Abstract
The current opioid overdose epidemic highlights the urgent need to develop safer and more effective treatments for chronic pain1. Cannabinoid receptor type 1 (CB1) is a promising non-opioid target for pain relief, but its clinical use has been limited by centrally mediated psychoactivity and tolerance. We overcame both issues by designing peripherally restricted CB1 agonists that minimize arrestin recruitment. We achieved these goals by computationally designing positively charged derivatives of the potent CB1 agonist MDMB-Fubinaca2. We designed these ligands to occupy a cryptic pocket identified through molecular dynamics simulations—an extended binding pocket that opens rarely and leads to the conserved signalling residue D2.50 (ref. 3). We used structure determination, pharmacological assays and molecular dynamics simulations to verify the binding modes of these ligands and to determine the molecular mechanism by which they achieve this dampening of arrestin recruitment. Our lead ligand, VIP36, is highly peripherally restricted and demonstrates notable efficacy in three mouse pain models, with 100-fold dose separation between analgesic efficacy and centrally mediated side effects. VIP36 exerts analgesic efficacy through peripheral CB1 receptors and shows limited analgesic tolerance. These results show how targeting a cryptic pocket in a G-protein-coupled receptor can lead to enhanced peripheral selectivity, biased signalling, desired in vivo pharmacology and reduced adverse effects. This has substantial implications for chronic pain treatment but could also revolutionize the design of drugs targeting other G-protein-coupled receptors.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The cryo-EM maps and corresponding coordinates will be deposited in the Electron Microscopy Data Bank under accession codes EMD-44199 (VIP36–CB1–Gi–scFv16) and EMD-44247 (VIP2.33–CB1–Gi) and in the PDB under accession codes 9b54 (VIP36–CB1–Gi–scFv16) and 9b65 (VIP2.33–CB1–Gi). The authors declare that all data supporting the findings of this study are available within the article, extended data and supplementary information files. All compounds can be made available upon reasonable request from the authors. Source data are provided with this paper.
References
Ciccarone, D. The rise of illicit fentanyls, stimulants and the fourth wave of the opioid overdose crisis. Curr. Opin. Psychiatry 34, 344–350 (2021).
Gamage, T. F. et al. Molecular and behavioral pharmacological characterization of abused synthetic cannabinoids MMB- and MDMB-FUBINACA, MN-18, NNEI, CUMYL-PICA, and 5-Fluoro-CUMYL-PICA. J. Pharmacol. Exp. Ther. 365, 437–446 (2018).
Zarzycka, B., Zaidi, S. A., Roth, B. L. & Katritch, V. Harnessing ion-binding sites for GPCR pharmacology. Pharmacol. Rev. 71, 571–595 (2019).
Nahin, R. L. Estimates of pain prevalence and severity in adults: United States, 2012. J. Pain 16, 769–780 (2015).
Cohen, S. P., Vase, L. & Hooten, W. M. Chronic pain: an update on burden, best practices, and new advances. Lancet 397, 2082–2097 (2021).
Volkow, N. D. & Blanco, C. The changing opioid crisis: development, challenges and opportunities. Mol. Psychiatry 26, 218–233 (2021).
Pertwee, R. G. Pharmacology of cannabinoid CB1 and CB2 receptors. Pharmacol. Ther. 74, 129–180 (1997).
Woodhams, S. G., Chapman, V., Finn, D. P., Hohmann, A. G. & Neugebauer, V. The cannabinoid system and pain. Neuropharmacology 124, 105–120 (2017).
Finlay, D. B. et al. Do toxic synthetic cannabinoid receptor agonists have signature in vitro activity profiles? A case study of AMB-FUBINACA. ACS Chem. Neurosci. 10, 4350–4360 (2019).
Banister, S. D. et al. Pharmacology of indole and indazole synthetic cannabinoid designer drugs AB-FUBINACA, ADB-FUBINACA, AB-PINACA, ADB-PINACA, 5F-AB-PINACA, 5F-ADB-PINACA, ADBICA, and 5F-ADBICA. ACS Chem. Neurosci. 6, 1546–1559 (2015).
Banister, S. D. et al. Pharmacology of valinate and tert-leucinate synthetic cannabinoids 5F-AMBICA, 5F-AMB, 5F-ADB, AMB-FUBINACA, MDMB-FUBINACA, MDMB-CHMICA, and their analogues. ACS Chem. Neurosci. 7, 1241–1254 (2016).
