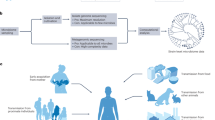
Abstract
Human microbiomes are essential to health throughout the lifespan and are increasingly recognized and studied for their roles in metabolic, immunological and neurological processes. Although the full complexity of these microbial communities is not fully understood, their clinical and industrial exploitation is well advanced and expanding, needing greater oversight guided by a consensus from the research community. One of the most controversial issues in microbiome research is the definition of a ‘healthy’ human microbiome. This concept is complicated by the microbial variability over different spatial and temporal scales along with the challenge of applying a unified definition to the spectrum of healthy microbiome configurations. In this Perspective, we examine the progress made and the key gaps that remain to be addressed to fully harness the benefits of the human microbiome. We propose a road map to expand our knowledge of the microbiome–health relationship, incorporating epidemiological approaches informed by the unique ecological characteristics of these communities.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Change history
16 December 2024
A Correction to this paper has been published: https://doi.org/10.1038/s41579-024-01145-8
References
Berg, G. et al. Microbiome definition re-visited: old concepts and new challenges. Microbiome 8, 103 (2020).
Lynch, S. V. & Pedersen, O. The human intestinal microbiome in health and disease. N. Engl. J. Med. 375, 2369–2379 (2016).
Turnbaugh, P. J. et al. The Human Microbiome Project. Nature 449, 804–810 (2007).
Larsen, N. et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS ONE 5, e9085 (2010).
Qin, N. et al. Alterations of the human gut microbiome in liver cirrhosis. Nature 513, 59–64 (2014).
Wang, T. et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 6, 320–329 (2012).
Gevers, D. et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 15, 382–392 (2014).
Jeffery, I. B. et al. Differences in fecal microbiomes and metabolomes of people with vs without irritable bowel syndrome and bile acid malabsorption. Gastroenterology 158, 1016–1028.e8 (2020).
McGuinness, A. J. et al. A systematic review of gut microbiota composition in observational studies of major depressive disorder, bipolar disorder and schizophrenia. Mol. Psychiatry 27, 1920–1935 (2022).
Nikolova, V. L. et al. Perturbations in gut microbiota composition in psychiatric disorders: a review and meta-analysis. JAMA Psychiatry 78, 1343–1354 (2021).
Zhang, X., Chen, B., Zhao, L. & Li, H. The gut microbiota: emerging evidence in autoimmune diseases. Trends Mol. Med. 26, 862–873 (2020).
Jansen, D. et al. Community types of the human gut virome are associated with endoscopic outcome in ulcerative colitis. J. Crohns Colitis 17, 1504–1513 (2023).
Salosensaari, A. et al. Taxonomic signatures of cause-specific mortality risk in human gut microbiome. Nat. Commun. 12, 2671 (2021).
US Food and Drug Administration. Approval letter—REBYOTA. FDA https://www.fda.gov/vaccines-blood-biologics/vaccines/rebyota (2022).
Walter, J. & Shanahan, F. Fecal microbiota-based treatment for recurrent Clostridioides difficile infection. Cell 186, 1087 (2023).
Bibbò, S. et al. Fecal microbiota transplantation: screening and selection to choose the optimal donor. J. Clin. Med. 9, 1–14 (2020).
van Duijkeren, E. et al. Long-term carriage of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in the general population in the Netherlands. Clin. Infect. Dis. 66, 1368–1376 (2018).
Piewngam, P. et al. Composition of the intestinal microbiota in extended-spectrum β-lactamase-producing Enterobacteriaceae carriers and non-carriers in Thailand. Int. J. Antimicrob. Agents 53, 435–441 (2019).
Vandeputte, D. et al. Temporal variability in quantitative human gut microbiome profiles and implications for clinical research. Nat. Commun. 12, 6740 (2021).
Abdill, R. J., Adamowicz, E. M. & Blekhman, R. Public human microbiome data are dominated by highly developed countries. PLoS Biol. 20, e3001536 (2022).
Keohane, D. M. et al. Microbiome and health implications for ethnic minorities after enforced lifestyle changes. Nat. Med. 26, 1089–1095 (2020).
Makhalanyane, T. P. et al. African microbiomes matter. Nat. Rev. Microbiol. 21, 479–481 (2023).
