Abstract
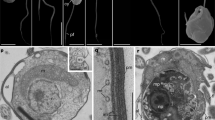
The eukaryote Tree of Life (eToL) depicts the relationships among all eukaryotic organisms; its root represents the Last Eukaryotic Common Ancestor (LECA) from which all extant complex lifeforms are descended1. Locating this root is crucial for reconstructing the features of LECA, both as the endpoint of eukaryogenesis and the start point for the evolution of the myriad complex traits underpinning the diversification of living eukaryotes. However, the position of the root remains contentious due to pervasive phylogenetic artefacts stemming from inadequate evolutionary models, poor taxon sampling and limited phylogenetic signal1. Here we estimate the root of the eToL with unprecedented resolution on the basis of a new, much larger, dataset of mitochondrial proteins that includes all known eukaryotic supergroups. Our analyses of a 100 taxon × 93 protein dataset with state-of-the-art phylogenetic models and an extensive evaluation of alternative hypotheses show that the eukaryotic root lies between two multi-supergroup assemblages: ‘Opimoda+’ and ‘Diphoda+’. This position is consistently supported across different models and robustness analyses. Notably, groups containing ‘typical excavates’ are placed on both sides of the root, suggesting the complex features of the ‘excavate’ cell architecture trace back to LECA. This study sheds light on the ancestral cells from which extant eukaryotes arose and provides a crucial framework for investigating the origin and evolution of canonical eukaryotic features.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All sequence alignments and phylogenetic trees are available at Figshare (https://figshare.com/s/59b28ecc0056dc8d0d03)34 and the accession codes for publicly available data used in these analyses are given in Supplementary Table 7. The data included in the accession codes can be found in the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov), the MMETSP database on the iMicrobe website https://www.imicrobe.us/#/projects/104 or the PhyloFisher v.1 dataset available at Figshare (https://doi.org/10.6084/m9.figshare.15141900.v1)72 as indicated in Supplementary Table 7.
Code availability
Custom scripts have been deposited at Figshare (https://figshare.com/s/59b28ecc0056dc8d0d03)34.
References
Burki, F., Roger, A. J., Brown, M. W. & Simpson, A. G. B. The new tree of eukaryotes. Trends Ecol. Evol. 35, 43–55 https://doi.org/10.1016/j.tree.2019.08.008 (2020).
Tikhonenkov, D. V. et al. Microbial predators form a new supergroup of eukaryotes. Nature 612, 714–719 (2022).
Eme, L., Sharpe, S. C., Brown, M. W. & Roger, A. J. On the age of eukaryotes: evaluating evidence from fossils and molecular clocks. Cold Spring Harb. Perspect. Biol. 6, a016139 (2014).
Betts, H. C. et al. Integrated genomic and fossil evidence illuminates life’s early evolution and eukaryote origin. Nat. Ecol. Evol. 2, 1556–1562 (2018).
Strassert, J. F. H., Irisarri, I., Williams, T. A. & Burki, F. A molecular timescale for eukaryote evolution with implications for the origin of red algal-derived plastids. Nat. Commun. 12, 1879 (2021).
Katz, L. A., Grant, J. R., Parfrey, L. W. & Burleigh, J. G. Turning the crown upside down: gene tree parsimony roots the eukaryotic tree of life. Syst. Biol. 61, 653–660 (2012).
Cerón-Romero, M. A., Fonseca, M. M., De Oliveira Martins, L., Posada, D. & Katz, L. A. Phylogenomic analyses of 2,786 genes in 158 lineages support a root of the eukaryotic tree of life between Opisthokonts and all other lineages. Genome Biol. Evol. 14, evac119 (2022).
Cavalier-Smith, T. Kingdoms Protozoa and Chromista and the eozoan root of the eukaryotic tree. Biol. Lett. 6, 342–345 (2010).
Stechmann, A. & Cavalier-Smith, T. Rooting the eukaryote tree by using a derived gene fusion. Science 297, 89–91 (2002).
Richards, T. A. & Cavalier-Smith, T. Myosin domain evolution and the primary divergence of eukaryotes. Nature 436, 1113–1118 (2005).