Wiley, J. L. et al. AB-CHMINACA, AB-PINACA, and FUBIMINA: affinity and potency of novel synthetic cannabinoids in producing Δ9-tetrahydrocannabinol-like effects in mice. J. Pharmacol. Exp. Ther. 354, 328–339 (2015).
Slivicki, R. A., Xu, Z., Mali, S. S. & Hohmann, A. G. Brain permeant and impermeant inhibitors of fatty-acid amide hydrolase suppress the development and maintenance of paclitaxel-induced neuropathic pain without producing tolerance or physical dependence in vivo and synergize with paclitaxel to reduce tumor cell line viability in vitro. Pharmacol. Res. 142, 267–282 (2019).
Henderson-Redmond, A. N. et al. c-Jun N terminal kinase signaling pathways mediate cannabinoid tolerance in an agonist-specific manner. Neuropharmacology 164, 107847 (2020).
Metna-Laurent, M., Mondésir, M., Grel, A., Vallée, M. & Piazza, P.-V. Cannabinoid-induced tetrad in mice. Curr. Protoc. Neurosci. 80, 9.59.1–9.59.10 (2017).
Clapper, J. R. et al. Anandamide suppresses pain initiation through a peripheral endocannabinoid mechanism. Nat. Neurosci. 13, 1265–1270 (2010).
Slivicki, R. A. et al. Brain-permeant and -impermeant inhibitors of fatty acid amide hydrolase synergize with the opioid analgesic morphine to suppress chemotherapy-induced neuropathic nociception without enhancing effects of morphine on gastrointestinal transit. J. Pharmacol. Exp. Ther. 367, 551–563 (2018).
Slivicki, R. A., Yi, J., Brings, V. E., Huynh, P. N. & Gereau, R. W. The cannabinoid agonist CB-13 produces peripherally mediated analgesia in mice but elicits tolerance and signs of central nervous system activity with repeated dosing. Pain 163, 1603–1621 (2022).
Gardin, A., Kucher, K., Kiese, B. & Appel-Dingemanse, S. Cannabinoid receptor agonist 13, a novel cannabinoid agonist: first in human pharmacokinetics and safety. Drug Metab. Dispos. 37, 827–833 (2009).
Ford, N. C. et al. Role of primary sensory neurone cannabinoid type-1 receptors in pain and the analgesic effects of the peripherally acting agonist CB-13 in mice. Br. J. Anaesth. 128, 159–173 (2022).
Seltzman, H. H. et al. Peripherally selective cannabinoid 1 receptor (CB1R) agonists for the treatment of neuropathic pain. J. Med. Chem. 59, 7525–7543 (2016).
Mulpuri, Y. et al. Synthetic peripherally-restricted cannabinoid suppresses chemotherapy-induced peripheral neuropathy pain symptoms by CB1 receptor activation. Neuropharmacology 139, 85–97 (2018).
Piscura, M. K. et al. Mechanisms of cannabinoid tolerance. Biochem. Pharmacol. 214, 115665 (2023).
Nguyen, P. T. et al. β-Arrestin2 regulates cannabinoid CB 1 receptor signaling and adaptation in a central nervous system region-dependent manner. Biol. Psychiatry 71, 714–724 (2012).
Ford, B. M. et al. Characterization of structurally novel G protein biased CB1 agonists: implications for drug development. Pharmacol. Res. 125, 161–177 (2017).
Liao, Y. Y. et al. Snapshot of the cannabinoid receptor 1-arrestin complex unravels the biased signaling mechanism. Cell 186, 5784–5797.e17 (2023).
Raehal, K. M. & Bohn, L. M. β-Arrestins: regulatory role and therapeutic potential in opioid and cannabinoid receptor-mediated analgesia. Handb. Exp. Pharmacol. 219, 427–443 (2014).
Breivogel, C. S., Lambert, J. M., Gerfin, S., Huffman, J. W. & Razdan, R. K. Sensitivity to Δ9-tetrahydrocannabinol is selectively enhanced in beta-arrestin2-/- mice. Behav. Pharmacol. 19, 298–307 (2008).
Wouters, E. et al. Assessment of biased agonism among distinct synthetic cannabinoid receptor agonist scaffolds. ACS Pharmacol. Transl. Sci. 3, 285–295 (2020).
