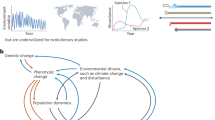
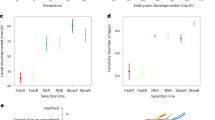
Abstract
From experimental evolution in the laboratory to sustained measurements of natural selection in the wild, long-term studies have revolutionized our understanding of evolution. By directly investigating evolutionary dynamics in real time, these approaches have provided unparallelled insights into the complex interplay between evolutionary process and pattern. These approaches can reveal oscillations, stochastic fluctuations and systematic trends that unfold over extended periods, expose critical time lags between environmental shifts and population responses, and illuminate how subtle effects may accumulate into significant evolutionary patterns. Long-term studies can also reveal otherwise cryptic trends that unfold over extended periods, and offer the potential for serendipity: observing rare events that spur new evolutionary hypotheses and research directions. Despite the challenges of conducting long-term research, exacerbated by modern funding landscapes favouring short-term projects, the contributions of long-term studies to evolutionary biology are indispensable. This is particularly true in our rapidly changing, human-dominated world, where such studies offer a crucial window into how environmental changes and altered species interactions shape evolutionary trajectories. In this Review article, we showcase the groundbreaking discoveries of long-term evolutionary studies, underscoring their crucial role in advancing our understanding of the complex nature of evolution across multiple systems and timescales.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Grant, P. R. & Grant, B. R. Unpredictable evolution in a 30-year study of Darwin’s finches. Science 296, 707–711 (2002).
Bozdag, G. O. et al. De novo evolution of macroscopic multicellularity. Nature 617, 747–754 (2023).
Good, B. H., McDonald, M. J., Barrick, J. E., Lenski, R. E. & Desai, M. M. The dynamics of molecular evolution over 60,000 generations. Nature 551, 45–50 (2017).
Kassen, R. Experimental Evolution and the Nature of Biodiversity (Oxford Univ. Press, 2024).
Endler, J. A. Natural Selection in the Wild (Princeton Univ. Press, 1986).
Grant, P. R. & Grant, B. R. 40 Years of Evolution: Darwin’s Finches on Daphne Major Island (Princeton Univ. Press, 2014). A seminal book summarizing compelling evidence for rapid evolution by natural selection, based on the meticulous long-term field studies by the authors, spanning four decades of morphological changes in Galápagos finches in response to environmental pressures.
Losos, J. B. Improbable Destinies: Fate, Chance, and the Future of Evolution (Penguin, 2018).
Exciting times for evolutionary biology. Nat. Ecol. Evol. 8, 593–594 (2024).
Lamichhaney, S. et al. Rapid hybrid speciation in Darwin’s finches. Science 359, 224–228 (2018). This long-term field study documents the rapid formation of a new species in the wild through hybridization between two distinct species of Darwin’s finches in the Galápagos, demonstrating that hybridization can be a powerful mechanism driving rapid evolutionary diversification.
Lenski, R. E. Revisiting the design of the long-term evolution experiment with Escherichia coli. J. Mol. Evol. 91, 241–253 (2023). This paper offers valuable insights into the design and implementation of the LTEE, highlighting the key features that have made this study so successful for studying evolutionary dynamics, potential improvements to experimental design and future directions for this ongoing experiment.
Barrick, J. E. et al. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461, 1243–1247 (2009).
Blount, Z. D., Borland, C. Z. & Lenski, R. E. Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proc. Natl Acad. Sci. USA 105, 7899–7906 (2008). This article demonstrates the crucial role of historical contingency in the evolution of novel traits, using the LTEE with E. coli to show that the emergence of a key innovation was dependent on the specific sequence of previous mutations, highlighting the importance of chance events and the order of mutations in shaping evolutionary outcomes.
Silvertown, J. et al. The Park Grass Experiment 1856–2006: its contribution to ecology. J. Ecol. 94, 801–814 (2006).
Snaydon, R. & Davies, M. Rapid population differentiation in a mosaic environment. Heredity 37, 9–25 (1976).
Siepielski, A. M. et al. No evidence that warmer temperatures are associated with selection for smaller body sizes. Proc. R. Soc. B 286, 20191332 (2019).
Travisano, M. in Experimental Evolution: Concepts, Methods, and Applications of Selection Experiments (eds Garland, T. & Rose, M. R.) 111–133 (2009).