Rogozin, I. B., Basu, M. K., Csürös, M. & Koonin, E. V. Analysis of rare genomic changes does not support the Unikont–Bikont phylogeny and suggests cyanobacterial symbiosis as the point of primary radiation of eukaryotes. Genome Biol. Evol. 1, 99–113 (2009).
Leonard, G. & Richards, T. A. Genome-scale comparative analysis of gene fusions, genefissions, and the fungal tree of life. Proc. Natl Acad. Sci. USA 109, 21402–21407 (2012).
Vosseberg, J. et al. Timing the origin of eukaryotic cellular complexity with ancient duplications. Nat. Ecol. Evol. 5, 92–100 (2021).
Eme, L. et al. Inference and reconstruction of the heimdallarchaeial ancestry of eukaryotes. Nature 618, 992–999 (2023).
Pittis, A. A. & Gabaldón, T. Late acquisition of mitochondria by a host with chimaeric prokaryotic ancestry. Nature 531, 101–104 (2016).
Roger, A. J., Muñoz-Gómez, S. A. & Kamikawa, R. The origin and diversification of mitochondria. Curr. Biol. 27, R1177–R1192 (2017).
Muñoz-Gómez, S. A. et al. Site-and-branch-heterogeneous analyses of an expanded dataset favour mitochondria as sister to known Alphaproteobacteria. Nat. Ecol. Evolution 6, 253–262 (2022).
Al Jewari, C. & Baldauf, S. L. An excavate root for the eukaryote Tree of Life. Sci. Adv. 9, eade4973 (2023).
Derelle, R. & Lang, B. F. Rooting the eukaryotic tree with mitochondrial and bacterial proteins. Mol. Biol. Evol. 29, 1277–1289 (2012).
Derelle, R. et al. Bacterial proteins pinpoint a single eukaryotic root. Proc. Natl Acad. Sci. USA 112, E693–E699 (2015).
He, D., Fiz-Palacios, O., Fu, C. J., Tsai, C. C. & Baldauf, S. L. An alternative root for the eukaryote Tree of Life. Curr. Biol. 24, 465–470 (2014).
Al Jewari, C. & Baldauf, S. L. Conflict over the eukaryote root resides in strong outliers, mosaics and missing data sensitivity of site-specific (CAT) mixture models. Syst. Biol. 0, 1–16 (2022).
Lax, G. et al. Hemimastigophora is a novel supra-kingdom-level lineage of eukaryotes. Nature 564, 410–414 (2018).
Kapli, P., Yang, Z. & Telford, M. J. Phylogenetic tree building in the genomic age. Nat. Rev. Genet. 21, 428–444 (2020).
Susko, E., Lincker, L. & Roger, A. J. Accelerated estimation of frequency classes in site-heterogeneous profile mixture models. Mol. Biol. Evol. 35, 1266–1283 (2018).
Crotty, S. M. et al. GHOST: recovering historical signal from heterotachously evolved sequence alignments. Syst. Biol. 69, 249–264 (2019).
Gaston, D., Susko, E. & Roger, A. J. A phylogenetic mixture model for the identification of functionally divergent protein residues. Bioinformatics 27, 2655–2663 (2011).
Quang, L. S., Gascuel, O. & Lartillot, N. Empirical profile mixture models for phylogenetic reconstruction. Bioinformatics 24, 2317–2323 (2008).
Szánthó, L. L., Lartillot, N., Szöllősi, G. J. & Schrempf, D. Compositionally constrained sites drive long-branch attraction. Syst. Biol. https://doi.org/10.1093/SYSBIO/SYAD013 (2023).
Jerlström-Hultqvist, J. et al. A unique symbiosome in an anaerobic single-celled eukaryote. Nat. Commun. 15, 9726 (2024).
Baños, H., Susko, E. & Roger, A. J. Is over-parameterization a problem for profile mixture models? Syst. Biol. https://doi.org/10.1093/SYSBIO/SYAD063 (2023).
Brown, M. W. et al. Phylogenomics places orphan protistan lineages in a novel eukaryotic super-group. Genome Biol. Evol. 10, 427–433 (2018).