Reynoso-García, J. et al. A complete guide to human microbiomes: body niches, transmission, development, dysbiosis, and restoration. Front. Syst. Biol. 2, 951403 (2022).
Huttenhower, C. et al. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Vemuri, R., Shankar, E. M., Chieppa, M., Eri, R. & Kavanagh, K. Beyond just bacteria: functional biomes in the gut ecosystem including virome, mycobiome, archaeome and helminths. Microorganisms 8, 483 (2020).
Stockdale, S. R. & Hill, C. Progress and prospects of the healthy human gut virome. Curr. Opin. Virol. 51, 164–171 (2021).
Laforest-Lapointe, I. & Arrieta, M.-C. Microbial eukaryotes: a missing link in gut microbiome studies. mSystems 3, e00201–e00217 (2018).
Lloyd-Price, J., Abu-Ali, G. & Huttenhower, C. The healthy human microbiome. Genome Med. 8, 51 (2016).
Shanahan, F., Ghosh, T. S. & O’Toole, P. W. The healthy microbiome—what is the definition of a healthy gut microbiome? Gastroenterology 160, 483–494 (2021).
Najmanová, L., Vídeňská, P. & Cahová, M. Healthy microbiome—a mere idea or a sound concept? Physiol. Res. 71, 719–738 (2022).
Vandeputte, D., Tito, R. Y., Vanleeuwen, R., Falony, G. & Raes, J. Practical considerations for large-scale gut microbiome studies. FEMS Microbiol. Rev. 41, S154–S167 (2017).
McBurney, M. I. et al. Establishing what constitutes a healthy human gut microbiome: state of the science, regulatory considerations, and future directions. J. Nutr. 149, 1882–1895 (2019).
Constitution of the World Health Organization. A. J. Public Health Nations Health 36, 1315–1323 (1946).
Qin, J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010).
Herd, P., Palloni, A., Rey, F. & Dowd, J. B. Social and population health science approaches to understand the human microbiome. Nat. Hum. Behav. 2, 808–815 (2018).
Barton, W. et al. The microbiome of professional athletes differs from that of more sedentary subjects in composition and particularly at the functional metabolic level. Gut 67, 625–633 (2018).
Raygoza Garay, J. A. et al. Gut microbiome composition is associated with future onset of Crohn’s disease in healthy first-degree relatives. Gastroenterology 165, 670–681 (2023).
Ferreiro, A. L. et al. Gut microbiome composition may be an indicator of preclinical Alzheimer’s disease. Sci. Transl. Med. 15, eabo2984 (2023).
Ashton, J. J., Seaby, E. G., Beattie, R. M. & Ennis, S. NOD2 in Crohn’s disease—unfinished business. J. Crohns Colitis 17, 450–458 (2023).
Chowdhury, S. R., Chandra Das, D., Sunna, T. C., Beyene, J. & Hossain, A. Global and regional prevalence of multimorbidity in the adult population in community settings: a systematic review and meta-analysis. EClinicalMedicine 57, 101860 (2023).
Khezrian, M., McNeil, C. J., Murray, A. D. & Myint, P. K. An overview of prevalence, determinants and health outcomes of polypharmacy. Ther. Adv. Drug. Saf. 11, 2042098620933741 (2020).
Sonnenburg, E. D. & Sonnenburg, J. L. The ancestral and industrialized gut microbiota and implications for human health. Nat. Rev. Microbiol. 17, 383–390 (2019).
Le Bastard, Q., Vangay, P., Batard, E., Knights, D. & Montassier, E. US immigration is associated with rapid and persistent acquisition of antibiotic resistance genes in the gut. Clin. Infect. Dis. 71, 419–421 (2020).
Pang, S. et al. Longevity of centenarians is reflected by the gut microbiome with youth-associated signatures. Nat. Aging 3, 436–449 (2023).
Foxman, B. & Riley, L. Molecular epidemiology of infection foxman and riley molecular epidemiology: focus on infection. Am. J. Epidemiol. 153, 1135–1141 (2001).
Midha, S., Chawla, S. & Garg, P. K. Modifiable and non-modifiable risk factors for pancreatic cancer: a review. Cancer Lett. 381, 269–277 (2016).