Oleinikovas, V., Saladino, G., Cossins, B. P. & Gervasio, F. L. Understanding cryptic pocket formation in protein targets by enhanced sampling simulations. J. Am. Chem. Soc. 138, 14257–14263 (2016).
Wassman, C. D. et al. Computational identification of a transiently open L1/S3 pocket for reactivation of mutant p53. Nat. Commun. 4, 1407 (2013).
Ramos-Gonzalez, N., Paul, B. & Majumdar, S. IUPHAR themed review: opioid efficacy, bias, and selectivity. Pharmacol. Res. 197, 106961 (2023).
Manning, J. J., Rawcliffe, G., Finlay, D. B. & Glass, M. Cannabinoid 1 (CB1) receptor arrestin subtype-selectivity and phosphorylation dependence. Br. J. Pharmacol. 180, 369–382 (2023).
Faouzi, A. et al. Structure-based design of bitopic ligands for the µ-opioid receptor. Nature 613, 767–774 (2023).
Ople, R. S. et al. Signaling modulation mediated by ligand water interactions with the sodium site at μOR. ACS Cent. Sci. 10, 1490–1503 (2024).
Krishna Kumar, K. et al. Structure of a signaling cannabinoid receptor 1-G protein complex. Cell 176, 448–458.e12 (2019).
Krishna Kumar, K. et al. Structural basis for activation of CB1 by an endocannabinoid analog. Nat. Commun. 14, 2672 (2023).
Uprety, R. et al. Controlling opioid receptor functional selectivity by targeting distinct subpockets of the orthosteric site. eLife 10, 1–58 (2021).
Chakraborty, S. et al. A novel mitragynine analog with low-efficacy mu opioid receptor agonism displays antinociception with attenuated adverse effects. J. Med. Chem. 64, 13873–13892 (2021).
Qu, Q. et al. Insights into distinct signaling profiles of the µOR activated by diverse agonists. Nat. Chem. Biol. 19, 423–430 (2023).
Suomivuori, C.-M. et al. Molecular mechanism of biased signaling in a prototypical G protein-coupled receptor. Science 367, 881–887 (2020).
Wingler, L. M., McMahon, C., Staus, D. P., Lefkowitz, R. J. & Kruse, A. C. Distinctive activation mechanism for angiotensin receptor revealed by a synthetic nanobody. Cell 176, 479–490.e12 (2019).
Dziadulewicz, E. K. et al. Naphthalen-1-yl-(4-pentyloxynaphthalen-1-yl)methanone: a potent, orally bioavailable human CB1/CB2 dual agonist with antihyperalgesic properties and restricted central nervous system penetration. J. Med. Chem. 50, 3851–3856 (2007).
Lichtman, A. H. & Martin, B. R. Cannabinoid tolerance and dependence. Handb. Exp. Pharmacol. 168, 691–717 (2005).
Pradhan, A. A. et al. Characterization of a novel model of chronic migraine. Pain 155, 269–274 (2014).
Yamamoto, T. et al. Selective targeting of peripheral cannabinoid receptors prevents behavioral symptoms and sensitization of trigeminal neurons in mouse models of migraine and medication overuse headache. Pain 162, 2246–2262 (2021).
Besnard, J. et al. Automated design of ligands to polypharmacological profiles. Nature 492, 215–220 (2012).
Ramaekers, J. G., Mason, N. L. & Theunissen, E. L. Blunted highs: pharmacodynamic and behavioral models of cannabis tolerance. Eur. Neuropsychopharmacol. 36, 191–205 (2020).
Bass, C. E. & Martin, B. R. Time course for the induction and maintenance of tolerance to Delta(9)-tetrahydrocannabinol in mice. Drug Alcohol Depend. 60, 113–119 (2000).
Henderson-Redmond, A. N. et al. Sex differences in tolerance to delta-9-tetrahydrocannabinol in mice with cisplatin-evoked chronic neuropathic pain. Front. Mol. Biosci. 8, 684115 (2021).
Slivicki, R. A. et al. Impact of Δ9-tetrahydrocannabinol and oxycodone co-administration on measures of antinociception, dependence, circadian activity, and reward in mice. Preprint at bioRxiv https://doi.org/10.1101/2023.12.04.569809 (2023).
Colizzi, M. & Bhattacharyya, S. Cannabis use and the development of tolerance: a systematic review of human evidence. Neurosci. Biobehav. Rev. 93, 1–25 (2018).
