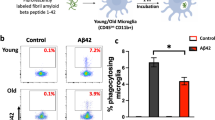
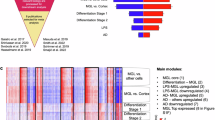
Abstract
Microglia are the resident immune cells in the brain and have pivotal roles in neurodevelopment and neuroinflammation1,2. This study investigates the function of the immune-checkpoint molecule TIM-3 (encoded by HAVCR2) in microglia. TIM-3 was recently identified as a genetic risk factor for late-onset Alzheimer’s disease3, and it can induce T cell exhaustion4. However, its specific function in brain microglia remains unclear. We demonstrate in mouse models that TGFβ signalling induces TIM-3 expression in microglia. In turn, TIM-3 interacts with SMAD2 and TGFBR2 through its carboxy-terminal tail, which enhances TGFβ signalling by promoting TGFBR-mediated SMAD2 phosphorylation, and this process maintains microglial homeostasis. Genetic deletion of Havcr2 in microglia leads to increased phagocytic activity and a gene-expression profile consistent with the neurodegenerative microglial phenotype (MGnD), also referred to as disease-associated microglia (DAM). Furthermore, microglia-targeted deletion of Havcr2 ameliorates cognitive impairment and reduces amyloid-β pathology in 5×FAD mice (a transgenic model of Alzheimer’s disease). Single-nucleus RNA sequencing revealed a subpopulation of MGnD microglia in Havcr2-deficient 5×FAD mice characterized by increased pro-phagocytic and anti-inflammatory gene expression alongside reduced pro-inflammatory gene expression. These transcriptomic changes were corroborated by single-cell RNA sequencing data across most microglial clusters in Havcr2-deficient 5×FAD mice. Our findings reveal that TIM-3 mediates microglia homeostasis through TGFβ signalling and highlight the therapeutic potential of targeting microglial TIM-3 in Alzheimer’s disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The RNA-seq and snRNA-seq data were deposited into the GEO at the National Center for Biotechnology Information with the accession number GSE223424. The accession number for the scRNA-seq data in the GEO is GSE267764. Public mouse microglial microarray data were obtained from table S1 of a previous publication21. Public bulk RNA-seq datasets were used in the analysis of the Tgfbr2cKO (GSE111385) and Clec7a+ data (GSE101689). Public scRNA-seq and snRNA-seq datasets were used for the analyses of P9 and P28 cortical microglia (Fig. 1; GSE127449), mouse brain atlas (Extended Data Fig. 1; http://mousebrain.org/), 6-month-old mouse brain (Extended Data Fig. 10; GSE98969), 7-month-old mouse brain (Extended Data Fig. 10, GSE140510), IFN-responsive MGnD (Extended Data Fig. 11; GSE205569) and human brain (Extended Data Fig. 1; Broad Institute single-cell portal SCP371). Source data are provided with this paper.
Code availability
All code used in the analysis of bulk and scRNA-seq datasets and generation of figures are available from GitHub (https://github.com/ayshwaryas/tim3_microglia/).
References
Colonna, M. & Butovsky, O. Microglia function in the central nervous system during health and neurodegeneration. Annu. Rev. Immunol. 35, 441–468 (2017).
Butovsky, O. & Weiner, H. L. Microglial signatures and their role in health and disease. Nat. Rev. Neurosci. 19, 622–635 (2018).
Wightman, D. P. et al. A genome-wide association study with 1,126,563 individuals identifies new risk loci for Alzheimer’s disease. Nat. Genet. 53, 1276–1282 (2021).
Wolf, Y., Anderson, A. C. & Kuchroo, V. K. TIM3 comes of age as an inhibitory receptor. Nat. Rev. Immunol. 20, 173–185 (2020).
Jansen, I. E. et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet. 51, 404–413 (2019).
Kelley, K. W., Nakao-Inoue, H., Molofsky, A. V. & Oldham, M. C. Variation among intact tissue samples reveals the core transcriptional features of human CNS cell classes. Nat. Neurosci. 21, 1171–1184 (2018).
