Abstract
Bipolar disorder is a leading contributor to the global burden of disease1. Despite high heritability (60–80%), the majority of the underlying genetic determinants remain unknown2. We analysed data from participants of European, East Asian, African American and Latino ancestries (n = 158,036 cases with bipolar disorder, 2.8 million controls), combining clinical, community and self-reported samples. We identified 298 genome-wide significant loci in the multi-ancestry meta-analysis, a fourfold increase over previous findings3, and identified an ancestry-specific association in the East Asian cohort. Integrating results from fine-mapping and other variant-to-gene mapping approaches identified 36 credible genes in the aetiology of bipolar disorder. Genes prioritized through fine-mapping were enriched for ultra-rare damaging missense and protein-truncating variations in cases with bipolar disorder4, highlighting convergence of common and rare variant signals. We report differences in the genetic architecture of bipolar disorder depending on the source of patient ascertainment and on bipolar disorder subtype (type I or type II). Several analyses implicate specific cell types in the pathophysiology of bipolar disorder, including GABAergic interneurons and medium spiny neurons. Together, these analyses provide additional insights into the genetic architecture and biological underpinnings of bipolar disorder.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Genome-wide association summary statistics for these analyses are available at https://www.med.unc.edu/pgc/download-results/. The full GWAS summary statistics for the 23andMe datasets will be made available through 23andMe to qualified researchers under an agreement with 23andMe that protects the privacy of the 23andMe participants. Please visit https://research.23andme.com/collaborate/#dataset-access for more information and to apply to access the data. After applying with 23andMe, the full summary statistics including all analysed SNPs and samples in the GWAS meta-analyses will be accessible to the approved researchers. Genotype data are available for a subset of cohorts, including dbGAP accession numbers and/or restrictions, as described in the ‘Cohort descriptions’ section of the supplementary materials.
Code availability
No custom code was developed for this study. All software and tools used for the analyses presented are publicly available and referenced within the respective sections in the Methods of the article.
References
Carvalho, A. F., Firth, J. & Vieta, E. Bipolar disorder. N. Engl. J. Med. 383, 58–66 (2020).
Lichtenstein, P. et al. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet 373, 234–239 (2009).
Mullins, N. et al. Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat. Genet. 53, 817–829 (2021).
Palmer, D. S. et al. Exome sequencing in bipolar disorder identifies AKAP11 as a risk gene shared with schizophrenia. Nat. Genet. 54, 541–547 (2022).
McIntyre, R. S. et al. Bipolar disorders. Lancet 396, 1841–1856 (2020).
Aghababaie-Babaki, P. et al. Global, regional, and national burden and quality of care index (QCI) of bipolar disorder: a systematic analysis of the Global Burden of Disease Study 1990 to 2019. Int. J. Soc. Psychiatry 69, 1958–1970 (2023).
Geddes, J. R. & Miklowitz, D. J. Treatment of bipolar disorder. Lancet 381, 1672–1682 (2013).
American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders (DSM-5®) (American Psychiatric Association, 2013).
Reed, G. M. et al. Innovations and changes in the ICD-11 classification of mental, behavioural and neurodevelopmental disorders. World Psychiatry 18, 3–19 (2019).
Psychiatric GWAS Consortium Bipolar Disorder Working Group. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat. Genet. 43, 977–983 (2011).
Stahl, E. A. et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat. Genet. 51, 793–803 (2019).
Ching, C. R. K. et al. What we learn about bipolar disorder from large-scale neuroimaging: findings and future directions from the ENIGMA Bipolar Disorder Working Group. Hum. Brain Mapp. 43, 56–82 (2022).
Bulik-Sullivan, B. K. et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
McGrath, J. J. et al. Age of onset and cumulative risk of mental disorders: a cross-national analysis of population surveys from 29 countries. Lancet Psychiatry 10, 668–681 (2023).
Holland, D. et al. Beyond SNP heritability: polygenicity and discoverability of phenotypes estimated with a univariate Gaussian mixture model. PLoS Genet. 16, e1008612 (2020).
Frei, O. et al. Bivariate causal mixture model quantifies polygenic overlap between complex traits beyond genetic correlation. Nat. Commun. 10, 2417 (2019).
Merikangas, K. R. et al. Prevalence and correlates of bipolar spectrum disorder in the World Mental Health Survey Initiative. Arch. Gen. Psychiatry 68, 241–251 (2011).
