleviates non-alcoholic fatty liver disease through modulating the gut-liver axis. Cell Metab. 35, 1752–1766.e8 (2023).
Nie, Q. et al. Gut symbionts alleviate MASH through a secondary bile acid biosynthetic pathway. Cell 187, 2717–2734.e33 (2024).
Canfora, E. E., Meex, R. C. R., Venema, K. & Blaak, E. E. Gut microbial metabolites in obesity, NAFLD and T2DM. Nat. Rev. Endocrinol. 15, 261–273 (2019).
Chambers, E. S. et al. Effects of targeted delivery of propionate to the human colon on appetite regulation, body weight maintenance and adiposity in overweight adults. Gut 64, 1744–1754 (2015).
Aoki, R. et al. Commensal microbe-derived acetate suppresses NAFLD/NASH development via hepatic FFAR2 signalling in mice. Microbiome 9, 188 (2021).
Yang, X. et al. Inulin-enriched Megamonas funiformis ameliorates metabolic dysfunction-associated fatty liver disease by producing propionic acid. npj Biofilms Microbiomes 9, 84 (2023).
Gao, Z. et al. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes 58, 1509–1517 (2009).
Zhao, Z. H. et al. Sodium butyrate supplementation inhibits hepatic steatosis by stimulating liver kinase B1 and insulin-induced gene. Cell Mol. Gastroenterol. Hepatol. 12, 857–871 (2021).
Xia, J. K. et al. Roles and regulation of histone acetylation in hepatocellular carcinoma. Front. Genet. 13, 982222 (2022).
Olaniyi, K. S. & Amusa, O. A. Sodium acetate-mediated inhibition of histone deacetylase alleviates hepatic lipid dysregulation and its accompanied injury in streptozotocin-nicotinamide-induced diabetic rats. Biomed. Pharmacother. 128, 110226 (2020).
Sarkar, A. et al. Butyrate limits inflammatory macrophage niche in NASH. Cell Death Dis. 14, 332 (2023).
Song, Q. et al. Bifidobacterium pseudolongum-generated acetate suppresses non-alcoholic fatty liver disease-associated hepatocellular carcinoma. J. Hepatol. 79, 1352–1365 (2023).
Lau, H. C. et al. Lactobacillus acidophilus suppresses non-alcoholic fatty liver disease-associated hepatocellular carcinoma through producing valeric acid. EBioMedicine 100, 104952 (2024).
Kim, M. H., Kang, S. G., Park, J. H., Yanagisawa, M. & Kim, C. H. Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology 145, 396–406.e10 (2013).
Belcheva, A. et al. Gut microbial metabolism drives transformation of MSH2-deficient colon epithelial cells. Cell 158, 288–299 (2014).
Singh, V. et al. Microbiota-dependent hepatic lipogenesis mediated by stearoyl CoA desaturase 1 (SCD1) promotes metabolic syndrome in TLR5-deficient mice. Cell Metab. 22, 983–996 (2015).
Singh, V. et al. Dysregulated microbial fermentation of soluble fiber induces cholestatic liver cancer. Cell 175, 679–694.e22 (2018).
Samuel, B. S. et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc. Natl Acad. Sci. USA 105, 16767–16772 (2008).
Bjursell, M. et al. Improved glucose control and reduced body fat mass in free fatty acid receptor 2-deficient mice fed a high-fat diet. Am. J. Physiol. Endocrinol. Metab. 300, E211–E220 (2011).
Chu, H., Duan, Y., Yang, L. & Schnabl, B. Small metabolites, possible big changes: a microbiota-centered view of non-alcoholic fatty liver disease. Gut 68, 359–370 (2019).
Rau, M. et al. Fecal SCFAs and SCFA-producing bacteria in gut microbiome of human NAFLD as a putative link to systemic T-cell activation and advanced disease. United European Gastroenterol. J. 6, 1496–1507 (2018).
Michail, S. et al. Altered gut microbial energy and metabolism in children with non-alcoholic fatty liver disease. FEMS Microbiol. Ecol. 91, 1–9 (2015).
Deng, K. et al. Comparison of fecal and blood metabolome reveals inconsistent associations of the gut microbiota with cardiometabolic diseases. Nat. Commun. 14, 571 (2023).