Yang, A. C. et al. A human brain vascular atlas reveals diverse mediators of Alzheimer’s risk. Nature 603, 885–892 (2022).
Wang, Y. et al. TREM2-mediated early microglial response limits diffusion and toxicity of amyloid plaques. J. Exp. Med. 213, 667–675 (2016).
Yuan, P. et al. TREM2 haplodeficiency in mice and humans impairs the microglia barrier function leading to decreased amyloid cmpaction and severe axonal dystrophy. Neuron 90, 724–739 (2016).
Krasemann, S. et al. The TREM2–APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity 47, 566–581 (2017).
Butovsky, O. et al. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat. Neurosci. 17, 131–143 (2014).
Olah, M. et al. A transcriptomic atlas of aged human microglia. Nat. Commun. 9, 539 (2018).
Zhang, Y. et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 34, 11929–11947 (2014).
Chihara, N. et al. Induction and transcriptional regulation of the co-inhibitory gene module in T cells. Nature 558, 454–459 (2018).
Schnell, A., Bod, L., Madi, A. & Kuchroo, V. K. The yin and yang of co-inhibitory receptors: toward anti-tumor immunity without autoimmunity. Cell Res. 30, 285–299 (2020).
Gayden, T. et al. Germline HAVCR2 mutations altering TIM-3 characterize subcutaneous panniculitis-like T cell lymphomas with hemophagocytic lymphohistiocytic syndrome. Nat. Genet. 50, 1650–1657 (2018).
Polprasert, C. et al. Frequent germline mutations of HAVCR2 in sporadic subcutaneous panniculitis-like T-cell lymphoma. Blood Adv. 3, 588–595 (2019).
Dixon, K. O. et al. TIM-3 restrains anti-tumour immunity by regulating inflammasome activation. Nature 595, 101–106 (2021).
Zeisel, A. et al. Molecular architecture of the mouse nervous system. Cell 174, 999–1014 (2018).
Gaublomme, J. T. et al. Nuclei multiplexing with barcoded antibodies for single-nucleus genomics. Nat. Commun. 10, 2907 (2019).
Thion, M. S. et al. Microbiome influences prenatal and adult microglia in a sex-specific manner. Cell 172, 500–516 (2018).
Attaai, A. et al. Postnatal maturation of microglia is associated with alternative activation and activated TGFβ signaling. Glia 66, 1695–1708 (2018).
Gosselin, D. et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell 159, 1327–1340 (2014).
He, D. et al. Disruption of the IL-33–ST2–AKT signaling axis impairs neurodevelopment by inhibiting microglial metabolic adaptation and phagocytic function. Immunity 55, 159–173 (2022).
Buttgereit, A. et al. Sall1 is a transcriptional regulator defining microglia identity and function. Nat. Immunol. 17, 1397–1406 (2016).
Lund, H. et al. Fatal demyelinating disease is induced by monocyte-derived macrophages in the absence of TGF-β signaling. Nat. Immunol. 19, 1–7 (2018).
Qin, Y. et al. A milieu molecule for TGF-β required for microglia function in the nervous system. Cell 174, 156–171 (2018).
Arnold, T. D. et al. Impaired αVβ8 and TGFβ signaling lead to microglial dysmaturation and neuromotor dysfunction. J. Exp. Med. 216, 900–915 (2019).
Zoller, T. et al. Silencing of TGFβ signalling in microglia results in impaired homeostasis. Nat. Commun. 9, 4011 (2018).
Yona, S. et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38, 79–91 (2013).
Koyama, S. et al. Adaptive resistance to therapeutic PD-1 blockade is associated with upregulation of alternative immune checkpoints. Nat. Commun. 7, 10501 (2016).
Huang, R. Y., Francois, A., McGray, A. R., Miliotto, A. & Odunsi, K. Compensatory upregulation of PD-1, LAG-3, and CTLA-4 limits the efficacy of single-agent checkpoint blockade in metastatic ovarian cancer. Oncoimmunology 6, e1249561 (2017).