Ikeda, M. et al. A genome-wide association study identifies two novel susceptibility loci and trans population polygenicity associated with bipolar disorder. Mol. Psychiatry 23, 639–647 (2018).
Frei, O. et al. Improved functional mapping of complex trait heritability with GSA-MiXeR implicates biologically specific gene sets. Nat. Genet. https://doi.org/10.1038/s41588-024-01771-1 (2024).
Cuéllar-Partida, G. et al. Complex-Traits Genetics Virtual Lab: a community-driven web platform for post-GWAS analyses. Preprint at bioRxiv https://doi.org/10.1101/518027 (2019).
Ge, T., Chen, C.-Y., Ni, Y., Feng, Y.-C. A. & Smoller, J. W. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat. Commun. 10, 1776 (2019).
de Leeuw, C. A., Mooij, J. M., Heskes, T. & Posthuma, D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 11, e1004219 (2015).
Siletti, K. et al. Transcriptomic diversity of cell types across the adult human brain. Science 382, eadd7046 (2023).
Finucane, H. K. et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 47, 1228–1235 (2015).
Yao, S. et al. Connecting genomic results for psychiatric disorders to human brain cell types and regions reveals convergence with functional connectivity. Preprint at medRxiv https://doi.org/10.1101/2024.01.18.24301478 (2024).
Weissbrod, O. et al. Functionally informed fine-mapping and polygenic localization of complex trait heritability. Nat. Genet. 52, 1355–1363 (2020).
McLaren, W. et al. The Ensembl Variant Effect Predictor. Genome Biol. 17, 122 (2016).
Singh, T. et al. Rare coding variants in ten genes confer substantial risk for schizophrenia. Nature 604, 509–516 (2022).
Olfson, E. et al. Rare de novo damaging DNA variants are enriched in attention-deficit/hyperactivity disorder and implicate risk genes. Nat. Commun. 15, 5870 (2024).
Gusev, A. et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 48, 245–252 (2016).
Bhattacharya, A. et al. Isoform-level transcriptome-wide association uncovers genetic risk mechanisms for neuropsychiatric disorders in the human brain. Nat. Genet. 55, 2117–2128 (2023).
Fulco, C. P. et al. Activity-by-contact model of enhancer–promoter regulation from thousands of CRISPR perturbations. Nat. Genet. 51, 1664–1669 (2019).
Nasser, J. et al. Genome-wide enhancer maps link risk variants to disease genes. Nature 593, 238–243 (2021).
Koopmans, F. et al. SynGO: an evidence-based, expert-curated knowledge base for the synapse. Neuron 103, 217–234.e4 (2019).
Kang, H. J. et al. Spatio-temporal transcriptome of the human brain. Nature 478, 483–489 (2011).
Gaspar, H. A. & Breen, G. Drug enrichment and discovery from schizophrenia genome-wide association results: an analysis and visualisation approach. Sci. Rep. 7, 12460 (2017).
Cannon, M. et al. DGIdb 5.0: rebuilding the drug-gene interaction database for precision medicine and drug discovery platforms. Nucleic Acids Res. 52, D1227–D1235 (2024).
Reigstad, C. S. et al. Gut microbes promote colonic serotonin production through an effect of short-chain fatty acids on enterochromaffin cells. FASEB J. 29, 1395–1403 (2015).
Ortega, M. A. et al. Microbiota–gut–brain axis mechanisms in the complex network of bipolar disorders: potential clinical implications and translational opportunities. Mol. Psychiatry 28, 2645–2673 (2023).
Huang, S. et al. Lithium carbonate alleviates colon inflammation through modulating gut microbiota and Treg cells in a GPR43-dependent manner. Pharmacol. Res. 175, 105992 (2022).
Trubetskoy, V. et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature 604, 502–508 (2022).
Kloiber, S. et al. Neurodevelopmental pathways in bipolar disorder. Neurosci. Biobehav. Rev. 112, 213–226 (2020).
Jonsson, L., Song, J., Joas, E., Pålsson, E. & Landén, M. Polygenic scores for psychiatric disorders associate with year of first bipolar disorder diagnosis: a register-based study between 1972 and 2016. Psychiatry Res. 339, 116081 (2024).
Zimmerman, M., Ruggero, C. J., Chelminski, I. & Young, D. Is bipolar disorder overdiagnosed? J. Clin. Psychiatry 69, 935–940 (2008).