Clutton-Brock, T. & Sheldon, B. C. Individuals and populations: the role of long-term, individual-based studies of animals in ecology and evolutionary biology. Trends Ecol. Evol. 25, 562–573 (2010). This paper emphasizes the critical importance of long-term, individual-based studies in understanding the complex interactions between ecological and evolutionary processes, as well as the role of individual variation in shaping population dynamics and evolutionary trajectories.
Sheldon, B. C., Kruuk, L. E. & Alberts, S. C. The expanding value of long-term studies of individuals in the wild. Nat. Ecol. Evol. 6, 1799–1801 (2022).
Coulson, T. et al. Age, sex, density, winter weather, and population crashes in Soay sheep. Science 292, 1528–1531 (2001).
Ozgul, A. et al. The dynamics of phenotypic change and the shrinking sheep of St. Kilda. Science 325, 464–467 (2009).
Moose, S. P., Dudley, J. W. & Rocheford, T. R. Maize selection passes the century mark: a unique resource for 21st century genomics. Trends Plant Sci. 9, 358–364 (2004).
Reznick, D. N. & Travis, J. Experimental studies of evolution and eco-evo dynamics in guppies (Poecilia reticulata). Annu. Rev. Ecol. Evol. Syst. 50, 335–354 (2019). This article summarizes decades of field experiments of Trinidadian guppies investigating the complex interplay between ecological and evolutionary processes, focusing on how predation, resource availability and other environmental factors can drive adaptive changes in life history traits, morphology and behaviour.
Reznick, D. N., Ghalambor, C. K. & Crooks, K. Experimental studies of evolution in guppies: a model for understanding the evolutionary consequences of predator removal in natural communities. Mol. Ecol. 17, 97–107 (2008).
Heckley, A. M., Pearce, A. E., Gotanda, K. M., Hendry, A. P. & Oke, K. B. Compiling forty years of guppy research to investigate the factors contributing to (non) parallel evolution. J. Evol. Biol. 35, 1414–1431 (2022).
Schoener, T. W., Kolbe, J. J., Leal, M., Losos, J. B. & Spiller, D. A. A multigenerational field experiment on eco-evolutionary dynamics of the influential lizard Anolis sagrei: a mid-term report. Copeia 105, 543–549 (2017). This study provides valuable insights into the interplay between ecological and evolutionary processes by demonstrating rapid adaptive changes in Anolis lizard populations in response to experimentally manipulated environmental conditions, highlighting the importance of long-term field studies in understanding the dynamics of eco-evolutionary feedbacks in real time.
Travis, J. et al. in Advances in Ecological Research Vol. 50 (eds MoyaLarano, J. et al.) 1–40 (Elsevier, 2014).
Philiptschenko, J. Variabilität Und Variation (Gebrüder Borntraeger, 1927).
Rolland, J. et al. Conceptual and empirical bridges between micro-and macroevolution. Nat. Ecol. Evol. 7, 1181–1193 (2023).
Herron, M. D., Conlin, P. L. & Ratcliff, W. C. The Evolution of Multicellularity (CRC Press, 2022).
Jacobeen, S. et al. Cellular packing, mechanical stress and the evolution of multicellularity. Nat. Phys. 14, 286–290 (2018).
Ratcliff, W. C., Fankhauser, J. D., Rogers, D. W., Greig, D. & Travisano, M. Origins of multicellular evolvability in snowflake yeast. Nat. Commun. 6, 6102 (2015).
Montrose, K. et al. Proteostatic tuning underpins the evolution of novel multicellular traits. Sci. Adv. 10, eadn2706 (2024).
Zamani-Dahaj, S. A. et al. Spontaneous emergence of multicellular heritability. Genes 14, 1635 (2023).
Coyne, J. A. & Orr, H. A. Speciation (Sinauer Associates, 2004).
Grant, P. R. & Grant, B. R. How and Why Species Multiply: The Radiation of Darwin’s Finches (Princeton Univ. Press, 2007).
Cracraft, J. in Evolution Innovation (ed. Nitecki, M. H.) 21–44 (1990).
Miller, A. H. & Stroud, J. T. Novel tests of the key innovation hypothesis: adhesive toepads in arboreal lizards. Syst. Biol. 71, 139–152 (2022).