Strassert, J. F. H., Jamy, M., Mylnikov, A. P., Tikhonenkov, D. V. & Burki, F. New phylogenomic analysis of the enigmatic phylum Telonemia further resolves the eukaryote Tree of Life. Mol. Biol. Evol. 36, 757 (2019).
Williamson, K. et al. A robustly rooted tree of eukaryotes reveals their excavate ancestry [Data]. Figshare https://doi.org/10.6084/m9.figshare.26863594.v1 (2025).
Cavalier-Smith, T. Ciliary transition zone evolution and the root of the eukaryote tree: implications for opisthokont origin and classification of kingdoms Protozoa, Plantae, and Fungi. Protoplasma 259, 487–593 (2022).
Cavalier-Smith, T. & Chao, E. E. Y. Multidomain ribosomal protein trees and the planctobacterial origin of neomura (eukaryotes, archaebacteria). Protoplasma 257, 621–753 (2020).
Baker, B. A. et al. Expanded phylogeny of extremely halophilic archaea shows multiple independent adaptations to hypersaline environments. Nat. Microbiol. 9, 964–975 (2024).
Susko, E. Tests for two trees using likelihood methods. Mol. Biol. Evol. 31, 1029–1039 (2014).
Markowski, E. & Susko, E. Performance of topology tests under extreme selection bias. Mol. Biol. Evol. 41, msad280 (2024).
Heiss, A. A. et al. Combined morphological and phylogenomic re-examination of malawimonads, a critical taxon for inferring the evolutionary history of eukaryotes. R. Soc. Open Sci. 5, 171707 (2018).
Susko, E. & Roger, A. J. Long branch attraction biases in phylogenetics. Syst. Biol. 70, 838–843 (2021).
Kapli, P. & Telford, M. J. Topology-dependent asymmetry in systematic errors affects phylogenetic placement of Ctenophora and Xenacoelomorpha. Sci. Adv. 6, 5162–5173 (2020).
Inagaki, Y., Susko, E., Fast, N. M. & Roger, A. J. Covarion shifts cause a long-branch attraction artifact that unites Microsporidia and Archaebacteria in EF-1α phylogenies. Mol. Biol. Evol. 21, 1340–1349 (2004).
Eglit, Y. et al. Meteora sporadica, a protist with incredible cell architecture, is related to Hemimastigophora. Curr. Biol. 34, 451–459.e6 (2024).
Yubuki, N. & Leander, B. S. Evolution of microtubule organizing centers across the tree of eukaryotes. Plant J. 75, 230–244 (2013).
Heiss, A. A., Walker, G. & Simpson, A. G. B. The microtubular cytoskeleton of the apusomonad Thecamonas, a sister lineage to the opisthokonts. Protist 164, 598–621 (2013).
Suzuki-Tellier, S., Kiørboe, T. & Simpson, A. G. B. The function of the feeding groove of ‘typical excavate’ flagellates. J. Eukaryot. Microbiol. 71, e13016 (2024).
Takishita, K. et al. Multigene phylogenies of diverse carpediemonas-like organisms identify the closest relatives of ‘amitochondriate’ diplomonads and retortamonads. Protist 163, 344–355 (2012).
Leger, M. M. et al. Organelles that illuminate the origins of Trichomonas hydrogenosomes and Giardia mitosomes. Nat. Ecol. Evol. 1, 0092 (2017).
Heiss, A. A., Walker, G. & Simpson, A. G. B. The ultrastructure of ancyromonas, a eukaryote without supergroup affinities. Protist 162, 373–393 (2011).
Brugerolle, G. Description of a new freshwater heterotrophic flagellate Sulcomonas lacustris affiliated to the collodictyonids. Acta Protozool. 45, 175–182 (2006).
Brugerolle, G., Bricheux, G., Philippe, H. & Coffe, G. Collodictyon triciliatum and Diphylleia rotans (=Aulacomonas submarina) form a new family of flagellates (Collodictyonidae) with tubular mitochondrial cristae that is phylogenetically distant from other flagellate groups. Protist 153, 59–70 (2002).
Tikhonenkov, D. V. et al. Description of Colponema vietnamica sp.n. and Acavomonas peruviana n. gen. n. sp., two new alveolate Phyla (Colponemidia nom. nov. and Acavomonidia nom. nov.) and their contributions to reconstructing the ancestral state of alveolates and eukaryotes. PLoS ONE 9, e95467 (2014).