Zimmerman, M., Ruggero, C. J., Chelminski, I. & Young, D. Psychiatric diagnoses in patients previously overdiagnosed with bipolar disorder. J. Clin. Psychiatry 71, 26–31 (2010).
Sollis, E. et al. The NHGRI-EBI GWAS Catalog: knowledgebase and deposition resource. Nucleic Acids Res. 51, D977–D985 (2023).
Li, X. & Zhao, H. Automated feature extraction from population wearable device data identified novel loci associated with sleep and circadian rhythms. PLoS Genet. 16, e1009089 (2020).
Kichaev, G. et al. Leveraging polygenic functional enrichment to improve GWAS power. Am. J. Hum. Genet. 104, 65–75 (2019).
Thalamuthu, A. et al. Genome-wide interaction study with major depression identifies novel variants associated with cognitive function. Mol. Psychiatry 27, 1111–1119 (2022).
Bryc, K., Durand, E. Y., Macpherson, J. M., Reich, D. & Mountain, J. L. The genetic ancestry of African Americans, Latinos, and European Americans across the United States. Am. J. Hum. Genet. 96, 37–53 (2015).
Duncan, L. et al. Analysis of polygenic risk score usage and performance in diverse human populations. Nat. Commun. 10, 3328 (2019).
Lewis, C. M. & Vassos, E. Polygenic risk scores: from research tools to clinical instruments. Genome Med. 12, 44 (2020).
Yengo, L. et al. A saturated map of common genetic variants associated with human height. Nature 610, 704–712 (2022).
Wray, N. R. et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet. 50, 668–681 (2018).
Cai, N. et al. Minimal phenotyping yields genome-wide association signals of low specificity for major depression. Nat. Genet. 52, 437–447 (2020).
Legge, S. E. et al. Genetic and phenotypic features of schizophrenia in the UK Biobank. JAMA Psychiatry https://doi.org/10.1001/jamapsychiatry.2024.0200 (2024).
Lam, M. et al. RICOPILI: rapid imputation for consortias pipeline. Bioinformatics 36, 930–933 (2020).
Price, A. L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. https://doi.org/10.1038/ng1847 (2006).
Loh, P.-R. et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat. Genet. 48, 1443–1448 (2016).
Das, S. et al. Next-generation genotype imputation service and methods. Nat. Genet. 48, 1284–1287 (2016).
McCarthy, S. et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 48, 1279–1283 (2016).
Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015).
Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Chen, W. et al. Improved analyses of GWAS summary statistics by reducing data heterogeneity and errors. Nat. Commun. 12, 7117 (2021).
Yang, J., Lee, S. H., Goddard, M. E. & Visscher, P. M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82 (2011).
Yang, J. et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 44, 369–375 (2012).
Demontis, D. et al. Genome-wide analyses of ADHD identify 27 risk loci, refine the genetic architecture and implicate several cognitive domains. Nat. Genet. 55, 198–208 (2023).
Brown, B. C., Asian Genetic Epidemiology Network Type 2 Diabetes Consortium, Ye, C. J., Price, A. L. & Zaitlen, N. Transethnic genetic-correlation estimates from summary statistics. Am. J. Hum. Genet. 99, 76–88 (2016).
Hübel, C. et al. Genomics of body fat percentage may contribute to sex bias in anorexia nervosa. Am. J. Med. Genet. B Neuropsychiatr. Genet. 180, 428–438 (2019).
Zhu, Z. et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat. Commun. 9, 224 (2018).
Shadrin, A. A. et al. Dissecting the genetic overlap between three complex phenotypes with trivariate MiXeR. Preprint at medRxiv https://doi.org/10.1101/2024.02.23.24303236 (2024).
R Core Team. R: A Language and Environment for Statistical Computing. R Foundation http://www.R-project.org/ (2013).
Lee, S. H., Goddard, M. E., Wray, N. R. & Visscher, P. M. A better coefficient of determination for genetic profile analysis. Genet. Epidemiol. 36, 214–224 (2012).
Ashburner, M. et al. Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 (2000).
Gazal, S. et al. Linkage disequilibrium-dependent architecture of human complex traits shows action of negative selection. Nat. Genet. 49, 1421–1427 (2017).
Kingma, D. P. & Ba, J. Adam: a method for stochastic optimization. Preprint at https://arxiv.org/abs/1412.6980 (2014).