Shi, Y. & Massague, J. Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell 113, 685–700 (2003).
Rangachari, M. et al. Bat3 promotes T cell responses and autoimmunity by repressing Tim-3-mediated cell death and exhaustion. Nat. Med. 18, 1394–1400 (2012).
Huang, Y. H. et al. CEACAM1 regulates TIM-3-mediated tolerance and exhaustion. Nature 517, 386–390 (2015).
Miyazono, K. I. et al. Structural basis for transcriptional coactivator recognition by SMAD2 in TGF-β signaling. Sci. Signal. 13, eabb9043 (2020).
Macias-Silva, M. et al. MADR2 is a substrate of the TGFβ receptor and its phosphorylation is required for nuclear accumulation and signaling. Cell 87, 1215–1224 (1996).
Stevens, B. et al. The classical complement cascade mediates CNS synapse elimination. Cell 131, 1164–1178 (2007).
Paolicelli, R. C. et al. Synaptic pruning by microglia is necessary for normal brain development. Science 333, 1456–1458 (2011).
Schafer, D. P. et al. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron 74, 691–705 (2012).
Ueno, M. et al. Layer V cortical neurons require microglial support for survival during postnatal development. Nat. Neurosci. 16, 543–551 (2013).
Hagemeyer, N. et al. Microglia contribute to normal myelinogenesis and to oligodendrocyte progenitor maintenance during adulthood. Acta Neuropathol. 134, 441–458 (2017).
Wlodarczyk, A. et al. A novel microglial subset plays a key role in myelinogenesis in developing brain. EMBO J. 36, 3292–3308 (2017).
Griciuc, A. et al. Alzheimer’s disease risk gene CD33 inhibits microglial uptake of amyloid β. Neuron 78, 631–643 (2013).
Scheltens, P. et al. Alzheimer’s disease. Lancet 397, 1577–1590 (2021).
Margeta, M. A. et al. Apolipoprotein E4 impairs the response of neurodegenerative retinal microglia and prevents neuronal loss in glaucoma. Immunity 55, 1627–1644 (2022).
Griciuc, A. et al. TREM2 acts downstream of CD33 in modulating microglial pathology in Alzheimer’s disease. Neuron 103, 820–835 (2019).
Oakley, H. et al. Intraneuronal β-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: potential factors in amyloid plaque formation. J. Neurosci. 26, 10129–10140 (2006).
Goldmann, T. et al. A new type of microglia gene targeting shows TAK1 to be pivotal in CNS autoimmune inflammation. Nat. Neurosci. 16, 1618–1626 (2013).
Bero, A. W. et al. Bidirectional relationship between functional connectivity and amyloid-β deposition in mouse brain. J. Neurosci. 32, 4334–4340 (2012).
Wang, S. et al. Anti-human TREM2 induces microglia proliferation and reduces pathology in an Alzheimer’s disease model. J. Exp. Med. 217, e20200785 (2020).
Boza-Serrano, A. et al. Galectin-3, a novel endogenous TREM2 ligand, detrimentally regulates inflammatory response in Alzheimer’s disease. Acta Neuropathol. 138, 251–273 (2019).
Siew, J. J. et al. Galectin-3 is required for the microglia-mediated brain inflammation in a model of Huntington’s disease. Nat. Commun. 10, 3473 (2019).
Condello, C., Yuan, P., Schain, A. & Grutzendler, J. Microglia constitute a barrier that prevents neurotoxic protofibrillar Aβ42 hotspots around plaques. Nat. Commun. 6, 6176 (2015).
Gratuze, M. et al. Activated microglia mitigate Aβ-associated tau seeding and spreading. J. Exp. Med. 218, e20210542 (2021).
Huang, Y. et al. Microglia use TAM receptors to detect and engulf amyloid β plaques. Nat. Immunol. 22, 586–594 (2021).
Keren-Shaul, H. et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell 169, 1276–1290 (2017).