Janouškovec, J. et al. A new lineage of eukaryotes illuminates early mitochondrial genome reduction. Curr. Biol. 27, 3717–3724.e5 (2017).
Leander, B. S. Eukaryotic evolution: deep phylogeny does not imply morphological novelty. Curr. Biol. 33, R112–R114 (2023).
Cavalier-Smith, T. Early evolution of eukaryote feeding modes, cell structural diversity, and classification of the protozoan phyla Loukozoa, Sulcozoa, and Choanozoa. Eur. J. Protistol. 49, 115–178 (2013).
Tice, A. K. et al. PhyloFisher: a phylogenomic package for resolving eukaryotic relationships. PLoS Biol. 19, e3001365 (2021).
Buchfink, B., Reuter, K. & Drost, H. G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods 18, 366–368 (2021).
Fu, L., Niu, B., Zhu, Z., Wu, S. & Li, W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150 (2012).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Capella-Gutierrez, S., Silla-Martinez, J. M. & Gabaldon, T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009).
Criscuolo, A. & Gribaldo, S. BMGE (block mapping and gathering with entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol. 10, 210 (2010).
Minh, B. Q. et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534 (2020).
Hoang, D. T., Chernomor, O., Von Haeseler, A., Minh, B. Q. & Vinh, L. S. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522 (2018).
Tukey, J. W. Exploratory Data Analysis (Addison-Wesley Publishing Company, 1977).
Nguyen, L. T., Schmidt, H. A., Von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268 (2015).
Menardo, F. et al. Treemmer: a tool to reduce large phylogenetic datasets with minimal loss of diversity. BMC Bioinf. 19, 164 (2018).
Susko, E. & Roger, A. J. On the use of information criteria for model selection in phylogenetics. Mol. Biol. Evol. 37, 549–562 (2020).
Lartillot, N., Lepage, T. & Blanquart, S. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25, 2286–2288 (2009).
Guindon, S. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010).
Reynolds, D. in Encyclopedia of Biometrics (ed. Li, S. Z.) 659–663 (Springer, 2009).
Brown, M. Data associated with PhyloFisher. Figshare https://doi.org/10.6084/m9.figshare.15141900.v1 (2021).
Acknowledgements
We thank N. Ly-Trong for assistance with model implementation in IQ-TREE and J. Jerlström-Hultqvist for assistance with data acquisition and processing. We also thank J. Ross for assistance with figure design. This work was supported by the Moore-Simons Project on the Origin of the Eukaryotic Cell, Simons Foundation grant no. 735923LPI (https://doi.org/10.46714/735923LPI) awarded to A.J.R., E.S. and L.E. and by NSERC Discovery grants awarded to A.J.R. (grant no. RGPIN-2022-05430), A.G.B.S. (grant no. 298366-2019) and E.S. K.W. was supported by a graduate scholarship from the Killam Foundation.
Author information
Authors and Affiliations
Contributions
Ancestral sequence reconstruction analyses, expansion and curation of the final datasets, and all phylogenomic analyses were performed by K.W. in consultation with A.J.R., A.G.B.S. and L.E. K.W. and H.B. performed simulation analyses. H.B. developed and implemented the MEOW model and performed cross-validation testing. C.G.P.M. and E.S. performed GFmix analyses. E.S. performed topology testing. B.Q.M. implemented the FunDi model in IQ-TREE2. S.A.M.-G., R.K. and R.J.S.O. provided molecular data. K.W., A.J.R., A.G.B.S., H.B., C.G.P.M. and E.S. wrote, and all authors edited and approved, the manuscript. A.J.R. and L.E. initially conceived the study.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Thomas Richards and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Entropy and posterior probability of the predicted ancestral sequence at the root of Alphaproteobacteria as ingroup taxa are removed.
A. The mean, median, and standard deviation of the posterior probability as taxa are removed. B. The mean, median, and standard deviation of the entropy as taxa are removed.
Extended Data Fig. 2 Bayesian consensus tree estimated from Anae+ dataset.