Habib, N. et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat. Methods 14, 955–958 (2017).
Lake, B. B. et al. Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nat. Biotechnol. 36, 70–80 (2017).
Skene, N. G. et al. Genetic identification of brain cell types underlying schizophrenia. Nat. Genet. 50, 825–833 (2018).
Zeisel, A. et al. Molecular architecture of the mouse nervous system. Cell 174, 999–1014.e22 (2018).
Saunders, A. et al. Molecular diversity and specializations among the cells of the adult mouse brain. Cell 174, 1015–1030 (2018).
Watanabe, K., Umićević Mirkov, M., de Leeuw, C. A., van den Heuvel, M. P. & Posthuma, D. Genetic mapping of cell type specificity for complex traits. Nat. Commun. 10, 3222 (2019).
Zhu, Z. et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 48, 481–487 (2016).
Qi, T. et al. Genetic control of RNA splicing and its distinct role in complex trait variation. Nat. Genet. 54, 1355–1363 (2022).
Qi, T. et al. Identifying gene targets for brain-related traits using transcriptomic and methylomic data from blood. Nat. Commun. 9, 2282 (2018).
Shrine, N. et al. Multi-ancestry genome-wide association analyses improve resolution of genes and pathways influencing lung function and chronic obstructive pulmonary disease risk. Nat. Genet. 55, 410–422 (2023).
Roth, B. L., Lopez, E., Patel, S. & Kroeze, W. K. The multiplicity of serotonin receptors: uselessly diverse molecules or an embarrassment of riches? Neuroscientist 6, 252–262 (2000).
Mendez, D. et al. ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res. 47, D930–D940 (2019).
Sheils, T. K. et al. TCRD and Pharos 2021: mining the human proteome for disease biology. Nucleic Acids Res. 49, D1334–D1346 (2021).
Yoo, M. et al. DSigDB: drug signatures database for gene set analysis. Bioinformatics 31, 3069–3071 (2015).
Morselli Gysi, D. & Barabási, A.-L. Noncoding RNAs improve the predictive power of network medicine. Proc. Natl Acad. Sci. USA 120, e2301342120 (2023).
Menche, J. et al. Disease networks. Uncovering disease–disease relationships through the incomplete interactome. Science 347, 1257601 (2015).
Acknowledgements
We thank the participants who donated their time, life experiences and DNA to this research, the clinical and scientific teams that worked with them, and the investigators who comprise the PGC. The PGC has received major funding from the US National Institute of Mental Health (PGC4: R01 MH124839, PGC3: U01 MH109528, PGC2: U01 MH094421 and PGC1: U01 MH085520). Statistical analyses were carried out on the NL Genetic Cluster Computer (http://www.geneticcluster.org) hosted by SURFsara. The content is solely the responsibility of the authors and does not necessarily represent the official views of the US National Institutes of Health. Individual and cohort-specific funding acknowledgements are detailed in the Supplementary Information.
Author information
Authors and Affiliations
Consortia
Contributions