Pradhan, A. D. et al. Inflammatory biomarkers, hormone replacement therapy, and incident coronary heart disease prospective analysis from the women’s health initiative observational study. JAMA 288, 980–987 (2002).
Zhernakova, A. et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 352, 565–569 (2016).
Gacesa, R. et al. Environmental factors shaping the gut microbiome in a Dutch population. Nature 604, 732–739 (2022).
Winter, S. E. & Bäumler, A. J. Gut dysbiosis: ecological causes and causative effects on human disease. Proc. Natl Acad. Sci. USA 120, e2316579120 (2023).
Le Chatelier, E. et al. Richness of human gut microbiome correlates with metabolic markers. Nature 500, 541–546 (2013).
Vandeputte, D. et al. Quantitative microbiome profiling links gut community variation to microbial load. Nature 551, 507–511 (2017).
Vieira-Silva, S. et al. Statin therapy is associated with lower prevalence of gut microbiota dysbiosis. Nature 581, 310–315 (2020).
France, M. T. et al. VALENCIA: a nearest centroid classification method for vaginal microbial communities based on composition. Microbiome 8, 166 (2020).
Wang, N. & Fang, J. Y. Fusobacterium nucleatum, a key pathogenic factor and microbial biomarker for colorectal cancer. Trends Microbiol. 31, 159–172 (2023).
Pleguezuelos-Manzano, C. et al. Mutational signature in colorectal cancer caused by genotoxic pks+ E. coli. Nature 580, 269–273 (2020).
Wang, Z. et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472, 57–65 (2011).
Fu, J. et al. The gut microbiome contributes to a substantial proportion of the variation in blood lipids. Circ. Res. 117, 817–824 (2015).
Manor, O. et al. Health and disease markers correlate with gut microbiome composition across thousands of people. Nat. Commun. 11, 5206 (2020).
Frioux, C. et al. Enterosignatures define common bacterial guilds in the human gut microbiome. Cell Host Microbe 31, 1111–1125.e6 (2023).
Anthamatten, L. et al. Stratification of human gut microbiomes by succinotype is associated with inflammatory bowel disease status. Preprint at bioRxiv https://doi.org/10.1101/2023.11.21.568118v1 (2023).
Falony, G. et al. Population-level analysis of gut microbiome variation. Science 352, 560–564 (2016).
Cotillard, A. et al. A posteriori dietary patterns better explain variations of the gut microbiome than individual markers in the American Gut Project. Am. J. Clin. Nutr. 115, 432–443 (2022).
Walker, R. L. et al. Population study of the gut microbiome: associations with diet, lifestyle, and cardiometabolic disease. Genome Med. 13, 188 (2021).
Lu, J. et al. Chinese gut microbiota and its associations with staple food type, ethnicity, and urbanization. NPJ Biofilms Microbiomes 7, 71 (2021).
Armet, A. M. et al. Rethinking healthy eating in light of the gut microbiome. Cell Host Microbe 30, 764–785 (2022).
Kates, A. E. et al. Household pet ownership and the microbial diversity of the human gut microbiota. Front. Cell Infect. Microbiol. 10, 73 (2020).
Tun, H. M. et al. Exposure to household furry pets influences the gut microbiota of infants at 3–4 months following various birth scenarios. Microbiome 5, 40 (2017).
Aasmets, O., Krigul, K. L., Lüll, K., Metspalu, A. & Org, E. Gut metagenome associations with extensive digital health data in a volunteer-based Estonian microbiome cohort. Nat. Commun. 13, 869 (2022).
Procházková, N. et al. Advancing human gut microbiota research by considering gut transit time. Gut 72, 180–191 (2023).
Roager, H. M. et al. Colonic transit time is related to bacterial metabolism and mucosal turnover in the gut. Nat. Microbiol. 1, 16093 (2016).
Vandeputte, D. et al. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut 65, 57–62 (2016).
He, Y. et al. Regional variation limits applications of healthy gut microbiome reference ranges and disease models. Nat. Med. 24, 1532–1535 (2018).
Schnorr, S. L. et al. Gut microbiome of the Hadza hunter-gatherers. Nat. Commun. 5, 3654 (2014).
Yatsunenko, T. et al. Human gut microbiome viewed across age and geography. Nature 486, 222–227 (2012).