Badawy, A. A. Kynurenine pathway of tryptophan metabolism: regulatory and functional aspects. Int. J. Tryptophan Res. 10, 1178646917691938 (2017).
Agus, A., Planchais, J. & Sokol, H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe 23, 716–724 (2018).
Sui, G., Jia, L., Quan, D., Zhao, N. & Yang, G. Activation of the gut microbiota-kynurenine-liver axis contributes to the development of nonalcoholic hepatic steatosis in nondiabetic adults. Aging 13, 21309–21324 (2021).
Arto, C. et al. Metabolic profiling of tryptophan pathways: implications for obesity and metabolic dysfunction-associated steatotic liver disease. Eur. J. Clin. Invest. 54, e14279 (2024).
Wang, L. et al. Gut-derived serotonin contributes to the progression of non-alcoholic steatohepatitis via the liver HTR2A/PPARγ2 pathway. Front. Pharmacol. 11, 553 (2020).
Wegermann, K. et al. Serum bile acid, vitamin E, and serotonin metabolites are associated with future liver-related events in nonalcoholic fatty liver disease. Hepatol. Commun. 5, 608–617 (2021).
Opitz, C. A. et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature 478, 197–203 (2011).
Fujigaki, S. et al. Lipopolysaccharide induction of indoleamine 2,3-dioxygenase is mediated dominantly by an IFN-γ-independent mechanism. Eur. J. Immunol. 31, 2313–2318 (2001).
Laurans, L. et al. Genetic deficiency of indoleamine 2,3-dioxygenase promotes gut microbiota-mediated metabolic health. Nat. Med. 24, 1113–1120 (2018).
Choi, W. et al. Serotonin signals through a gut-liver axis to regulate hepatic steatosis. Nat. Commun. 9, 4824 (2018).
Crane, J. D. et al. Inhibiting peripheral serotonin synthesis reduces obesity and metabolic dysfunction by promoting brown adipose tissue thermogenesis. Nat. Med. 21, 166–172 (2015).
Gao, K., Mu, C. L., Farzi, A. & Zhu, W. Y. Tryptophan metabolism: a link between the gut microbiota and brain. Adv. Nutr. 11, 709–723 (2020).
Rothhammer, V. et al. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat. Med. 22, 586–597 (2016).
Stockinger, B., Shah, K. & Wincent, E. AHR in the intestinal microenvironment: safeguarding barrier function. Nat. Rev. Gastroenterol. Hepatol. 18, 559–570 (2021).
Ma, L. et al. Indole alleviates diet-induced hepatic steatosis and inflammation in a manner involving myeloid cell 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3. Hepatology 72, 1191–1203 (2020).
Yu, J. S. et al. Lactobacillus lactis and Pediococcus pentosaceus-driven reprogramming of gut microbiome and metabolome ameliorates the progression of non-alcoholic fatty liver disease. Clin. Transl Med. 11, e634 (2021).
Ji, Y., Gao, Y., Chen, H., Yin, Y. & Zhang, W. Indole-3-acetic acid alleviates nonalcoholic fatty liver disease in mice via attenuation of hepatic lipogenesis, and oxidative and inflammatory stress. Nutrients 11, 2062 (2019).
Zhao, Z. H. et al. Indole-3-propionic acid inhibits gut dysbiosis and endotoxin leakage to attenuate steatohepatitis in rats. Exp. Mol. Med. 51, 1–14 (2019).
Zhu, B. et al. Indole supplementation ameliorates MCD-induced NASH in mice. J. Nutr. Biochem. 107, 109041 (2022).
Mbaye, B. et al. A catalog of ethanol-producing microbes in humans. Future Microbiol. 19, 697–714 (2024).
Yuan, J. et al. Fatty liver disease caused by high-alcohol-producing Klebsiella pneumoniae. Cell Metab. 30, 675–688.e7 (2019).
Meijnikman, A. S. et al. Microbiome-derived ethanol in nonalcoholic fatty liver disease. Nat. Med. 28, 2100–2106 (2022).
Mbaye, B. et al. Endogenous ethanol and triglyceride production by gut Pichia kudriavzevii, Candida albicans and Candida glabrata yeasts in non-alcoholic steatohepatitis. Cells 11, 3390 (2022).