D’Souza, D. C. et al. Blunted psychotomimetic and amnestic effects of delta-9-tetrahydrocannabinol in frequent users of cannabis. Neuropsychopharmacology 33, 2505–2516 (2008).
Jones, R. T., Benowitz, N. L. & Herning, R. I. Clinical relevance of cannabis tolerance and dependence. J. Clin. Pharmacol. 21, 143S–152S (1981).
Cuttler, C., Mischley, L. K. & Sexton, M. Sex differences in cannabis use and effects: a cross-sectional survey of cannabis users. Cannabis Cannabinoid Res. 1, 166–175 (2016).
Metna-Laurent, M., Mondésir, M., Grel, A., Vallée, M. & Piazza, P. V. Cannabinoid-induced tetrad in mice. Curr. Protoc. Neurosci. 2017, 9.59.1–9.59.10 (2017).
Zhang, H. et al. Peripherally restricted cannabinoid 1 receptor agonist as a novel analgesic in cancer-induced bone pain. Pain 159, 1814–1823 (2018).
Do, T. P., Hougaard, A., Dussor, G., Brennan, K. C. & Amin, F. M. Migraine attacks are of peripheral origin: the debate goes on. J. Headache Pain 24, 3 (2023).
Acknowledgements
This study was supported by the NIH through the NIH HEAL initiative under grant no. R34NS126036 (to S.M., R.O.D. and R.W.G.) and grant nos. F32DA051160 and K99DA056691 (to R.A.S.). N.R.-G. thanks the PhRMA Foundation Postdoctoral Fellowship (ISNI ID: 0000 0000 9959 8153; Crossref Funder ID: 100001797). The cryo-EM data were collected at the Stanford Cryo-Electron Microscopy Center with the help of E. Montabana. We thank B. White for providing the G proteins. Receptor-binding profiles and PRESTO-Tango were generously provided by the National Institute of Mental Health’s Psychoactive Drug Screening Program, contract no. HHSN-271-2018-00023-C. We thank V. Katritch and S. Zaidi from the University of Southern California for extensive discussions on the sodium site in GPCRs. We thank N. Lambert of Augusta University for sharing CB1-Rluc constructs with us.
Author information
Authors and Affiliations
Contributions
S.M. and R.W.G. conceived the study. V.A.R., M.B.U. and A.K. synthesized the compounds and aided in their chemical and pharmacological characterization under the supervision of S.M. E.S.O. prepared the CB1–Gi complex; obtained and processed the cryo-EM data; refined the structure from the cryo-EM density maps; and aided in signalling, structure-based ligand design and interpretation of bias under the supervision of K.K.K. and B.K.K. K.K.K. supervised the structural biology and signalling aspects of this project, in addition to the interpretation of CB1 bias and its correlation with structures. A.S.P. and D.A. performed ligand design and docking under the supervision of R.O.D. A.S.P., D.A. and B.L.S. performed and analysed molecular dynamics simulations under the supervision of R.O.D. K.A., B.R.V. and N.R.-G. performed the profiling studies under the supervision of S.M. Z.B., R.A.S., J.M., J.F.-H. and M.P. carried out the behavioural assays under the supervision of R.W.G. E.M., Y.A.-A. and J.A. carried out chronic migraine model and locomotor studies under the supervision of A.A.P. Y.S. performed the GTP turnover assay under the supervision of K.K.K. and B.K.K. T.N.P. carried out the radioligand binding affinity experiment under the supervision of K.K.K. and B.K.K. L.L. carried out the PK studies under the supervision of M.D.C. X.-P.H. carried out 45 CNS receptor radioligand binding and PRESTO-Tango assays. S.M., V.A.R., E.S.O., A.S.P., Z.B., R.A.S., K.K.K. and R.O.D. wrote the paper with contributions from other authors.
Corresponding authors
Ethics declarations
Competing interests
S.M. is a founder of Sparian Biosciences. B.K.K. is a founder of and consultant for ConfometRx. R.O.D. holds equity in Septerna, Inc. S.M., V.A.R., R.W.G., Z.B., R.A.S., A.S.P., D.A. and R.O.D. have filed a patent related to the lead compounds disclosed in this paper.
Peer review
Peer review information
Nature thanks Javier García-Nafría, Igor Spigelman and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Computational ligand design.