De Filippo, C. et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl Acad. Sci. USA 107, 14691–14696 (2010).
Tamburini, F. B. et al. Short- and long-read metagenomics of urban and rural South African gut microbiomes reveal a transitional composition and undescribed taxa. Nat. Commun. 13, 926 (2022).
Shad, N. S., Shaikh, N. I. & Cunningham, S. A. Migration spurs changes in the human microbiome: a review. J. Racial Ethn. Health Disparities https://doi.org/10.1007/s40615-023-01813-0 (2023).
Copeland, J. K. et al. The impact of migration on the gut metagenome of South Asian Canadians. Gut Microbes 13, 1–29 (2021).
Kaplan, R. C. et al. Gut microbiome composition in the Hispanic Community Health Study/Study of Latinos is shaped by geographic relocation, environmental factors, and obesity. Genome Biol. 20, 219 (2019).
Vangay, P. et al. US immigration westernizes the human gut microbiome. Cell 175, 962–972.e10 (2018).
Blaser, M. J. The theory of disappearing microbiota and the epidemics of chronic diseases. Nat. Rev. Immunol. 17, 461–463 (2017).
Browne, H. P. et al. Boosting microbiome science worldwide could save millions of children’s lives. Nature 625, 237–240 (2024).
Wang, H. et al. Global, regional, and national under-5 mortality, adult mortality, age-specific mortality, and life expectancy, 1970–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet 390, 1084–1150 (2017).
Brewster, R. et al. Surveying gut microbiome research in Africans: toward improved diversity and representation. Trends Microbiol. 27, 824–835 (2019).
Smith, M. I. et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science 339, 548–554 (2013).
Deehan, E. C. et al. Precision microbiome modulation with discrete dietary fiber structures directs short-chain fatty acid production. Cell Host Microbe 27, 389–404.e6 (2020).
Tebani, A. et al. Integration of molecular profiles in a longitudinal wellness profiling cohort. Nat. Commun. 11, 4487 (2020).
Dekkers, K. F. et al. An online atlas of human plasma metabolite signatures of gut microbiome composition. Nat. Commun. 13, 5370 (2022).
Gou, W. et al. Westlake Gut Project: a consortium of microbiome epidemiology for the gut microbiome and health research in China. Med. Microecol. 14, 100064 (2022).
Cernava, T. et al. Metadata harmonization—standards are the key for a better usage of omics data for integrative microbiome analysis. Environ. Microbiomes 17, 33 (2022).
Nagata, N. et al. Population-level metagenomics uncovers distinct effects of multiple medications on the human gut microbiome. Gastroenterology 163, 1038–1052 (2022).
Pasolli, E. et al. Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell 176, 649–662.e20 (2019).
Allali, I. et al. Human microbiota research in Africa: a systematic review reveals gaps and priorities for future research. Microbiome 9, 241 (2021).
Stewart, C. J. et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 562, 583–588 (2018).
Claesson, M. J. et al. Gut microbiota composition correlates with diet and health in the elderly. Nature 488, 178–184 (2012).
Amato, K. R. et al. The human gut microbiome and health inequities. Proc. Natl Acad. Sci. USA 118, e2017947118 (2021).
Dixon, M., Dunlop, A. L., Corwin, E. J. & Kramer, M. R. Joint effects of individual socioeconomic status and residential neighborhood context on vaginal microbiome composition. Front. Public. Health 11, 1029741 (2023).
Bowyer, R. C. E. et al. Socioeconomic status and the gut microbiome: a TwinsUK cohort study. Microorganisms 7, 17 (2019).
Kaisanlahti, A. et al. Maternal microbiota communicates with the fetus through microbiota-derived extracellular vesicles. Microbiome 11, 249 (2023).
Mueller, N. T., Differding, M. K., Østbye, T., Hoyo, C. & Benjamin-Neelon, S. E. Association of birth mode of delivery with infant faecal microbiota, potential pathobionts, and short chain fatty acids: a longitudinal study over the first year of life. BJOG 128, 1293–1303 (2021).
Coker, M. O. et al. Specific class of intrapartum antibiotics relates to maturation of the infant gut microbiota: a prospective cohort study. BJOG 127, 217–227 (2020).