Stroud, J. T. & Losos, J. B. Ecological opportunity and adaptive radiation. Annu. Rev. Ecol. Evol. Syst. 47, 507–532 (2016).
Simpson, G. G. in The Major Features of Evolution (Columbia Univ. Press, 1953).
Erwin, D. H. A conceptual framework of evolutionary novelty and innovation. Biol. Rev. 96, 1–15 (2021).
Miller, A. H., Stroud, J. T. & Losos, J. B. The ecology and evolution of key innovations. Trends Ecol. Evol. 38, 122–131 (2023).
Rabosky, D. L. Phylogenetic tests for evolutionary innovation: the problematic link between key innovations and exceptional diversification. Phil. Trans. R. Soc. B 372, 20160417 (2017).
Blount, Z. D., Barrick, J. E., Davidson, C. J. & Lenski, R. E. Genomic analysis of a key innovation in an experimental Escherichia coli population. Nature 489, 513–518 (2012).
Hall, B. G. Chromosomal mutation for citrate utilization by Escherichia coli K-12. J. Bacteriol. 151, 269–273 (1982).
Turner, C. B., Blount, Z. D., Mitchell, D. H. & Lenski, R. E. Evolution of a cross-feeding interaction following a key innovation in a long-term evolution experiment with Escherichia coli. Microbiology 169, 001390 (2023).
Eldredge, N. et al. The dynamics of evolutionary stasis. Paleobiology 31, 133–145 (2005).
Siepielski, A. M., DiBattista, J. D. & Carlson, S. M. It’s about time: the temporal dynamics of phenotypic selection in the wild. Ecol. Lett. 12, 1261–1276 (2009).
Siepielski, A. M. et al. Precipitation drives global variation in natural selection. Science 355, 959–962 (2017).
Wake, D. B., Roth, G. & Wake, M. H. On the problem of stasis in organismal evolution. J. Theor. Biol. 101, 211–224 (1983).
Estes, S. & Arnold, S. J. Resolving the paradox of stasis: models with stabilizing selection explain evolutionary divergence on all timescales. Am. Nat. 169, 227–244 (2007).
Charlesworth, B., Lande, R. & Slatkin, M. A neo-Darwinian commentary on macroevolution. Evolution 36, 474–498 (1982).
Stroud, J. T., Moore, M., Langerhans, R. B. & Losos, J. B. Fluctuating selection maintains distinct species phenotypes in an ecological community in the wild. Proc. Natl Acad. Sci. USA 120, e2222071120 (2023).
Gibbs, H. L. & Grant, P. R. Oscillating selection on Darwin’s finches. Nature 327, 511–513 (1987).
Eldredge, N. Macroevolution Dynamics (McGraw-Hill, 1989).
Svensson, E. & Calsbeek, R. The Adaptive Landscape in Evolutionary Biology (Oxford Univ. Press, 2012).
Wadgymar, S. M., Daws, S. C. & Anderson, J. T. Integrating viability and fecundity selection to illuminate the adaptive nature of genetic clines. Evol. Lett. 1, 26–39 (2017).
Grant, P. R. & Grant, B. R. From microcosm to macrocosm: adaptive radiation of Darwin’s finches. Evol. J. Linn. Soc. 3, kzae006 (2024).
Burga, A., Ben-David, E., Lemus Vergara, T., Boocock, J. & Kruglyak, L. Fast genetic mapping of complex traits in C. elegans using millions of individuals in bulk. Nat. Commun. 10, 2680 (2019).
Huxley, J. Evolution: The Modern Synthesis (Allen and Unwin, 1942).
Gould, S. J. The Structure of Evolutionary Theory (Harvard Univ. Press, 2002).
Dobzhansky, T. Genetics and the Origin of Species (Columbia Univ. Press, 1982).
Fisher, R. A. The Genetical Theory of Natural Selection (Clarendon Press, 1930).
Wright, S. Evolution in Mendelian populations. Genetics 16, 97–159 (1931).
Haldane, J. B. The Causes of Evolution Vol. 5 (Princeton Univ. Press, 1990).
Wadgymar, S. M., DeMarche, M. L., Josephs, E. B., Sheth, S. N. & Anderson, J. T. Local adaptation: causal agents of selection and adaptive trait divergence. Annu. Rev. Ecol. Evol. Syst. 53, 87–111 (2022).