Consensus phylogeny estimated under the CAT + GTR model in PhyloBayes from 4 chains after 29,000 cycles, with a burn-in of 1000. Posterior probabilities for each bipartition are indicated. Note that these 4 chains included one in which Hemimastigophora branched on the opposite side of the root, separate from its previously inferred close relative Meteora, with this non-convergence resulting in posterior probabilities ~0.75 (0.74) for several deep nodes in the Diphoda+ side of the tree.
Extended Data Fig. 3 Phylogeny estimated from Anae+ dataset with CAT-PMSF model.
Amino acid exchangeability matrix and site frequency profiles were estimated on the alignment and a guide tree in PhyloBayes. Rate variation was modelled with the Free Rate model with four classes. Support values indicated are from 100 replicates of non-parametric bootstrapping.
Extended Data Fig. 4 Bayesian consensus tree estimated from Anae- dataset.
Consensus phylogeny estimated under the CAT + GTR model in PhyloBayes from 4 chains after 16,500 cycles, with a burn-in of 1000. Posterior probabilities for each bipartition are indicated.
Extended Data Fig. 5 Phylogeny estimated from Anae- dataset with CAT-PMSF model.
Amino acid exchangeability matrix and site frequency profiles were estimated on the alignment and a guide tree in PhyloBayes. Rate variation was modelled with the Free Rate model with four classes. Support values indicated are from 100 replicates of non-parametric bootstrapping.
Extended Data Fig. 6 Phylogeny estimated from Anae+ dataset with only nuclear-encoded proteins.
Phylogeny was estimated in IQ-TREE2 under the LG + MEOW80 + G4 model with 1000 replicates each of SH-aLRT and UFBOOT2. Support values are displayed on branches as SH-aLRT/UFBOOT2.
Extended Data Fig. 7 Phylogeny estimated from Anae- dataset with only nuclear-encoded proteins.
Phylogeny was estimated in IQ-TREE2 under the LG + MEOW80 + G4 model with 1000 replicates each of SH-aLRT and UFBOOT2. Support values are displayed on branches as SH-aLRT/UFBOOT2.
Extended Data Fig. 8 Evaluating change in support for the root position as fastest-evolving sites are removed.
A. Phylogenies were estimated in IQ-TREE2 under the LG + MEOW80 + G4 model with 1000 replicates each of SH-aLRT and UFBOOT2. Support values are indicated for the bipartitions relevant to the position of the eukaryote root.
Extended Data Fig. 9 Phylogeny estimated from Anae- dataset after removal of fastest evolving taxa.
Phylogeny was estimated in IQ-TREE2 under the LG + MEOW80 + G4 model with 1000 replicates each of SH-aLRT and UFBOOT2. Support values are displayed on branches as SH-aLRT/UFBOOT2.
Extended Data Fig. 10 Phylogeny estimated from Anae- dataset after removal of most divergent genes.
Alignment after gene removal consisted of 19324 sites from 80 concatenated genes. The phylogeny was estimated in IQ-TREE2 under the LG + MEOW80 + G4 model with 1000 replicates each of SH-aLRT and UFBOOT2. Support values are displayed on branches as SH-aLRT/UFBOOT2.
Supplementary information
Supplementary Information
This file contains Supplementary Methods and Refs.
Supplementary Tables
This file contains Supplementary Tables 1–11.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Williamson, K., Eme, L., Baños, H. et al. A robustly rooted tree of eukaryotes reveals their excavate ancestry. Nature 640, 974–981 (2025). https://doi.org/10.1038/s41586-025-08709-5
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41586-025-08709-5
This article is cited by
-
Revisiting giant virus-host dynamics in brown algae: old stories and new perspectives
The EMBO Journal (2026)
-
The fate of artificial transgenes in Acanthamoeba castellanii
BMC Genomics (2025)
-
The role of gene duplication and paralog specialisation in the evolution of the mammalian PRPS complex
Nature Communications (2025)
-
Evolution, composition and functions of cullin E3 ubiquitin ligases in trypanosomes
Scientific Reports (2025)
-
Identification of a deep-branching lineage of algae using environmental plastid genomes
Nature Communications (2025)