Yang, B. et al. Bifidobacterium and Lactobacillus composition at species level and gut microbiota diversity in infants before 6 weeks. Int. J. Mol. Sci. 20, 3306 (2019).
Beller, L. et al. Successional stages in infant gut microbiota maturation. mBio 12, e0185721 (2021).
Laursen, M. F. et al. Maternal milk microbiota and oligosaccharides contribute to the infant gut microbiota assembly. ISME Commun. 1, 21 (2021).
Duranti, S. et al. Maternal inheritance of bifidobacterial communities and bifidophages in infants through vertical transmission. Microbiome 5, 66 (2017).
Feehily, C. et al. Detailed mapping of Bifidobacterium strain transmission from mother to infant via a dual culture-based and metagenomic approach. Nat. Commun. 14, 3015 (2023).
McKeen, S. et al. Adaptation of the infant gut microbiome during the complementary feeding transition. PLoS ONE 17, e0270213 (2022).
Lim, E. S. et al. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat. Med. 21, 1228–1234 (2015).
Laursen, M. F., Bahl, M. I. & Licht, T. R. Settlers of our inner surface-factors shaping the gut microbiota from birth to toddlerhood. FEMS Microbiol. Rev. 45, fuab001 (2021).
Warmink-Perdijk, W. D. B. et al. Lifelines NEXT: as prospective birth cohort adding the next generation to the three-generation Lifelines cohort study. Eur. J. Epidemiol. 35, 157–168 (2020).
Korpela, K. et al. Cohort profile: Finnish Health and Early Life Microbiota (HELMi) longitudinal birth cohort. BMJ Open. 9, e028500 (2019).
Jeffery, I. B., Lynch, D. B. & O’Toole, P. W. Composition and temporal stability of the gut microbiota in older persons. ISME J. 10, 170–182 (2016).
Wilmanski, T., Gibbons, S. M. & Price, N. D. Healthy aging and the human gut microbiome: why we cannot just turn back the clock. Nat. Aging 2, 869–871 (2022).
Si, J. et al. Long-term life history predicts current gut microbiome in a population-based cohort study. Nat. Aging 2, 885–895 (2022).
Korpela, K. et al. Gut microbiota develop towards an adult profile in a sex-specific manner during puberty. Sci. Rep. 11, 23297 (2021).
McVey Neufeld, K. A., Luczynski, P., Dinan, T. G. & Cryan, J. F. Reframing the teenage wasteland: adolescent microbiota–gut–brain axis. Can. J. Psychiatry 61, 214–221 (2016).
Park, M. G., Cho, S. & Oh, M. M. Menopausal changes in the microbiome—a review focused on the genitourinary microbiome. Diagnostics 13, 1193 (2023).
Peters, B. A. et al. Menopause is associated with an altered gut microbiome and estrobolome, with implications for adverse cardiometabolic risk in the hispanic community health study/study of Latinos. mSystems 7, e0027322 (2022).
McDonald, D. et al. American Gut: an open platform for citizen science microbiome research. mBio 3, e00031-18 (2018).
McCallum, G. & Tropini, C. The gut microbiota and its biogeography. Nat. Rev. Microbiol. 22, 105–118 (2023).
Kennedy, K. M. et al. Questioning the fetal microbiome illustrates pitfalls of low-biomass microbial studies. Nature 613, 639–649 (2023).
Hayes, R. B. et al. Association of oral microbiome with risk for incident head and neck squamous cell cancer. JAMA Oncol. 4, 358–365 (2018).
Fan, X. et al. Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case–control study. Gut 67, 120–127 (2018).
McClelland, R. S. et al. Evaluation of the association between the concentrations of key vaginal bacteria and the increased risk of HIV acquisition in African women from five cohorts: a nested case–control study. Lancet Infect. Dis. 18, 554–564 (2018).
Jie, Z. et al. A transomic cohort as a reference point for promoting a healthy human gut microbiome. Med. Microecol. 8, 100039 (2021).
Liu, X. et al. Metagenome–genome-wide association studies reveal human genetic impact on the oral microbiome. Cell Discov. 7, 117 (2021).
Jie, Z. et al. Life history recorded in the vagino-cervical microbiome along with multi-omes. Genomics Proteom. Bioinforma. 20, 304–321 (2022).