Crawley, M. et al. Determinants of species richness in the Park Grass Experiment. Am. Nat. 165, 179–192 (2005).
Snaydon, R. Rapid population differentiation in a mosaic environment. I. The response of Anthoxanthum odoratum populations to soils. Evolution 24, 257–269 (1970).
Clausen, J., Keck, D. D. & Hiesey, W. M. Regional differentiation in plant species. Am. Nat. 75, 231–250 (1941).
Clausen, J. & Hiesey, W. M. Experimental Studies on the Nature of Species. IV. Genetic Structure of Ecological Races (Carnegie Institute, 1958).
Couce, A. et al. Changing fitness effects of mutations through long-term bacterial evolution. Science 383, eadd1417 (2024). This article provides a groundbreaking demonstration of how the fitness effects of mutations can change over the course of long-term evolution, using the LTEE with E. coli to show that mutations that were initially beneficial can become neutral or even deleterious as the genetic background evolves through time.
Wiser, M. J., Ribeck, N. & Lenski, R. E. Long-term dynamics of adaptation in asexual populations. Science 342, 1364–1367 (2013).
Kryazhimskiy, S., Rice, D. P., Jerison, E. R. & Desai, M. M. Global epistasis makes adaptation predictable despite sequence-level stochasticity. Science 344, 1519–1522 (2014).
Lenski, R. E. Experimental evolution and the dynamics of adaptation and genome evolution in microbial populations. ISME J. 11, 2181–2194 (2017).
Couce, A. & Tenaillon, O. A. The rule of declining adaptability in microbial evolution experiments. Front. Genet. 6, 128797 (2015).
Chou, H.-H., Chiu, H.-C., Delaney, N. F., Segrè, D. & Marx, C. J. Diminishing returns epistasis among beneficial mutations decelerates adaptation. Science 332, 1190–1192 (2011).
Burke, M. K. et al. Genome-wide analysis of a long-term evolution experiment with Drosophila. Nature 467, 587–590 (2010).
Bonnet, T. et al. Genetic variance in fitness indicates rapid contemporary adaptive evolution in wild animals. Science 376, 1012–1016 (2022). This paper compiles long-term datasets from evolutionary field studies to assess rapid adaptive evolution in a wide range of wild animal populations, highlighting the importance of considering contemporary evolutionary changes in conservation and management strategies.
Kruuk, L. E. et al. Antler size in red deer: heritability and selection but no evolution. Evolution 56, 1683–1695 (2002).
Kruuk, L. E., Merilä, J. & Sheldon, B. C. Phenotypic selection on a heritable size trait revisited. Am. Nat. 158, 557–571 (2001).
Milner, J. M., Albon, S. D., Illius, A. W., Pemberton, J. M. & Clutton-Brock, T. H. Repeated selection of morphometric traits in the Soay sheep on St Kilda. J. Anim. Ecol. 68, 472–488 (1999).
Larsson, K., Van der Jeugd, H. P., Van der Veen, I. T. & Forslund, P. Body size declines despite positive directional selection on heritable size traits in a barnacle goose population. Evolution 52, 1169–1184 (1998).
Merilä, J., Kruuk, L. & Sheldon, B. Cryptic evolution in a wild bird population. Nature 412, 76–79 (2001).
Stroud, J. T. et al. Observing character displacement from process to pattern in a novel vertebrate community. Nat. Commun. 15, 9862 (2024).
Gauzere, J. et al. Maternal effects do not resolve the paradox of stasis in birth weight in a wild red deer populaton. Evolution 76, 2605–2617 (2022).
Kruuk, L. E., Clutton-Brock, T. & Pemberton, J. M. in Quantitative Genetics in the Wild Vol. 10 (eds Charmantier, A. et al.) 160–176 (Oxford Univ. Press, 2014).
Pemberton, J. M. Evolution of quantitative traits in the wild: mind the ecology. Phil. Trans. R. Soc. B 365, 2431–2438 (2010).
Conner, J. K. Quantitative genetic approaches to evolutionary constraint: how useful? Evolution 66, 3313–3320 (2012).
Price, T., Kirkpatrick, M. & Arnold, S. J. Directional selection and the evolution of breeding date in birds. Science 240, 798–799 (1988).
Etterson, J. R. & Shaw, R. G. Constraint to adaptive evolution in response to global warming. Science 294, 151–154 (2001).