Seki, E. et al. TLR4 enhances TGF-β signaling and hepatic fibrosis. Nat. Med. 13, 1324–1332 (2007).
Rivera, C. A. et al. Toll-like receptor-4 signaling and Kupffer cells play pivotal roles in the pathogenesis of non-alcoholic steatohepatitis. J. Hepatol. 47, 571–579 (2007).
Yu, J. et al. Hepatocyte TLR4 triggers inter-hepatocyte Jagged1/Notch signaling to determine NASH-induced fibrosis. Sci. Transl Med. 13, eabe1692 (2021).
Swanson, K. V., Deng, M. & Ting, J. P. The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat. Rev. Immunol. 19, 477–489 (2019).
Csak, T. et al. Fatty acid and endotoxin activate inflammasomes in mouse hepatocytes that release danger signals to stimulate immune cells. Hepatology 54, 133–144 (2011).
Vandanmagsar, B. et al. The NLRP3 inflammasome instigates obesity-induced inflammation and insulin resistance. Nat. Med. 17, 179–188 (2011).
Gaul, S. et al. Hepatocyte pyroptosis and release of inflammasome particles induce stellate cell activation and liver fibrosis. J. Hepatol. 74, 156–167 (2021).
Henao-Mejia, J. et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature 482, 179–185 (2012).
Pierantonelli, I. et al. Lack of NLRP3-inflammasome leads to gut-liver axis derangement, gut dysbiosis and a worsened phenotype in a mouse model of NAFLD. Sci. Rep. 7, 12200 (2017).
Kaufmann, B. et al. Cell-specific deletion of NLRP3 inflammasome identifies myeloid cells as key drivers of liver inflammation and fibrosis in murine steatohepatitis. Cell Mol. Gastroenterol. Hepatol. 14, 751–767 (2022).
Remetic, J. et al. Loss of bile salt export pump aggravates lipopolysaccharide-induced liver injury in mice due to impaired hepatic endotoxin clearance. Hepatology 75, 1095–1109 (2022).
Levine, D. M., Parker, T. S., Donnelly, T. M., Walsh, A. & Rubin, A. L. In vivo protection against endotoxin by plasma high density lipoprotein. Proc. Natl Acad. Sci. USA 90, 12040–12044 (1993).
Han, Y. H. et al. Enterically derived high-density lipoprotein restrains liver injury through the portal vein. Science 373, eabe6729 (2021).
Nikeghbalian, S. et al. Correlation between HDL2, HDL3 and serum ferritin levels with fatty liver and NAFLD activity score (NAS) in liver histology of organ donors. BMC Gastroenterol. 21, 405 (2021).
Jing, Y. Y. et al. Toll-like receptor 4 signaling promotes epithelial-mesenchymal transition in human hepatocellular carcinoma induced by lipopolysaccharide. BMC Med. 10, 98 (2012).
Liu, W. T. et al. Toll like receptor 4 facilitates invasion and migration as a cancer stem cell marker in hepatocellular carcinoma. Cancer Lett. 358, 136–143 (2015).
Dapito, D. H. et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell 21, 504–516 (2012).
Liu, W. T. et al. Lipopolysaccharide induces the differentiation of hepatic progenitor cells into myofibroblasts constitutes the hepatocarcinogenesis-associated microenvironment. Cell Death Differ. 27, 85–101 (2020).
Ansari, S. R. et al. Risk factors for infections with multidrug-resistant Stenotrophomonas maltophilia in patients with cancer. Cancer 109, 2615–2622 (2007).
Safdar, A. & Rolston, K. V. Stenotrophomonas maltophilia: changing spectrum of a serious bacterial pathogen in patients with cancer. Clin. Infect. Dis. 45, 1602–1609 (2007).
Barrow, F. et al. Microbiota-driven activation of intrahepatic B cells aggravates NASH through innate and adaptive signaling. Hepatology 74, 704–722 (2021).
Kotsiliti, E. et al. Intestinal B cells license metabolic T-cell activation in NASH microbiota/antigen-independently and contribute to fibrosis by IgA-FcR signalling. J. Hepatol. 79, 296–313 (2023).