Kurushima, Y. et al. Host genotype links to salivary and gut microbiota by periodontal status. J. Dent. Res. 102, 146–156 (2023).
Nearing, J. T., DeClercq, V., Van Limbergen, J. & Langille, M. G. I. Assessing the variation within the oral microbiome of healthy adults. mSphere 5, e00451-20 (2020).
Willis, J. R. et al. Citizen-science reveals changes in the oral microbiome in Spain through age and lifestyle factors. NPJ Biofilms Microbiomes 8, 38 (2022).
Lebeer, S. et al. A citizen-science-enabled catalogue of the vaginal microbiome and associated factors. Nat. Microbiol. 8, 2183–2195 (2023).
Correia, G. D., Marchesi, J. R. & MacIntyre, D. A. Moving beyond DNA: towards functional analysis of the vaginal microbiome by non-sequencing-based methods. Curr. Opin. Microbiol. 73, 102292 (2023).
Byrd, A. L., Belkaid, Y. & Segre, J. A. The human skin microbiome. Nat. Rev. Microbiol. 16, 143–155 (2018).
Jo, S. et al. Oral and gut dysbiosis leads to functional alterations in Parkinson’s disease. NPJ Parkinsons Dis. 8, 87 (2022).
De Filippis, F. et al. Distinct genetic and functional traits of human intestinal Prevotella copri strains are associated with different habitual diets. Cell Host Microbe 25, 444–453.e3 (2019).
Valles-Colomer, M. et al. The person-to-person transmission landscape of the gut and oral microbiomes. Nature 614, 125–135 (2023).
Zhernakova, D. V. et al. Host genetic regulation of human gut microbial structural variation. Nature 625, 813–821 (2024).
Zahavi, L. et al. Bacterial SNPs in the human gut microbiome associate with host BMI. Nat. Med. 29, 2785–2792 (2023).
Tedersoo, L., Albertsen, M., Anslan, S. & Callahan, B. Perspectives and benefits of high-throughput long-read sequencing in microbial ecology. Appl. Environ. Microbiol. 87, 1–19 (2021).
Chen, L. et al. The long-term genetic stability and individual specificity of the human gut microbiome. Cell 184, 2302–2315.e12 (2021).
Jovel, J. et al. Characterization of the gut microbiome using 16S or shotgun metagenomics. Front. Microbiol. 7, 459 (2016).
Hu, D., Fuller, N. R., Caterson, I. D., Holmes, A. J. & Reeves, P. R. Single-gene long-read sequencing illuminates Escherichia coli strain dynamics in the human intestinal microbiome. Cell Rep. 38, 1102339 (2022).
Lloyd-Price, J. et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 550, 61–66 (2017).
Carrow, H. C., Batachari, L. E. & Chu, H. Strain diversity in the microbiome: lessons from Bacteroides fragilis. PLoS Pathog. 16, e1009056 (2020).
Magnúsdóttir, S. et al. Generation of genome-scale metabolic reconstructions for 773 members of the human gut microbiota. Nat. Biotechnol. 35, 81–89 (2017).
Gehrig, J. L. et al. Finding the right fit: evaluation of short-read and long-read sequencing approaches to maximize the utility of clinical microbiome data. Microb. Genom. 8, 000794 (2022).
Pascal Andreu, V. et al. gutSMASH predicts specialized primary metabolic pathways from the human gut microbiota. Nat. Biotechnol. 41, 1416–1423 (2023).
Lind, A. L. & Pollard, K. S. Accurate and sensitive detection of microbial eukaryotes from whole metagenome shotgun sequencing. Microbiome 9, 58 (2021).
Shkoporov, A. N. et al. The human gut virome is highly diverse, stable, and individual specific. Cell Host Microbe 26, 527–541.e5 (2019).
Begum, N. et al. Host–mycobiome metabolic interactions in health and disease. Gut Microbes 14, 2121576 (2022).
Usyk, M. et al. Comprehensive evaluation of shotgun metagenomics, amplicon sequencing, and harmonization of these platforms for epidemiological studies. Cell Rep. Methods 3, 100391 (2023).
Pollak, S. Plant DNA in feces as a nutritional crystal ball. Proc. Natl Acad. Sci. USA 120, e2309172120 (2023).