The management group for this paper was led by O.A.A. The management group comprised a subset of authors responsible for the study design, conduct, primary and final interpretation, and included A.M., A.J.F., N.M., A.D.F., R.A.O., H.J.E., K.S.O. and O.A.A; this group was also responsible for primary drafting and editing of the manuscript. The analytical team, led by K.S.O., was responsible for the main analyses presented in the paper, and included M. Koromina, T.v.d.V., T.B., F.S.D., J.M.K.Y., K.-H.L., X.W., J.R.I.C., B.L.M., C.C.M, A.V.R., P.A.L., E. Koch, A. Harder, N.P., J.B. and K.S.O. Imputation, quality control and GWAS were conducted by K.S.O., M. Koromina, B.L.M., K.-H.L., X.W. and J.M.K.Y. Heritability and genetic correlation analyses were performed by K.S.O. MiXeR was done by K.S.O. and A. Shadrin. Polygenic association was conducted by T.v.d.V., T.B., B.L.M. and P.A.L. Gene and gene set analyses were done by K.S.O., C.C.M. and A.V.R. Cell-type-specific analyses were performed by F.S.D. Single-nucleus RNA sequencing enrichment was done by A. Harder and J.H.-L. Fine-mapping was conducted by M. Koromina. Rare variant analyses were performed by C. Liao. QTL integrative analyses were done by M. Koromina, T.B. and F.S.D. Enhancer–promoter interactions were analysed by J.B. Credible gene prioritization was performed by K.S.O. and M. Koromina. Temporal clustering was done by N.P. Drug enrichment was conducted by J.R.I.C. and E. Koch. Clinical assessments were performed by A.A., A.C., A.C.-B., A.D.B., A.D.F., A.E.V., A.H.Y., A. Havdahl, A.M., A.M.M., A. Perry, A. Pfennig, A.R., A. Serretti, A.V., B. Carpiniello, B.E., B.S., B.T.B., C.A.M., C. Lavebratt, C. Loughland, C.N.P., C.O., C.S., D.D., D.H., D.J.M., D. J. Smith, D.M., D.Q., E.C.S., E.E.T., E.J.R., E. Kim, E. Sigurdsson, E.S.G., E. Stordal, E.V., E.Z.R., F. Senner, F.S.G., F. Stein, F. Streit, F.T.F., G.B., G.K., G.M., H.A., H.-J.L., H.M., H.V., H.Y.P., I.D.W., I.J., I.M., I.R.G., J.A.R.-Q., J.B.P., J.B.V., J.G.-P., J. Garnham, J. Grove, J.H., J.I.N., J. L. Kalman, J. L. Kennedy, J.L.S., J. Lawrence, J. Lissowska, J.M.P., J.P.R., J.R.D., J.W.S., K.A., K.D., K.G.-S., K.J.O., L.A.J., L.B., L.F., L. Martinsson, L. Sirignano, L.T., L.Z., M.A., M. Bauer, M. Brum, M. Budde, M.C., M.C.O., M.F., M.G., M.G.-S., M.G.M., M.H.R., M. Haraldsson, M. Hautzinger, M.I., M.J.G., M.J.O., M. Kogevinas, M. Landén, M. Lundberg, M. Manchia, M. Mattheisen, M.P.B., M.P.V., M. Rietschel, M. Tesfaye, M.T.P., M. Tesli, N.A.-R., N.B., N.B.-K., N.C., N.D., N.G.M., N.I., O.A.A., O.B.S., O.K.D., O.M., P. B. Mitchell, P. B. Mortensen, P.C., P.F., P.M.C., R.A., R.B., R.S.K., S.A.K., S. Bengeser, S.K.-S., S.L., S.L.M., S.P., T.G.S., T.H., T.H.H., T.K., T.M.K., T.O., T.S., T.W., T.W.W., U.D., U.H., V.M., W.B., W. Maier and W. Myung. Data processing and analyses were performed by A.C., A.D.B., A.F.P., A. Harder, A.J.F., A.M.D., A. Shadrin, A.V.R., A.X.M., B. Coombes, B.L.M., B.M.