Miyamoto, Y. et al. Periportal macrophages protect against commensal-driven liver inflammation. Nature 629, 901–909 (2024).
Pfister, D. et al. NASH limits anti-tumour surveillance in immunotherapy-treated HCC. Nature 592, 450–456 (2021).
Li, M. et al. Spatial proteomics of immune microenvironment in nonalcoholic steatohepatitis-associated hepatocellular carcinoma. Hepatology 79, 560–574 (2024).
Tincopa, M. A. & Loomba, R. Non-invasive diagnosis and monitoring of non-alcoholic fatty liver disease and non-alcoholic steatohepatitis. Lancet Gastroenterol. Hepatol. 8, 660–670 (2023).
Guha, I. N. et al. Noninvasive markers of fibrosis in nonalcoholic fatty liver disease: validating the European liver fibrosis panel and exploring simple markers. Hepatology 47, 455–460 (2008).
Harrison, S. A. et al. A blood-based biomarker panel (NIS4) for non-invasive diagnosis of non-alcoholic steatohepatitis and liver fibrosis: a prospective derivation and global validation study. Lancet Gastroenterol. Hepatol. 5, 970–985 (2020).
Vali, Y. et al. Biomarkers for staging fibrosis and non-alcoholic steatohepatitis in non-alcoholic fatty liver disease (the LITMUS project): a comparative diagnostic accuracy study. Lancet Gastroenterol. Hepatol. 8, 714–725 (2023).
Zhang, X. et al. A blood-based biomarker panel for non-invasive diagnosis of metabolic dysfunction-associated steatohepatitis. Cell Metab. 37, 59–68.e3 (2025).
Lee, P. C. et al. Gut microbiota and metabolites associate with outcomes of immune checkpoint inhibitor-treated unresectable hepatocellular carcinoma. J. Immunother. Cancer 10, e004779 (2022).
Ponziani, F. R. et al. Gut dysbiosis and fecal calprotectin predict response to immune checkpoint inhibitors in patients with hepatocellular carcinoma. Hepatol. Commun. 6, 1492–1501 (2022).
Derosa, L. et al. Intestinal Akkermansia muciniphila predicts clinical response to PD-1 blockade in patients with advanced non-small-cell lung cancer. Nat. Med. 28, 315–324 (2022).
Iida, N. et al. Overuse of antianaerobic drug is associated with poor postchemotherapy prognosis of patients with hepatocellular carcinoma. Int. J. Cancer 145, 2701–2711 (2019).
Zhou, D. et al. Total fecal microbiota transplantation alleviates high-fat diet-induced steatohepatitis in mice via beneficial regulation of gut microbiota. Sci. Rep. 7, 1529 (2017).
Xue, L., Deng, Z., Luo, W., He, X. & Chen, Y. Effect of fecal microbiota transplantation on non-alcoholic fatty liver disease: a randomized clinical trial. Front. Cell Infect. Microbiol. 12, 759306 (2022).
Craven, L. et al. Allogenic fecal microbiota transplantation in patients with nonalcoholic fatty liver disease improves abnormal small intestinal permeability: a randomized control trial. Am. J. Gastroenterol. 115, 1055–1065 (2020).
Witjes, J. J. et al. Donor fecal microbiota transplantation alters gut microbiota and metabolites in obese individuals with steatohepatitis. Hepatol. Commun. 4, 1578–1590 (2020).
Stols-Goncalves, D. et al. Faecal microbiota transplantation affects liver DNA methylation in non-alcoholic fatty liver disease: a multi-omics approach. Gut Microbes 15, 2223330 (2023).
Bajaj, J. S. et al. Association between intestinal microbiota collected at hospital admission and outcomes of patients with cirrhosis. Clin. Gastroenterol. Hepatol. 17, 756–765.e3 (2019).
Cammarota, G. et al. European consensus conference on faecal microbiota transplantation in clinical practice. Gut 66, 569–580 (2017).
Loguercio, C. et al. Beneficial effects of a probiotic VSL#3 on parameters of liver dysfunction in chronic liver diseases. J. Clin. Gastroenterol. 39, 540–543 (2005).
Alisi, A. et al. Randomised clinical trial: the beneficial effects of VSL#3 in obese children with non-alcoholic steatohepatitis. Aliment. Pharmacol. Ther. 39, 1276–1285 (2014).