Quinn, R. A. et al. Global chemical effects of the microbiome include new bile-acid conjugations. Nature 579, 123–129 (2020).
Guo, C. J. et al. Discovery of reactive microbiota-derived metabolites that inhibit host proteases. Cell 168, 517–526.e18 (2017).
Hajjar, G. et al. Scaling-up metabolomics: current state and perspectives. Trends Anal. Chem. 167, 117225 (2023).
Wang, K. et al. The gut microbiome modifies the associations of short- and long-term physical activity with body weight changes. Microbiome 11, 121 (2023).
Mehta, R. S. et al. Stability of the human faecal microbiome in a cohort of adult men. Nat. Microbiol. 3, 347–355 (2018).
Yap, M. et al. Evaluation of methods for the reduction of contaminating host reads when performing shotgun metagenomic sequencing of the milk microbiome. Sci. Rep. 10, 21665 (2020).
Spreckels, J. E. et al. Analysis of microbial composition and sharing in low-biomass human milk samples: a comparison of DNA isolation and sequencing techniques. ISME Commun. 3, 116 (2023).
Rao, C. et al. Multi-kingdom ecological drivers of microbiota assembly in preterm infants. Nature 591, 633–638 (2021).
Galazzo, G. et al. How to count our microbes? The effect of different quantitative microbiome profiling approaches. Front. Cell. Infect. Microbiol. 10, 403 (2020).
Tito, R. Y. et al. Microbiome confounders and quantitative profiling challenge predicted microbial targets in colorectal cancer development. Nat. Med. 30, 1339–1348 (2024).
Chadwick, R. & Zwart, H. From ELSA to responsible research and promisomics. Life Sci. Soc. Policy 9, 3 (2013).
Nishijima, S. et al. Extensive gut virome variation and its associations with host and environmental factors in a population-level cohort. Nat. Commun. 13, 5252 (2022).
Brandwein, M., Katz, I., Katz, A. & Kohen, R. Beyond the gut: skin microbiome compositional changes are associated with BMI. Hum. Microb. J. 13, 100063 (2019).
Hughes, D. A. et al. Genome-wide associations of human gut microbiome variation and implications for causal inference analyses. Nat. Microbiol. 5, 1079–1087 (2020).
Koponen, K. K. et al. Associations of healthy food choices with gut microbiota profiles. Am. J. Clin. Nutr. 114, 605–616 (2021).
Palmu, J. et al. Association between the gut microbiota and blood pressure in a population cohort of 6953 individuals. J. Am. Heart Assoc. 9, e016641 (2020).
Palmu, J. et al. Gut microbiome and atrial fibrillation—results from a large population-based study. EBioMedicine 91, 104583 (2023).
Peters, B. A. et al. Association of the gut microbiome with kidney function and damage in the Hispanic Community Health Study/Study of Latinos (HCHS/SOL). Gut Microbes 15, 2186685 (2023).
Nash, A. K. et al. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome 5, 153 (2017).
Nogal, A. et al. Genetic and gut microbiome determinants of SCFA circulating and fecal levels, postprandial responses and links to chronic and acute inflammation. Gut Microbes 15, 2240050 (2023).
Armstrong, A. J. S., Parmar, V. & Blaser, M. J. Assessing saliva microbiome collection and processing methods. NPJ Biofilms Microbiomes 7, 81 (2021).
Robinson, C. K., Brotman, R. M. & Ravel, J. Intricacies of assessing the human microbiome in epidemiologic studies. Ann. Epidemiol. 26, 311–321 (2016).
Hill, C. J. et al. Effect of room temperature transport vials on DNA quality and phylogenetic composition of faecal microbiota of elderly adults and infants. Microbiome 4, 19 (2016).
Tang, Q. et al. Current sampling methods for gut microbiota: a call for more precise devices. Front. Cell Infect. Microbiol. 10, 151 (2020).
Lourenço, M. et al. The spatial heterogeneity of the gut limits predation and fosters coexistence of bacteria and bacteriophages. Cell Host Microbe 28, 390–401.e5 (2020).
Badawy, R. et al. Metadata concepts for advancing the use of digital health technologies in clinical research. Digit. Biomark. 3, 116–132 (2019).