Sun, N. et al. Human microglial state dynamics in Alzheimer’s disease progression. Cell 186, 4386–4403 (2023).
Zhou, Y. et al. Human and mouse single-nucleus transcriptomics reveal TREM2-dependent and TREM2-independent cellular responses in Alzheimer’s disease. Nat. Med. 26, 131–142 (2020).
Yin, Z. et al. Identification of a protective microglial state mediated by miR-155 and interferon-γ signaling in a mouse model of Alzheimer’s disease. Nat. Neurosci. 26, 1196–1207 (2023).
Ellwanger, D. C. et al. Prior activation state shapes the microglia response to antihuman TREM2 in a mouse model of Alzheimer’s disease. Proc. Natl Acad. Sci. USA 118, e2017742118 (2021).
Deczkowska, A. et al. Mef2C restrains microglial inflammatory response and is lost in brain ageing in an IFN-I-dependent manner. Nat. Commun. 8, 717 (2017).
Udeochu, J. C. et al. Tau activation of microglial cGAS–IFN reduces MEF2C-mediated cognitive resilience. Nat. Neurosci. 26, 737–750 (2023).
Gulen, M. F. et al. cGAS–STING drives ageing-related inflammation and neurodegeneration. Nature 620, 374–380 (2023).
Ennerfelt, H. et al. SYK coordinates neuroprotective microglial responses in neurodegenerative disease. Cell 185, 4135–4152 (2022).
Wang, S. et al. TREM2 drives microglia response to amyloid-β via SYK-dependent and -independent pathways. Cell 185, 4153–4169 (2022).
March-Diaz, R. et al. Hypoxia compromises the mitochondrial metabolism of Alzheimer’s disease microglia via HIF1. Nat. Aging 1, 385–399 (2021).
Monney, L. et al. Th1-specific cell surface protein Tim-3 regulates macrophage activation and severity of an autoimmune disease. Nature 415, 536–541 (2002).
Yin, Z. et al. APOE4 impairs the microglial response in Alzheimer’s disease by inducing TGFβ-mediated checkpoints. Nat. Immunol. 24, 1839–1853 (2023).
Rosenzweig, N. et al. Sex-dependent APOE4 neutrophil–microglia interactions drive cognitive impairment in Alzheimer’s disease. Nat. Med. 30, 2990–3003 (2024).
Rosenzweig, N. et al. PD-1/PD-L1 checkpoint blockade harnesses monocyte-derived macrophages to combat cognitive impairment in a tauopathy mouse model. Nat. Commun. 10, 465 (2019).
Baruch, K. et al. PD-1 immune checkpoint blockade reduces pathology and improves memory in mouse models of Alzheimer’s disease. Nat. Med. 22, 135–137 (2016).
Chen, X. et al. Microglia-mediated T cell infiltration drives neurodegeneration in tauopathy. Nature 615, 668–677 (2023).
Wang, T. W. et al. Blocking PD-L1–PD-1 improves senescence surveillance and ageing phenotypes. Nature 611, 358–364 (2022).
Su, W. et al. CXCR6 orchestrates brain CD8+ T cell residency and limits mouse Alzheimer’s disease pathology. Nat. Immunol. 24, 1735–1747 (2023).
Linnerbauer, M. et al. PD-L1 positive astrocytes attenuate inflammatory functions of PD-1 positive microglia in models of autoimmune neuroinflammation. Nat. Commun. 14, 5555 (2023).
Pertel, T. et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature 472, 361–365 (2011).
Zhang, Y., Feng, X. H. & Derynck, R. Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-β-induced transcription. Nature 394, 909–913 (1998).
McQuade, A. et al. Development and validation of a simplified method to generate human microglia from pluripotent stem cells. Mol. Neurodegener. 13, 67 (2018).
Myers, S. A. et al. Streamlined protocol for deep proteomic profiling of FAC-sorted cells and its application to freshly isolated murine immune cells. Mol. Cell. Proteomics 18, 995–1009 (2019).