Sheldon, B. C., Kruuk, L. E. B. & Merila, J. Natural selection and inheritance of breeding time and clutch size in the collared flycatcher. Evolution 57, 406–420 (2003).
Brookfield, J. F. Why are estimates of the strength and direction of natural selection from wild populations not congruent with observed rates of phenotypic change? BioEssays 38, 927–934 (2016).
Glądalski, M. et al. Extreme temperature drop alters hatching delay, reproductive success, and physiological condition in great tits. Int. J. Biometeorol. 64, 623–629 (2020).
Benton, T. G., Grant, A. & Clutton-Brock, T. H. Does environmental stochasticity matter? Analysis of red deer life-histories on Rum. Evol. Ecol. 9, 559–574 (1995).
Pemberton, J. M., Kruuk, L. E. & Clutton-Brock, T. The unusual value of long-term studies of individuals: the example of the Isle of Rum red deer project. Annu. Rev. Ecol. Evol. Syst. 53, 327–351 (2022).
Brown, W. L. & Wilson, E. O. Character displacement. Syst. Zool. 5, 49–64 (1956).
Grant, P. R. & Grant, B. R. The founding of a new population of Darwin’s finches. Evolution 49, 229–240 (1995).
Grant, P. R. & Grant, B. R. Evolution of character displacement in Darwin’s finches. Science 313, 224–226 (2006).
Boag, P. T. The heritability of external morphology in Darwin’s ground finches (Geospiza) on Isla Daphne Major, Galapagos. Evolution 37, 877–894 (1983).
Lamichhaney, S. et al. A beak size locus in Darwin’s finches facilitated character displacement during a drought. Science 352, 470–474 (2016).
Anderson, J. T., Inouye, D. W., McKinney, A. M., Colautti, R. I. & Mitchell-Olds, T. Phenotypic plasticity and adaptive evolution contribute to advancing flowering phenology in response to climate change. Proc. R. Soc. B Biol. Sci. 279, 3843–3852 (2012). This article highlights the importance of both phenotypic plasticity and adaptive evolution in enabling plant populations to respond to contemporary climate change by demonstrating that earlier flowering times in a long-term study of mountain wildflowers is driven by a combination of plastic responses and genetic changes.
Wadgymar, S. M., Ogilvie, J. E., Inouye, D. W., Weis, A. E. & Anderson, J. T. Phenological responses to multiple environmental drivers under climate change: insights from a long-term observational study and a manipulative field experiment. New Phytol. 218, 517–529 (2018).
Anderson, J. T. & Gezon, Z. J. Plasticity in functional traits in the context of climate change: a case study of the subalpine forb Boechera stricta (Brassicaceae). Glob. Chang. Biol. 21, 1689–1703 (2015).
Charmantier, A. et al. Adaptive phenotypic plasticity in response to climate change in a wild bird population. Science 320, 800–803 (2008).
Primack, R. B., Higuchi, H. & Miller-Rushing, A. J. The impact of climate change on cherry trees and other species in Japan. Biol. Conserv. 142, 1943–1949 (2009).
Bates, J. M. et al. Climate change affects bird nesting phenology: comparing contemporary field and historical museum nesting records. J. Anim. Ecol. 92, 263–272 (2023).
De Lisle, S. P., Mäenpää, M. I. & Svensson, E. I. Phenotypic plasticity is aligned with phenological adaptation on both micro- and macroevolutionary timescales. Ecol. Lett. 25, 790–801 (2022).
Bonnet, T. et al. The role of selection and evolution in changing parturition date in a red deer population. PLoS Biol. 17, e3000493 (2019).
Martin, R. A., da Silva, C. R., Moore, M. P. & Diamond, S. E. When will a changing climate outpace adaptive evolution? Wiley Interdiscip. Rev. Clim. Change 14, e852 (2023).
Simmonds, E. G., Cole, E. F., Sheldon, B. C. & Coulson, T. Phenological asynchrony: a ticking time-bomb for seemingly stable populations? Ecol. Lett. 23, 1766–1775 (2020).
Merilä, J. & Hendry, A. P. Climate change, adaptation, and phenotypic plasticity: the problem and the evidence. Evol. Appl. 7, 1–14 (2014).