Berry, S. E. et al. Human postprandial responses to food and potential for precision nutrition. Nat. Med. 26, 964–973 (2020).
Koonin, E. V., Dolja, V. V. & Krupovic, M. The healthy human virome: from virus–host symbiosis to disease. Curr. Opin. Virol. 47, 86–94 (2021).
Kumata, R., Ito, J., Takahashi, K., Suzuki, T. & Sato, K. A tissue level atlas of the healthy human virome. BMC Biol. 18, 55 (2020).
Dutilh, B. E. et al. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat. Commun. 5, 4498 (2014).
Yutin, N. et al. Analysis of metagenome-assembled viral genomes from the human gut reveals diverse putative CrAss-like phages with unique genomic features. Nat. Commun. 12, 1044 (2021).
Edwards, R. A. et al. Global phylogeography and ancient evolution of the widespread human gut virus crAssphage. Nat. Microbiol. 4, 1727–1736 (2019).
Clooney, A. G. et al. Whole-virome analysis sheds light on viral dark matter in inflammatory bowel disease. Cell Host Microbe 26, 764–778.e5 (2019).
Ma, Y., You, X., Mai, G., Tokuyasu, T. & Liu, C. A human gut phage catalog correlates the gut phageome with type 2 diabetes. Microbiome 6, 24 (2018).
Roux, S., Hallam, S. J., Woyke, T. & Sullivan, M. B. Viral dark matter and virus–host interactions resolved from publicly available microbial genomes. eLife 4, e08490 (2015).
Beller, L. & Matthijnssens, J. What is (not) known about the dynamics of the human gut virome in health and disease. Curr. Opin. Virol. 37, 52–57 (2019).
Chibani, C. M. et al. A catalogue of 1,167 genomes from the human gut archaeome. Nat. Microbiol. 7, 48–61 (2022).
Koskinen, K. et al. First insights into the diverse human archaeome: specific detection of archaea in the gastrointestinal tract, lung, and nose and on skin. mBio 8, e00824–17 (2017).
Beghini, F. et al. Large-scale comparative metagenomics of Blastocystis, a common member of the human gut microbiome. ISME J. 11, 2848–2863 (2017).
Scanlan, P. D. et al. The microbial eukaryote Blastocystis is a prevalent and diverse member of the healthy human gut microbiota. FEMS Microbiol. Ecol. 90, 326–330 (2014).
Marzano, V. et al. “Omic” investigations of protozoa and worms for a deeper understanding of the human gut “parasitome”. PLoS Negl. Trop. Dis. 11, e0005916 (2017).
Acknowledgements
The authors thank all those who helped facilitate the Human Microbiome Action (HMA) workshop on 6 October 2022, including workshop attendees, postdoctoral scribes, audiovisual experts and staff of the River Lee Hotel in Cork. This publication has been supported by the project entitled International Human Microbiome Coordination and Support Action (IHMCSA), which receives funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No. 964590.
Author information
Authors and Affiliations
Consortia
Contributions
R.J., K.B., A.L. and R.P.R. wrote the article. All authors contributed substantially to discussion of the content, and reviewed and edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
M.J.C. and P.D.C. are co-founders of SeqBiome Ltd. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Microbiology thanks Eugene B. Chang, who co-reviewed with Orlando DeLeon, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Joos, R., Boucher, K., Lavelle, A. et al. Examining the healthy human microbiome concept. Nat Rev Microbiol 23, 192–205 (2025). https://doi.org/10.1038/s41579-024-01107-0
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41579-024-01107-0
This article is cited by
-
Effector–host interactome map links type III secretion systems in healthy gut microbiomes to immune modulation
Nature Microbiology (2026)
-
Prospective association between the gut microbiota and incident pneumonia: a cohort study of 6419 individuals
Respiratory Research (2025)
-
Oral and fecal microbiota in Chinese adults with obesity reveal potential niche-specific microbiota associated with obesity
BMC Microbiology (2025)
-
Characteristics of gut and lung microbiota in patients with lung masses and their relationship with clinical features
BMC Microbiology (2025)
-
Microsporidian infection of mosquito larvae changes the host-associated microbiome towards the synthesis of antimicrobial factors
Parasites & Vectors (2025)