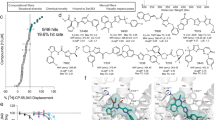
(a) Effect of 50 mM NaCl on CB1 agonist (FUB) and antagonist (SR) radioligand displacement assay against 3H-SR141716-bound CB1 expressing membranes. FUB-NaCl (IC50 = 26.2 nM (13.8-50.8 nM), FUB-No NaCl (IC50 = 21.3 nM (10.4-44.1 nM), SR-No NaCl (IC50 = 13.2 nM (8.1-21.7 nM), SR-NaCl (IC50 = 19.3 nM (9.9-40.5 nM). (b) Computational and structure-guided optimization of FUB analogs containing guanidinium group. Ligands with different linkers and attachment points were docked to the simulation frame containing the cryptic pocket using Schrodinger’s Glide (standard precision). VIP36 was the ligand with the best docking score and formed a tight interaction with the D2.50 sidechain. CI = Confidence interval.
Extended Data Fig. 2 Design of D2.50 site targeting ligand.
Design of novel ligands targeting CB1 D2.50 site. The linker is shown in blue and head group shown in red.
Extended Data Fig. 3 Representative chemical synthesis of ligands.
VIP36(8) used as an example.
Extended Data Fig. 4 CB1 selectivity with higher binding affinity.
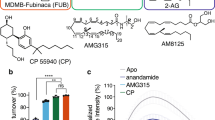
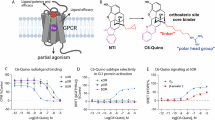
(a) VIP36 displays full efficacy for CB1-induced cAMP inhibition compared to CP55,940 and enhanced efficacy relative to FUB. Data is presented as a bar graph and median, maximum and minimum for each drug is: CB1-CP55,940: 100%, 103%, 97%, CB1-FUB: 85%, 85%, 78%, CB1-VIP36: 117%, 124%, 105% (CB1 n = 3 independent experiments done as quadruplets). One-way ANOVA followed by Tukey’s multiple comparison ***p = 0.0008. Data are represented as mean ± SEM. (b) Similarly, the biochemical GTP depletion assay shows VIP36 acts as a superagonist for CB1-mediated Gi turnover relative to FUB. Data is presented as a bar graph and median, maximum and minimum for each drug is: CB1 + FUB: 100%, 102%, 98%; CB1 + VIP36:108%, 110%, 107%; Gi:79%, 82%, 75%. n = 6 independent experiments, One-way ANOVA followed by Tukey’s multiple comparison ****p < 0.0001 for both. Data are represented as mean ± SEM. (c) VIP36 shows Gα-subtype efficacy bias for Gi-1 and Gi3 isoforms in comparison to both FUB and VIP2.33. Data is presented as a bar graph and median, maximum and minimum for each drug is; For Gi1 FUB: 94%, 111%, 89%; VIP36:173%, 176%, 162%; VIP2.33: 82%, 89%, 78%; Gi1 alone :39%, 40%, 37%; for Gi2 – FUB: 107%, 111%, 106%; VIP36:111%, 114%, 105%; VIP2.33:113%, 118%, 113%; Gi2 alone: 94%, 96%, 85%; for Gi3 - FUB: 128%, 133%, 122%; VIP36: 143%, 144%, 143%; VIP2.33: 128%, 128%, 127%; Gi3 alone: 96%, 98%, 88%; for Goa - FUB: 120%, 122%, 117%; VIP36: 118%, 118%, 116%; VIP2.33: 112%, 113%, 111%; Goa alone: 87%, 97%, 85%; for Gz - FUB: 105%, 110%, 103%; VIP36: 103%, 106%, 99%; VIP2.33: 106%, 110%, 103%; Gz alone: 21%, 47%, 9%. Data are represented as mean ± SEM. n = 3 independent experiments. One-way ANOVA followed by Tukey’s multiple comparison ****p < 0.0001 for all, VIP2.33 vs. Gi1 alone ***p = 0.0004, FUB vs. Gi2 alone **p = 0.0097, VIP36 vs. Gi2 alone **p = 0.0048, VIP2.33 vs. Gi2 alone ***p = 0.0006, FUB vs. Gi2 alone **p = 0.0097, VIP36 vs. Gi2 alone **p = 0.0048, VIP2.33 vs. Gi2 alone ***p = 0.0006, FUB vs. VIP36 *p = 0.0137, VIP36 vs. VIP2.33 *p = 0.0157, VIP2.33 vs. Goa alone ***p = 0.0003. (d). β-arrestin-1 profiling of lead bitopics VIP36 (EC50 = n.d, Emax < 20%) and VIP2.33 (EC50 = 4.2 nM, pEC50 ± SEM = 8.38 ± 0.2, EMax = 110 ± 10) compared with FUB (EC50 = 3.74 nM, pEC50 ± SEM = 8.43 ± 0.2, EMax = 96 ± 8), and CP55,940 (EC50 = n.d, Emax < 20%) controls. Note: Since CP55,940 did not signal through β-arrestin1 the data is normalized against FUB.(e) VIP36 (IC50 = 22.5 nM, CI 95% = 5.481e-008 to 1.046e-007) shows similar (even elevated) potency in a radioligand (3H-SR141716) displacement assay relative to the parent FUB (IC50 = 75.7 nM, CI 95% = 1.646e-008 to 3.068e-008). CI = Confidence interval. n values and primary statistics for all panels are provided in Supplementary Table S4.