Duseja, A. et al. High potency multistrain probiotic improves liver histology in non-alcoholic fatty liver disease (NAFLD): a randomised, double-blind, proof of concept study. BMJ Open Gastroenterol. 6, e000315 (2019).
Chong, P. L., Laight, D., Aspinall, R. J., Higginson, A. & Cummings, M. H. A randomised placebo controlled trial of VSL#3 probiotic on biomarkers of cardiovascular risk and liver injury in non-alcoholic fatty liver disease. BMC Gastroenterol. 21, 144 (2021).
Wong, V. W. et al. Treatment of nonalcoholic steatohepatitis with probiotics: a proof-of-concept study. Ann. Hepatol. 12, 256–262 (2013).
Aller, R. et al. Effect of a probiotic on liver aminotransferases in nonalcoholic fatty liver disease patients: a double blind randomized clinical trial. Eur. Rev. Med. Pharmacol. Sci. 15, 1090–1095 (2011).
Dao, M. C. et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology. Gut 65, 426–436 (2016).
Zhu, W., Yan, M., Cao, H., Zhou, J. & Xu, Z. Effects of Clostridium butyricum capsules combined with rosuvastatin on intestinal flora, lipid metabolism, liver function and inflammation in NAFLD patients. Cell Mol. Biol. 68, 64–69 (2022).
Everard, A., Matamoros, S., Geurts, L., Delzenne, N. M. & Cani, P. D. Saccharomyces boulardii administration changes gut microbiota and reduces hepatic steatosis, low-grade inflammation, and fat mass in obese and type 2 diabetic db/db mice. mBio 5, e01011-14 (2014).
Yang, A. M. et al. Saccharomyces boulardii ameliorates non-alcoholic steatohepatitis in mice induced by a methionine-choline-deficient diet through gut-liver axis. Front. Microbiol. 13, 887728 (2022).
Guslandi, M., Giollo, P. & Testoni, P. A. A pilot trial of Saccharomyces boulardii in ulcerative colitis. Eur. J. Gastroenterol. Hepatol. 15, 697–698 (2003).
Lang, S. & Schnabl, B. Microbiota and fatty liver disease-the known, the unknown, and the future. Cell Host Microbe 28, 233–244 (2020).
Hendrikx, T. et al. Bacteria engineered to produce IL-22 in intestine induce expression of REG3G to reduce ethanol-induced liver disease in mice. Gut 68, 1504–1515 (2019).
Huang, X. et al. Fructooligosaccharides attenuate non-alcoholic fatty liver disease by remodeling gut microbiota and association with lipid metabolism. Biomed. Pharmacother. 159, 114300 (2023).
Ni, Y. et al. Resistant starch decreases intrahepatic triglycerides in patients with NAFLD via gut microbiome alterations. Cell Metab. 35, 1530–1547.e8 (2023).
Zhang, X., Yu, J. & Wong, V. W. Vive la resistant starch: a potential treatment for metabolic dysfunction-associated steatohepatitis. Cell Metab. 35, 1491–1493 (2023).
Krawczyk, M. et al. Gut permeability might be improved by dietary fiber in individuals with nonalcoholic fatty liver disease (NAFLD) undergoing weight reduction. Nutrients 10, 1793 (2018).
Scorletti, E. et al. Synbiotics alter fecal microbiomes, but not liver fat or fibrosis, in a randomized trial of patients with nonalcoholic fatty liver disease. Gastroenterology 158, 1597–1610.e7 (2020).
Salminen, S. et al. The International Scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat. Rev. Gastroenterol. Hepatol. 18, 649–667 (2021).
Plovier, H. et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat. Med. 23, 107–113 (2017).
Depommier, C. et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat. Med. 25, 1096–1103 (2019).
Brandt, A. et al. Antibiotics attenuate diet-induced nonalcoholic fatty liver disease without altering intestinal barrier dysfunction. J. Nutr. Biochem. 123, 109495 (2024).
Jian, J. et al. Rifaximin ameliorates non-alcoholic steatohepatitis in mice through regulating gut microbiome-related bile acids. Front. Pharmacol. 13, 841132 (2022).