Valladares, F. et al. The effects of phenotypic plasticity and local adaptation on forecasts of species range shifts under climate change. Ecol. Lett. 17, 1351–1364 (2014).
Ford, E. B. Problems of heredity in the Lepidoptera. Biol. Rev. 12, 461–501 (1937).
Kettlewell, H. B. D. Selection experiments on industrial melanism in the Lepidoptera. Heredity 9, 323–342 (1955).
Cook, L. M., Grant, B. S., Saccheri, I. J. & Mallet, J. Selective bird predation on the peppered moth: the last experiment of Michael Majerus. Biol. Lett. 8, 609–612 (2012).
Kettlewell, H. B. D. The Evolution of Melanism (Oxford Univ. Press, 1973).
Cook, L. M., Dennis, R. L. & Dockery, M. The melanic form of the peppered moth, Biston betularia (Linnaeus, 1758)(Lepidoptera: Geometridae), in Manchester: the end of an era. Entomol. Gaz. 62, 91–99 (2011).
Czorlich, Y., Aykanat, T., Erkinaro, J., Orell, P. & Primmer, C. Rapid evolution in salmon life history induced by direct and indirect effects of fishing. Science 376, 420–423 (2022).
Heino, M., Díaz, Pauli, B. & Dieckmann, U. Fisheries-induced evolution. Annu. Rev. Ecol. Evol. Syst. 46, 461–480 (2015).
Donihue, C. M. & Lambert, M. R. Adaptive evolution in urban ecosystems. Ambio 44, 194–203 (2015).
Santangelo, J. S. et al. Global urban environmental change drives adaptation in white clover. Science 375, 1275–1281 (2022).
Lambert, M. R. & Donihue, C. M. Urban biodiversity management using evolutionary tools. Nat. Ecol. Evol. 4, 903–910 (2020).
Blount, Z. D., Lenski, R. E. & Losos, J. B. Contingency and determinism in evolution: replaying life’s tape. Science 362, eaam5979 (2018).
Reznick, D. & Travis, J. Is evolution predictable? Science 359, 738–739 (2018).
Conway Morris, S. Evolution: like any other science it is predictable. Phil. Trans. R. Soc. B 365, 133–145 (2010).
Beavan, A. J., Domingo-Sananes, M. R. & McInerney, J. O. Contingency, repeatability, and predictability in the evolution of a prokaryotic pangenome. Proc. Natl Acad. Sci. USA 121, e2304934120 (2024).
Morris, S. C. Life’s Solution: Inevitable Humans in a Lonely Universe (Cambridge Univ. Press, 2003).
Nosil, P., Flaxman, S. M., Feder, J. L. & Gompert, Z. Increasing our ability to predict contemporary evolution. Nat. Commun. 11, 5592 (2020).
Marques, D. A., Jones, F. C., Di Palma, F., Kingsley, D. M. & Reimchen, T. E. Experimental evidence for rapid genomic adaptation to a new niche in an adaptive radiation. Nat. Ecol. Evol. 2, 1128–1138 (2018).
Gompert, Z., Flaxman, S. M., Feder, J. L., Chevin, L.-M. & Nosil, P. Laplace’s demon in biology: models of evolutionary prediction. Evolution 76, 2794–2810 (2022).
Barrick, J. E. & Lenski, R. E. Genome dynamics during experimental evolution. Nat. Rev. Genet. 14, 827–839 (2013).
Jerison, E. R., Nguyen Ba, A. N., Desai, M. M. & Kryazhimskiy, S. Chance and necessity in the pleiotropic consequences of adaptation for budding yeast. Nat. Ecol. Evol. 4, 601–611 (2020).
Tenaillon, O. et al. The molecular diversity of adaptive convergence. Science 335, 457–461 (2012).
Lang, G. I. et al. Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature 500, 571–574 (2013).
Salverda, M. L., Koomen, J., Koopmanschap, B., Zwart, M. P. & de Visser, J. A. G. Adaptive benefits from small mutation supplies in an antibiotic resistance enzyme. Proc. Natl Acad. Sci. USA 114, 12773–12778 (2017).
Tenaillon, O. et al. Tempo and mode of genome evolution in a 50,000-generation experiment. Nature 536, 165–170 (2016).