-M., B.M.B., B.S.W., C.B.P., C. Cruceanu, C.C.M., C. Chatzinakos, C. Liao, C.M.N., C.S.W., C. Terao, C. Toma, D.A., D.M.H., D.W.M., E.A., E.A.S., E.C.B., E.C.C., E. Koch, E.M., E.S.G., F.D., F.J.M., F.S.D., G.A.R., G.B., G.P., G.T., H.-C.C., H. Stefansson, H. Sung, H.-H.W., I.C., J.B., J.C.-D., J.D.M., J.F., J.F.F., J.G.T., J. Grove, J.H.-L., J.K., J.M.B., J.M.F., J.M.K.Y., J.R.I.C., J.S.J., J.T.R.W., K.K., K.-H.L., K.S.O., L.G.S., L.J., L. Milani, L. Sindermann, M.-C.H., M.I., M.J.C., M. Koromina, M. Leber, M.M.N., M. Mattheisen, M. Ribasés, M. Rivera, M.S.A., M.S., M. Tesfaye, N.B.F., N.I., N.M., N.P., N.W.M., O.B.S., O.F., O.K.D., P.A.H., P.A.L., P.A.T., P.D.S., P.F.S., P.H., P.-H.K., P.M., P.P.Z., P.R., Q.S.L., R.J.S., R.M.M., R.Y., S.A., S. Børte, S. Cichon, S.D., S.D.G., S.E.M., S.H., S.H.W., S.J., S.R., S.-J.T., T.A.G., T.B., T.B.B., T.C., T.D.A., T.E.T., T.F.M.A., T.O., T.S., T.v.d.V., T.W., T.W.M., V.E.-P., W. Myung, X.W. and Y.K. Funding was obtained by A.C., A.D.B., A.H.Y., A.M.M., B.E., B.M.N., B.T.B., C.N.P., C. Pantelis, C.S.W., C. Terao, D. J. Stein, D.M., D.S., E.S.G., F.B., F.J.M., G.A.R., G.B., G.P.P., G.T., H.J.E., I.B.H., I.J., I.M., I.N.F., J.A.K., J.B.P., J.B.V., J.I.N., J.M.B., J.M.F., J.R.D., J.W.S., K.H., L.A., L.A.J., L.B., M.A., M. Boehnke, M.C.O., M.F., M.G.-S., M.H.R., M.I., M.J.G., M.J.O., M. Leboyer, M. Landén, M.M.N., M.N., M. Rietschel, M.S., M.T.P., N.C., N.G.M., N.I., O.A.A., O.M., P.A.T., P. B. Mitchell, P. B. Mortensen, P.P.Z., P.R.S., R.A.O., R.J.S., R.M.M., S.E.M., S.J., S.L., T.B.B., T.G.S., T.O., T.S., T.W., T.W.W., W.H.B. and Y.K. Recruitment and genotyping were performed by A.C., A.D.B., A.D.F., A.E.V., A.H.F., A.J.F., A.M.M., A.M., A.R., A. Serretti, A. Squassina, B. Carpiniello, B.-C.L., B.E., B.M.-M., B.M.N., B.T.B., C.A.M., C.B.P., C. Lochner, C.M.N., C.M.O., C.N.P., C. Pantelis, C. Pisanu, C.S.W., D.C.W., D.D., D.J.K., E.A., E.S.G., E. Stordal, E.V., E.Z.R., F.A.H., F.B., F.J.M., F.M., F.S.G., F. Stein, G.A.R., G.B., G.D.H., G.M., G.P.P., G.T., H.J.E., H. Stefansson, H.-H.W., I.B.H., I.D.W., I.J., J.A.R.-Q., J.B.V., J.H., J.H.K., J.H.-L., J.I.N., J.J.L., J. Lissowska, J.M.B., J.M.F., J.M.P., J.R.D., J.R.K., J.-W.K., J.W.S., J.-A.Z., K.H., K.J.O., K.S., L.A., L.A.J., L.J.S., L. Milani, L.T., M.A., M. Aslan, M.C.O., M.F., M.G., M.G.-S., M.I., M.J.C., M.J.G., M.J.O., M. Leboyer, M. Landén, M.M.N., M. Manchia, M.N., M. Ribasés, M. Rietschel, M.S., M.T.P., N.C., N.G.M., N.I., O.A.A., O.M., P.A.L., P. B. Mitchell, P. B. Mortensen, P.D.H., P.F., P.-H.K., P.R., P.R.S., Q.S.L., R.A., R.A.O., R.S.K., S.A.P., S. Bengesser, S. Cichon, S. Catts, S.E.M., S.L.M., S.R., T.G.S., T.H., T.K., T.S., T.W., T.W.W., U.D., U.S., V.J.C., W.H.B., W. Myung and Y.K.L. Numerous authors beyond the initial writing group contributed to data interpretation and provided edits, comments and suggestions to the paper. All authors reviewed the manuscript critically for important intellectual content and approved the final version of the manuscript for publication. The Chair of the PGC is P.F.S. The Bipolar Disorder Working Group of the PGC is led by O.A.A.