Abdel-Razik, A. et al. Rifaximin in nonalcoholic fatty liver disease: hit multiple targets with a single shot. Eur. J. Gastroenterol. Hepatol. 30, 1237–1246 (2018).
Wu, P. et al. Ursodeoxycholic acid alleviates nonalcoholic fatty liver disease by inhibiting apoptosis and improving autophagy via activating AMPK. Biochem. Biophys. Res. Commun. 529, 834–838 (2020).
Leuschner, U. F. et al. High-dose ursodeoxycholic acid therapy for nonalcoholic steatohepatitis: a double-blind, randomized, placebo-controlled trial. Hepatology 52, 472–479 (2010).
Zhang, W., Tang, Y., Huang, J. & Hu, H. Efficacy of ursodeoxycholic acid in nonalcoholic fatty liver disease: an updated meta-analysis of randomized controlled trials. Asia Pac. J. Clin. Nutr. 29, 696–705 (2020).
Patel, V. S. et al. Ursodeoxycholic acid’s effectiveness in the management of nonalcoholic fatty liver disease: a systematic review and meta-analysis. Euroasian J. Hepatogastroenterol. 14, 92–98 (2024).
Li, X. et al. Short-chain fatty acids in nonalcoholic fatty liver disease: new prospects for short-chain fatty acids as therapeutic targets. Heliyon 10, e26991 (2024).
Fang, W., Xue, H., Chen, X., Chen, K. & Ling, W. Supplementation with sodium butyrate modulates the composition of the gut microbiota and ameliorates high-fat diet-induced obesity in mice. J. Nutr. 149, 747–754 (2019).
Gan, L. et al. Bacteriophage targeting microbiota alleviates non-alcoholic fatty liver disease induced by high alcohol-producing Klebsiella pneumoniae. Nat. Commun. 14, 3215 (2023).
Federici, S. et al. Targeted suppression of human IBD-associated gut microbiota commensals by phage consortia for treatment of intestinal inflammation. Cell 185, 2879–2898.e24 (2022).
Hsu, C. L., Duan, Y., Fouts, D. E. & Schnabl, B. Intestinal virome and therapeutic potential of bacteriophages in liver disease. J. Hepatol. 75, 1465–1475 (2021).
Hampton, H. G., Watson, B. N. J. & Fineran, P. C. The arms race between bacteria and their phage foes. Nature 577, 327–336 (2020).
Gu, J. et al. A method for generation phage cocktail with great therapeutic potential. PLoS ONE 7, e31698 (2012).
Kirby, A. E. Synergistic action of gentamicin and bacteriophage in a continuous culture population of Staphylococcus aureus. PLoS ONE 7, e51017 (2012).
Verma, V., Harjai, K. & Chhibber, S. Restricting ciprofloxacin-induced resistant variant formation in biofilm of Klebsiella pneumoniae B5055 by complementary bacteriophage treatment. J. Antimicrob. Chemother. 64, 1212–1218 (2009).
Oechslin, F. Resistance development to bacteriophages occurring during bacteriophage therapy. Viruses 10, 351 (2018).
Shi, Y., Wang, G., Lau, H. C. & Yu, J. Metagenomic sequencing for microbial DNA in human samples: emerging technological advances. Int. J. Mol. Sci. 23, 2181 (2022).
Meslier, V. et al. Benchmarking second and third-generation sequencing platforms for microbial metagenomics. Sci. Data 9, 694 (2022).
Pereira, M. B., Wallroth, M., Jonsson, V. & Kristiansson, E. Comparison of normalization methods for the analysis of metagenomic gene abundance data. BMC Genom. 19, 274 (2018).
Lucchetti, M. et al. An organ-on-chip platform for simulating drug metabolism along the gut-liver Axis. Adv. Healthc. Mater. 13, e2303943 (2024).
Sun, Y. et al. Integrative plasma and fecal metabolomics identify functional metabolites in adenoma-colorectal cancer progression and as early diagnostic biomarkers. Cancer Cell 42, 1386–1400.e8 (2024).
Asnicar, F. et al. Blue poo: impact of gut transit time on the gut microbiome using a novel marker. Gut 70, 1665–1674 (2021).