Quandt, E. M., Deatherage, D. E., Ellington, A. D., Georgiou, G. & Barrick, J. E. Recursive genomewide recombination and sequencing reveals a key refinement step in the evolution of a metabolic innovation in Escherichia coli. Proc. Natl Acad. Sci. USA 111, 2217–2222 (2014).
Nosil, P. et al. Natural selection and the predictability of evolution in Timema stick insects. Science 359, 765–770 (2018). This article demonstrates that the direction and magnitude of natural selection can be used to predict evolutionary changes in wild populations using long-term field studies of natural selection in Californian Timema stick insects.
Nosil, P. et al. Evolution repeats itself in replicate long-term studies in the wild. Sci. Adv. 10, eadl3149 (2024).
Thurman, T. J. et al. The difficulty of predicting evolutionary change in response to novel ecological interactions: a field experiment with Anolis lizards. Am. Nat. 201, 537–556 (2023).
Chevin, L.-M., Gompert, Z. & Nosil, P. Frequency dependence and the predictability of evolution in a changing environment. Evol. Lett. 6, 21–33 (2022).
Hendry, A. P. Prediction in ecology and evolution. BioScience 73, 785–799 (2023).
Schluter, D. Variable success in linking micro and macroevolution. Evol. J. Linn. Soc. 3, kzae016 (2024).
Hendry, A. P. Eco-Evolutionary Dynamics (Princeton Univ. Press, 2017).
Schoener, T. W. The newest synthesis: understanding the interplay of evolutionary and ecological dynamics. Science 331, 426–429 (2011).
Post, D. M. & Palkovacs, E. P. Eco-evolutionary feedbacks in community and ecosystem ecology: interactions between the ecological theatre and the evolutionary play. Phil. Trans. R. Soc. B 364, 1629–1640 (2009).
Bassar, R. D., Coulson, T., Travis, J. & Reznick, D. N. Towards a more precise—and accurate—view of eco-evolution. Ecol. Lett. 24, 623–625 (2021).
Hendry, A. P. A critique for eco-evolutionary dynamics. Funct. Ecol. 33, 84–94 (2019).
Kokko, H. & López-Sepulcre, A. The ecogenetic link between demography and evolution: can we bridge the gap between theory and data? Ecol. Lett. 10, 773–782 (2007).
Yoshida, T., Jones, L. E., Ellner, S. P., Fussmann, G. F. & Hairston, N. G. Jr. Rapid evolution drives ecological dynamics in a predator–prey system. Nature 424, 303–306 (2003).
Mougi, A. Eco-evolutionary dynamics in microbial interactions. Sci. Rep. 13, 9042 (2023).
Hart, S. P., Turcotte, M. M. & Levine, J. M. Effects of rapid evolution on species coexistence. Proc. Natl Acad. Sci. USA 116, 2112–2117 (2019).
Rodríguez-Verdugo, A. & Ackermann, M. Rapid evolution destabilizes species interactions in a fluctuating environment. ISME J. 15, 450–460 (2021).
Kasada, M., Yamamichi, M. & Yoshida, T. Form of an evolutionary tradeoff affects eco-evolutionary dynamics in a predator–prey system. Proc. Natl Acad. Sci. USA 111, 16035–16040 (2014).
De Meester, L. et al. Analysing eco-evolutionary dynamics—the challenging complexity of the real world. Funct. Ecol. 33, 43–59 (2019).
Spiller, D. A., Schoener, T. W. & Piovia-Scott, J. in Ecology and Evolution of Plant–Herbivore Interactions on Islands (eds Moreira, X. & Abdala-Roberts, L.) 177–197 (Springer, 2024).
Reznick, D. N. et al. Eco-evolutionary feedbacks predict the time course of rapid life-history evolution. Am. Nat. 194, 671–692 (2019).
Gordon, S. P. et al. Adaptive changes in life history and survival following a new guppy introduction. Am. Nat. 174, 34–45 (2009).
Westrick, S. E., Broder, E. D., Reznick, D. N., Ghalambor, C. K. & Angeloni, L. Rapid evolution and behavioral plasticity following introduction to an environment with reduced predation risk. Ethology 125, 232–240 (2019).
Lapiedra, O., Schoener, T. W., Leal, M., Losos, J. B. & Kolbe, J. J. Predator-driven natural selection on risk-taking behavior in anole lizards. Science 360, 1017–1020 (2018).