Myers, S. A. et al. Discovery of proteins associated with a predefined genomic locus via dCas9–APEX-mediated proximity labeling. Nat. Methods 15, 437–439 (2018).
Becht, E. et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38–44 (2019).
Levine, J. H. et al. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell 162, 184–197 (2015).
Picelli, S. et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 10, 1096–1098 (2013).
Slyper, M. et al. A single-cell and single-nucleus RNA-seq toolbox for fresh and frozen human tumors. Nat. Med. 26, 792–802 (2020).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Graubert, A., Aguet, F., Ravi, A., Ardlie, K. G. & Getz, G. RNA-SeQC 2: efficient RNA-seq quality control and quantification for large cohorts. Bioinformatics 37, 3048–3050 (2021).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Gu, Z., Eils, R. & Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016).
Gu, Z., Gu, L., Eils, R., Schlesner, M. & Brors, B. circlize implements and enhances circular visualization in R. Bioinformatics 30, 2811–2812 (2014).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Zheng, G. X. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 (2021).
McGinnis, C. S., Murrow, L. M. & Gartner, Z. J. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 8, 329–337 (2019).
Zeisel, A. et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138–1142 (2015).
Di Bella, D. J. et al. Molecular logic of cellular diversification in the mouse cerebral cortex. Nature 595, 554–559 (2021).
Yao, Z. et al. A transcriptomic and epigenomic cell atlas of the mouse primary motor cortex. Nature 598, 103–110 (2021).
Van Hove, H. et al. A single-cell atlas of mouse brain macrophages reveals unique transcriptional identities shaped by ontogeny and tissue environment. Nat. Neurosci. 22, 1021–1035 (2019).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Liberzon, A. et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015).
Marsh, S. E. et al. Dissection of artifactual and confounding glial signatures by single-cell sequencing of mouse and human brain. Nat. Neurosci. 25, 306–316 (2022).
Liao, F. et al. Targeting of nonlipidated, aggregated apoE with antibodies inhibits amyloid accumulation. J. Clin. Invest. 128, 2144–2155 (2018).
Acknowledgements
We thank D. Holtzman50,104 for the HJ3.4b anti-Aβ antibody; M. Das for contributions to Co-IP–MS screening; S. Simmons for discussions with respect to snRNA-seq; staff at the NeuroTechnology Studio at Brigham and Women’s Hospital for providing access to instruments and consultation on data acquisition and data analyses. This work was supported by grants from the following organizations to the indicated authors: V.K.K.: NIH-NIAID (P01 AI073748, R01 AI144166 and P01 AI056299), NIH-NIA (R01 AG080992) and Cure Alzheimer’s Foundation Award ‘Role of checkpoint molecule TIM-3 in regulating microglia in Alzheimer’s disease’, ‘Role of checkpoint molecules TIM-3 and LAG-3 in microglial function in Alzheimer’s disease’ and ‘A new model of microglia genetic perturbation in vivo to screen all risk factors associated with Alzheimer’s diseases’; O.B.: NIH-NIA (R01AG054672, R01AG051812, R01AG075509 and R21AG076982), NIH-NINDS (R01NS088137), NIH-NEI (R01EY027921), Cure Alzheimer’s Fund (ApoE consortium), BrightFocus Foundation (2020A016806); M.B.-J.: NIH-NIA (R01 AG082704 and U19AG069701) and Cure Alzheimer’s Fund (Neuroimmune Consortium); and G.J.F.: NIAID (P01 AI056299). G.E.-S. is supported by fellowships from the Alzheimer Society of Canada (Discovery 23–10) and the Larry L. Hillblom Foundation (2024-A-023-FEL). The University of California, Irvine, Alzheimer’s Disease Research Center iPS cell core is funded by NIH AG066519. K.K. was supported by a Takeda Science Foundation Overseas Research Fellowship, a Japan Society for the Promotion of Science Overseas Research Fellowship, a Kanae Foundation for the Promotion of Medical Science Fellowship, a Mochida Memorial Foundation for Medical and Pharmaceutical Research Fellowship, The Kyoto University Foundation and a Konishi Foundation for International Exchange Fellowship. A. Suhail was supported by a postdoctoral fellowship from the National Multiple Sclerosis Society.