Corresponding authors
Ethics declarations
Competing interests
T.E.T., H. Stefansson and K.S. are employed by deCODE Genetics/Amgen. E.A.S. is an employee of Regeneron Genetics Center and owns stocks of Regeneron Pharmaceutical. K.-H.L. and X.W. are employed by 23andMe. Multiple additional authors work for pharmaceutical or biotechnology companies in a manner directly analogous to academic co-authors and collaborators. A.H.Y. has given paid lectures and served on advisory boards relating to drugs used in affective and related disorders for several companies (AstraZeneca, Eli Lilly, Lundbeck, Sunovion, Servier, Livanova, Janssen, Allergan, Bionomics and Sumitomo Dainippon Pharma), was Lead Investigator for the Embolden study (AstraZeneca), BCI Neuroplasticity study and Aripiprazole Mania study, and is an investigator for Janssen, Lundbeck, Livanova and Compass. J.I.N. is an investigator for Janssen. P.F.S. is on the advisory committee and a shareholder of Neumora Therapeutics. G.B. reports consultancy and speaker fees from Eli Lilly and Illumina, and grant funding from Eli Lilly. M. Landén has received speaker fees from Lundbeck. O.A.A. has served as a speaker for Janssen, Lundbeck and Sunovion, and as a consultant for Cortechs.ai. A.M.D. is a founder of and holds equity interest in CorTechs Labs and serves on its scientific advisory board; is a member of the scientific advisory board of Human Longevity and the Mohn Medical Imaging and Visualization Center; and has received research funding from General Electric Healthcare. E.V. has received grants and served as a consultant, advisor or CME speaker for the following entities: AB-Biotics, Abbott, Allergan, Angelini, AstraZeneca, Bristol Myers Squibb, Dainippon Sumitomo Pharma, Farmindustria, Ferrer, Forest Research Institute, Gedeon Richter, GlaxoSmithKline, Janssen, Lundbeck, Otsuka, Pfizer, Roche, SAGE, Sanofi-Aventis, Servier, Shire, Sunovion, Takeda, the Brain and Behaviour Foundation, the Catalan Government (AGAUR and PERIS), the Spanish Ministry of Science, Innovation, and Universities (AES and CIBERSAM), the Seventh European Framework Programme and Horizon 2020 and the Stanley Medical Research Institute. S.K.-S. received author’s, speaker’s and consultant honoraria from Janssen, Medice Arzneimittel Pütter GmbH and Takeda outside of the current work. A. Serretti is or has been a consultant and/or speaker for: Abbott, AbbVie, Angelini, AstraZeneca, Clinical Data, Boheringer, Bristol Myers Squibb, Eli Lilly, GlaxoSmithKline, Innovapharma, Italfarmaco, Janssen, Lundbeck, Naurex, Pfizer, Polifarma, Sanofi and Servier. J.R.D. has served as an unpaid consultant to Myriad-Neuroscience (formerly Assurex Health) in 2017 and 2019, and owns stock in CVS Health. B.M.N. is a member of the scientific advisory board at Deep Genomics, and consultant for Camp4 Therapeutics, Takeda Pharmaceutical and Biogen. B.-C.L., J.-W.K., Y.K.L., J.H.K., M. J. Cheon and D.J.K. are employed by Genoplan. I.B.H. is the Co-Director of Health and Policy at the Brain and Mind Centre (BMC) University of Sydney. The BMC operates an early-intervention youth services at Camperdown under contract to Headspace. I.B.H. is also the Chief Scientific Advisor to, and a 3.2% equity shareholder in, InnoWell. InnoWell was formed by the University of Sydney (45% equity) and PwC (Australia; 45% equity) to deliver the $30 M (AUD) Australian Government-funded Project Synergy (2017–2020; a 3-year program for the transformation of mental health services) and to lead transformation of mental health services internationally through the use of innovative technologies. M.J.O. and M.C.O. have received funding from Takeda Pharmaceuticals and Akrivia Health outside the scope of the current work. P. B. Mitchell. has received remuneration from Janssen (Australia) and Sanofi (Hangzhou) for lectures or advisory board membership. J.A.R.-Q. was on the speakers’ bureau and/or acted as consultant for Biogen, Idorsia, Casen-Recordati, Janssen-Cilag, Novartis, Takeda, Bial, Sincrolab, Neuraxpharm, Novartis, BMS, Medice, Rubió, Uriach, Technofarma and Raffo in the past 3 years; has also received travel awards (airplane tickets and hotel) for taking part in psychiatric meetings from Idorsia, Janssen-Cilag, Rubió, Takeda, Bial and Medice; and the Department of Psychiatry chaired by J.A.R.-Q. received unrestricted educational and research support from the following companies in the past 3 years: Exeltis, Idorsia, Janssen-Cilag, Neuraxpharm, Oryzon, Roche, Probitas and Rubió. All other authors declare no competing interests.
Peer review
Peer review information
Nature thanks Ditte Demontis, Veera Rajagopal and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Network diagram of the genetic correlations between BD ascertained from Clinical, Community and Self-report samples, as well as BD-subtypes (BDI and BDII).
The line widths are proportional to the strength of the correlations between pairs. BDI: bipolar disorder I, BDII: bipolar disorder II.