Losos, J. B., Schoener, T. W. & Spiller, D. A. Predator-induced behaviour shifts and natural selection in field-experimental lizard populations. Nature 432, 505–508 (2004).
Simon, T. N. et al. Local adaptation in Trinidadian guppies alters stream ecosystem structure at landscape scales despite high environmental variability. Copeia 105, 504–513 (2017).
Lapiedra, O. et al. Predator-driven behavioural shifts in a common lizard shape resource-flow from marine to terrestrial ecosystems. Ecol. Lett. 27, e14335 (2024).
Schoener, T. W., Spiller, D. A. & Losos, J. B. Predation on a common Anolis lizard: can the food-web effects of a devastating predator be reversed? Ecol. Monogr. 72, 383–407 (2002).
Hendry, A. P. Eco-evolutionary dynamics: an experimental demonstration in nature. Curr. Biol. 33, R814–R817 (2023).
Waide, R. B. & Kingsland, S. E. The Challenges of Long Term Ecological Research: A Historical Analysis (Springer, 2021).
Bono, J. M., Olesnicky, E. C. & Matzkin, L. M. Connecting genotypes, phenotypes and fitness: harnessing the power of CRISPR/Cas9 genome editing. Mol. Ecol. 24, 3810–3822 (2015).
Wang, Z. et al. Automated detection of an insect-induced keystone vegetation phenotype using airborne LiDAR. Methods Ecol. Evol. 15, 978–993 (2024).
Kays, R., Crofoot, M. C., Jetz, W. & Wikelski, M. Terrestrial animal tracking as an eye on life and planet. Science 348, aaa2478 (2015).
Nathan, R. et al. Big-data approaches lead to an increased understanding of the ecology of animal movement. Science 375, eabg1780 (2022).
Lande, R. & Arnold, S. J. The measurement of selection on correlated characters. Evolution 37, 1220–1226 (1983).
Svensson, E. I. Phenotypic selection in natural populations: what have we learned in 40 years? Evolution 77, 1493–1504 (2023).
Huang, X., Rymbekova, A., Dolgova, O., Lao, O. & Kuhlwilm, M. Harnessing deep learning for population genetic inference. Nat. Rev. Genet. 25, 61–78 (2024).
Borowiec, M. L. et al. Deep learning as a tool for ecology and evolution. Methods Ecol. Evol. 13, 1640–1660 (2022).
Stoddard, M. C., Kilner, R. M. & Town, C. Pattern recognition algorithm reveals how birds evolve individual egg pattern signatures. Nat. Commun. 5, 4117 (2014).
Grenfell, B. T. et al. Unifying the epidemiological and evolutionary dynamics of pathogens. Science 303, 327–332 (2004).
Faria, N. R. et al. The early spread and epidemic ignition of HIV-1 in human populations. Science 346, 56–61 (2014).
Dadonaite, B. et al. Spike deep mutational scanning helps predict success of SARS-CoV-2 clades. Nature 631, 617–626 (2024).
Bitter, M. C. et al. Continuously fluctuating selection reveals fine granularity of adaptation. Nature 634, 389–396 (2024).
Rudman, S. M. et al. Direct observation of adaptive tracking on ecological time scales in Drosophila. Science 375, eabj7484 (2022).
Acknowledgements
We thank J. Anderson, Z. Blount, S. Brown, T. Grainger, J. Losos, M. Moore and K. Thompson for valuable feedback on earlier versions of this manuscript; B. Sheldon, D. Schluter and an anonymous reviewer for critical feedback that greatly improved our manuscript; and M. Belan (ArtSci Studios) for the illustrations. This work was supported by grants from the US National Institutes of Health (grant no. 5R35GM138030) and the NSF Division of Environmental Biology (grant no. DEB-1845363) to W.C.R.
Author information
Authors and Affiliations
Contributions
J.T.S. conceptualized the manuscript. J.T.S. and W.C.R. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Ben Sheldon, Dolph Schluter and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Stroud, J.T., Ratcliff, W.C. Long-term studies provide unique insights into evolution. Nature 639, 589–601 (2025). https://doi.org/10.1038/s41586-025-08597-9
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41586-025-08597-9
This article is cited by
-
Priority effects inhibit the repeated evolution of phototrophy
npj Complexity (2026)
-
The heritability of spatial memory and caching behaviour in a food-storing bird
Animal Cognition (2025)