Extended Data Fig. 5 CryoEM structure determination for CB1-Gi bound to VIP36 or VIP-2.33.
(a) Schematic of CB1, Gi, and scFv16 purification and complexing protocol for subsequent cryoEM studies. (b,c) CryoEM data collection and processing pipeline for CB1-Gi-VIP36 (b) and CB1-Gi-VIP2.33 (c) showing representative reference-free 2D class averages and 3D classifications, along with the final density map colored by local resolution and the “gold standard” FSC curves calculated in Relion. Multiple views of the density surrounding VIP36 (b, green) and VIP2.33 (c, yellow) are shown.
Extended Data Fig. 6 G-protein bias through cryptic pocket engagement.
(a) In MD simulations, the VIP36 head group interacts with D2.50 through water-mediated interactions and occasionally through the formation of a direct salt-bridge and hydrogen-bonds. Images show representative simulation frames illustrating each type of interaction. Percentages indicate the percentage of simulation time that a particular type of interaction is formed, averaged over five independent simulations, each 2 µs in length. The direct interaction was calculated using frames where the positive nitrogen atoms on VIP36 were within 3 Å of the oxygen atoms of the D2.50 sidechain. (b) VIP36 and VIP2.33 stabilize distinct conformational ensembles of the intracellular end of the 6th transmembrane helix of CB1, as observed by bimane fluorescence experiments in various ligand-bound states. (c) Cryptic pocket opening is stabilized by bitopic ligands in simulations. Distributions show the distance between toggle switch residues in simulations with various ligands bound. Pocket opens rarely in simulations with FUB bound and is stabilized in the open conformation in simulations with either VIP36 bound or VIP2.33 bound. Five independent simulations, each 2 µs in length, were performed for each ligand. (d) Conformation of W356 is not significantly different between simulations with VIP36 and VIP2.33 bound. We compared multiple distance metrics describing the conformation of residue W356; the distances used are shown as red dashed lines in the inset image of the CB1R receptor with the atoms used labeled in red. Data presented as mean ± s.e.m. calculated from 5 independent simulations for VIP36 and and 6 independent simulations for VIP2.33. P-values calculated using two-sided Mann Whitney U-test: from left P = 0.53, 0.42, 0.94, 0.23.
Extended Data Fig. 7 Bias mechanism involves network of interaction between ligand, TM7 and D2.50.
(a) VIP36 binding pose is stable in MD simulations. Overlay of VIP36 pose from a representative MD simulation frame at 1000 ns (green) with the pose from the cryo-EM structure (grey), showing close agreement in ligand positioning within the receptor binding pocket. (b) Time evolution of RMSD (root-mean-square deviation) of VIP36 relative to its cryo-EM pose over 1000 ns of simulation for 5 independent simulations. We show both unsmoothed traces (thin, light green lines) and traces smoothed with a moving average (thick, dark green lines), using an averaging window of 30 ns. (c) Comparison of VIP36 and VIP2.33 interactions with TM7 residues. Left: simulation frame showing VIP36 (yellow sticks) bound to the receptor, highlighting key interacting residues on TM7 (C7.42, S7.46, N7.45, N7.49) and D2.50 on TM2. Dotted lines highlight a network of polar interactions between VIP36-TM7-D2.50. Right: Table quantifying the frequency of different types of interactions between specific TM7 residues and the ligands VIP36 and VIP2.33. Frequency is defined as the percentage of simulation frames in which a particular interaction is formed between the ligand and residue, calculated with the GetContacts software. The water-mediated hydrogen bond frequency corresponds to frames where a single water molecule engages both the ligand and the residue through hydrogen bonds. vdW corresponds to frames where at least one van der Waals interaction is formed between the ligand and residue atoms (the distance between a pair of atoms is less than the sum of van der Waals radius of each atom + 0.5 Å). Values represent the mean percentage of simulation time each interaction is formed, with s.e.m. in parentheses, calculated across independent simulations (N = 5 for VIP36, N = 6 for VIP2.33).