Extended Data Fig. 2 Univariate MiXeR estimates of the required effective sample size needed to capture 50% of the genetic variance (horizontal dashed line) associated with each BD ascertainment and subtype.
N and Sample size refer to the effective sample size. The estimated effective sample size (and standard errors) are given in the legend alongside each trait name.
Extended Data Fig. 3 Trivariate MiXeR estimates for the genetic overlap of BD from Clinical, Community and Self-report samples.
The percentages show the proportion of trait-influencing variants within each section of the Venn diagram relative to the sum of all trait-influencing variants across all samples. The size of the circles reflects the polygenicity of each trait.
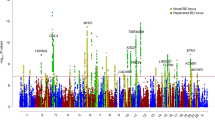
Extended Data Fig. 4 Miami plot for BD genome-wide meta-analyses, including all cohorts.
Upper panel: the multi-ancestry meta-analysis identified 298 genome-wide significant (GWS) loci. Lower panel: porcupine plot showing the results of the Latino (0 GWS loci), African American (0 GWS loci), East Asian (1 GWS locus) and European (229 GWS loci) meta-analyses. The x-axes show genomic position (chromosomes 1–22), and the y axes show statistical significance as –log10[p-value]. P-values are two-sided and based on an inverse-variance-weighted fixed-effects meta-analysis. The dashed black lines show the GWS threshold (P < 5 × 10−8). The star indicates the position of the East Asian GWS locus (rs117130410, 4:105734758, build GRCh37).
Extended Data Fig. 5 Cluster-level SNP-heritability enrichment for bipolar disorder.
The t-distributed stochastic neighbor embedding (tSNE) plot (left) (from Siletti et al.23) is coloured by the enrichment z-score. Grey indicates non-significantly enriched superclusters (FDR > 0.05). The barplot (right) shows the top 35 significantly enriched clusters. The numbers in parentheses on the y-axis indicate the cell type clusters as defined in Siletti et al.23.
Extended Data Fig. 6 Number of SNPs within the smallest 95% credible sets (CS) from meta-analysis of European and multi-ancestry meta-analyses when excluding and including self-report data.
Colours represent CS of varying size, with blue CS containing 0 SNPs and red CS containing 15+ SNPs. All fine-mapped SNPs regardless of their PIP were used to assess the size of the 95% credible sets.
Extended Data Fig. 8 Clustering of patterns of temporal variation in expression of 34 credible genes.
Cluster 1 (n = 21 genes) shows reduced prenatal gene expression, with gene expression peaking at birth and remaining stable over the life-course. Cluster 2 (n = 13 genes) includes genes with a peak gene expression during fetal development with a drop-off in expression before birth. Genes within each cluster are described in Supplementary Table 31. To illustrate the variability in gene expression within each cluster we plot each donor expression value in each sampled brain region for the 34 credible genes as individual points. Smoothing splines used to illustrate the age trajectory for each cluster is based on generalized additive models with the predicted 95% confidence interval in grey. We use age in days to plot the variation in gene expression with the x-axis on a log2 scale and labels for birth, 10, 18, and 65 years of age as reference points.
Supplementary information
Supplementary Information
The main supplementary information document, containing the Detailed description of TWAS, FOCUS and isoTWAS eQTL analyses, Supplementary Figures 1 and 2, Cohort descriptions, Detailed acknowledgements and Funding Sources, and Consortium/Group authors and affiliations.
Supplementary Tables
Supplementary Tables 1-35.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
O’Connell, K.S., Koromina, M., van der Veen, T. et al. Genomics yields biological and phenotypic insights into bipolar disorder. Nature 639, 968–975 (2025). https://doi.org/10.1038/s41586-024-08468-9
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41586-024-08468-9
This article is cited by
-
The Swedish bipolar collection (SWEBIC)
International Journal of Bipolar Disorders (2025)
-
Single-molecule DNA analysis implicates brain mitochondria pathology in bipolar disorder
Molecular Psychiatry (2025)
-
Canonical and non-canonical roles of oligodendrocyte precursor cells in mental disorders
npj Mental Health Research (2025)
-
Genomics of schizophrenia, bipolar disorder and major depressive disorder
Nature Reviews Genetics (2025)
-
Fine-mapping genomic loci refines bipolar disorder risk genes
Nature Neuroscience (2025)