Roager, H. M. et al. Colonic transit time is related to bacterial metabolism and mucosal turnover in the gut. Nat. Microbiol. 1, 16093 (2016).
Falony, G. et al. Population-level analysis of gut microbiome variation. Science 352, 560–564 (2016).
Prochazkova, N. et al. Advancing human gut microbiota research by considering gut transit time. Gut 72, 180–191 (2023).
Raba, G., Adamberg, S. & Adamberg, K. Acidic pH enhances butyrate production from pectin by faecal microbiota. FEMS Microbiol. Lett. 368, fnab042 (2021).
Ting, N. L., Lau, H. C. & Yu, J. Cancer pharmacomicrobiomics: targeting microbiota to optimise cancer therapy outcomes. Gut 71, 1412–1425 (2022).
Yang, Y. et al. Fecal microbiota transplantation: no longer cinderella in tumour immunotherapy. EBioMedicine 100, 104967 (2024).
Fong, W. et al. Lactobacillus gallinarum-derived metabolites boost anti-PD1 efficacy in colorectal cancer by inhibiting regulatory T cells through modulating IDO1/Kyn/AHR axis. Gut 72, 2272–2285 (2023).
Del Chierico, F. et al. Gut microbiota profiling of pediatric nonalcoholic fatty liver disease and obese patients unveiled by an integrated meta-omics-based approach. Hepatology 65, 451–464 (2017).
Boursier, J. et al. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology 63, 764–775 (2016).
Ratziu, V. et al. A randomized controlled trial of high-dose ursodesoxycholic acid for nonalcoholic steatohepatitis. J. Hepatol. 54, 1011–1019 (2011).
Rinella, M. E. et al. A multisociety Delphi consensus statement on new fatty liver disease nomenclature. J. Hepatol. 79, 1542–1556 (2023).
Younossi, Z. M. et al. The global epidemiology of NAFLD and NASH in patients with type 2 diabetes: a systematic review and meta-analysis. J. Hepatol. 71, 793–801 (2019).
Younossi, Z. M. et al. The global epidemiology of nonalcoholic fatty liver disease (NAFLD) and nonalcoholic steatohepatitis (NASH): a systematic review. Hepatology 77, 1335–1347 (2023).
Huang, D. Q., El-Serag, H. B. & Loomba, R. Global epidemiology of NAFLD-related HCC: trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 18, 223–238 (2021).
The Lancet Gastroenterology Hepatology. Resmetirom for NASH: balancing promise and prudence. Lancet Gastroenterol. Hepatol. 9, 273 (2024).
Newsome, P. N. et al. A placebo-controlled trial of subcutaneous semaglutide in nonalcoholic steatohepatitis. N. Engl. J. Med. 384, 1113–1124 (2021).
Loomba, R. et al. Randomized, controlled trial of the FGF21 analogue pegozafermin in NASH. N. Engl. J. Med. 389, 998–1008 (2023).
Harrison, S. A. et al. Safety and efficacy of once-weekly efruxifermin versus placebo in non-alcoholic steatohepatitis (HARMONY): a multicentre, randomised, double-blind, placebo-controlled, phase 2b trial. Lancet Gastroenterol. Hepatol. 8, 1080–1093 (2023).
Francque, S. M. et al. A randomized, controlled trial of the Pan-PPAR agonist lanifibranor in NASH. N. Engl. J. Med. 385, 1547–1558 (2021).
Acknowledgements
The authors were supported by the Noncommunicable Chronic Diseases-National Science and Technology Major Project (2023ZD0500200), the National Natural Science Foundation of China (82222901 and 82272619) and the Research Grants Council-General Research Fund (CUHK14117422, CUHK14119524 and CUHK14117123).
Author information
Authors and Affiliations
Contributions
H.C.-H.L. and X.Z. researched data for the article and wrote the manuscript. J.Y. made a substantial contribution to discussion of content and reviewed/edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Gastroenterology & Hepatology thanks Münevver Demir and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lau, H.CH., Zhang, X. & Yu, J. Gut microbiome in metabolic dysfunction-associated steatotic liver disease and associated hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol 22, 619–638 (2025). https://doi.org/10.1038/s41575-025-01089-1
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41575-025-01089-1