Extended Data Fig. 8 VIP36 is peripherally restricted and CB1 selective: (a-c). Pharmacokinetics of VIP36, FUB and CB13.
Brain and plasma pharmacokinetics were exposure of VIP36, CB13, and FUB at 3 mg/kg IP was carried out and concentration of drug was determined using LC-MS/MS. PK of VIP36 at 30 mg kg−1 IP was also carried out at 1 h and 3 h n = 3 biologically independent replicates in mice by time point. Data are represented as mean ± SEM. (d) Pharmacokinetics of VIP36 at 3 mg/kg IV was carried out and concentration of drug was determined using LC-MS/MS. Data are represented as mean ± SEM. (e) VIP36 displays full efficacy for CB1-induced cAMP inhibition compared to CP55,940 and enhanced efficacy relative to FUB, but shows no activity against CB2-induced cAMP inhibition in contrast to CP55,940 and FUB. Data is presented as a bar graph and median, maximum and minimum for each drug is: CB1-CP55,940: 100%, 103%, 97%; CB1-FUB: 85%, 85%, 78%; CB1-VIP36: 117%, 124%, 105%; CB2-CP55,940: 108%, 112%, 104%; CB2-FUB: 72%, 77%, 68%; CB2-VIP36: −3%, −1%, −4%; (CB1 n = 3, CB2 n = 2 independent experiments done as quadruplets). Data are represented as mean ± SEM. One-way ANOVA followed byTukey’s multiple comparisons test, CB2-CP55,940 vs. CB2-VIP36 ****p < 0.0001, CB2-FUB vs. CB2-VIP36 ****p < 0.0001, CB1- FUB vs. CB1-VIP36 ***p = 0.0008. (f) Similarly, the biochemical GTP depletion assay shows VIP36 acts as a superagonist for CB1-mediated Gi turnover relative to FUB but shows no activity against CB2-mediated Gi turnover. Data is presented as a bar graph and median, maximum and minimum for each drug is: CB1 + FUB: 100%, 102%, 98%; CB1 + VIP36:108%, 110%, 107%; Gi:79%, 82%, 75%; CB2 + FUB:100%, 101%, 98%; CB2 + VIP36:58%, 61%, 56%; Gi :60%, 65%, 55%. Data are represented as mean ± SEM. One-way ANOVA followed by Tukey’s multiple comparisons test ****p < 0.0001 for all. (g) The cryoEM structure of VIP36 bound to CB1 shows the necessity of a further outwards “open-active” conformation of the CB1 toggle switch residues. While CB1 has a relatively small L287 residue that can accommodate this new W356 rotamer state, CB2 has a bulky F287, blocking the formation of the “open-active” toggle switch. n values and primary statistics for all panels are provided in Supplementary Table S4.
Supplementary information
Supplementary Information
Supplementary Sections 1–4, including Supplementary Materials and Methods, Supplementary Tables 1–4, Supplementary Figs. 1–4, Ethics declarations and Supplementary References.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rangari, V.A., O’Brien, E.S., Powers, A.S. et al. A cryptic pocket in CB1 drives peripheral and functional selectivity. Nature 640, 265–273 (2025). https://doi.org/10.1038/s41586-025-08618-7
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41586-025-08618-7
This article is cited by
-
Enhancement of the endocannabinoid system through monoacylglycerol lipase inhibition relieves migraine-associated pain in mice
The Journal of Headache and Pain (2025)
-
Designer cannabinoids could be the key to pain relief without adverse effects
Nature (2025)
-
Cannabinoid targets hidden pocket for safer pain relief
Nature Reviews Drug Discovery (2025)