Author information
Authors and Affiliations
Contributions
K.K. primarily performed experiments with help from Z.Y., A.K., D.H., K.O.D., U.K.C., X.D. N.A., I.G., X.Z., R.T., A.A., V.S., L.L., J.-L.B., A.D., Y.C., S.S. and A. Suhail. A. Subramanian led, designed and performed the computational analyses with help from Y.W., who also contributed text for the methods. C.L. and N.G. ran preprocessing pipelines for selected bulk RNA-seq datasets with guidance from A. Subramanian (N.G.). T.P. and S.A.M. performed TIM-3 Co-IP–MS screening (data presented in Fig. 2c). M.N. generated snRNA-seq data with guidance from M.L.S. M.L.S. provided analytical and technical support for snRNA-seq. G.E.-S. and M.B.-J. generated the iPS cell-derived iMG. G.J.F. provided essential materials for Co-IP and WB from human cells described here. K.L.K., H.Z., T.D., O.R.-R., D.J.S., H.L.W., C.C. and A.R. provided input or other essential resources. K.K., Z.Y., O.B. and V.K.K. designed the experimental set-up and conceived the study. K.K. and A. Subramanian wrote the manuscript and prepared figures with input and edits from all authors.
Corresponding authors
Ethics declarations
Competing interests
G.J.F. has equity in Nextpoint, iTeos, IgM, Invaria, GV20, Bioentre and Geode. G.J.F. has served on advisory boards for iTeos, NextPoint, IgM, GV20, IOME, Bioentre, Santa Ana Bio, Simcere of America and Geode. G.J.F.’s interests were reviewed and managed by the Dana-Farber Cancer Institute in accordance with their conflict-of-interest policies. D.J.S. is a director of Prothena Biosciences and an ad hoc consultant to Roche and Eisai. M.B.-J. is a co-founder and consultant for Savanna Biotherapeutics (formally NovoGlia). A.R. is a cofounder and equity holder of Celsius Therapeutics, an equity holder in Immunitas and was a scientific advisory board member of ThermoFisher Scientific, Syros Pharmaceuticals, Neogene Therapeutics and Asimov until 31 July 2020. From 1 August 2020, A.R. is an employee of Genentech and has equity in Roche. M.L.S. is an equity holder, scientific cofounder and advisory board member of Immunity Therapeutics. O.B. is a cofounder of and has an ownership interest in Glial Therapeutics and Gliax. O.B. has a financial interest in Glial Therapeutics, a company developing a new therapy to target ITGB8–TGFβ signalling as a treatment for AD. O.B.’s interests were reviewed and are managed by Brigham and Women’s Hospital and Mass General Brigham in accordance with their conflict-of-interest policies and include collaboration with GSK and Regulus Therapeutics, receiving research funding from Sanofi and GSK, and honoraria for lectures and consultancy from UCB, Camp4 and Ono Pharma USA. V.K.K. has an ownership interest in Tizona Therapeutics, Trishula, Celsius Therapeutics, Bicara Therapeutics, Larkspur Therapeutics and Werewolf Therapeutics. V.K.K. has financial interests in Biocon Biologic, Compass, Elpiscience Biopharmaceutical, Equilium, PerkinElmer and Syngene. V.K.K. is a member of SABs for Cell Signaling Technology, Elpiscience Biopharmaceutical, Larkspur, Tizona Therapeutics, Tr1X and Werewolf. V.K.K.’s interests were reviewed and managed by Mass General Brigham in accordance with their conflict-of-interest policies.
Peer review
Peer review information
Nature thanks Michal Schwartz and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Havcr2 is highly expressed by microglia and induced by TGFβ.
a, Representative gating strategy for microglia. b, Representative gating strategy for immune cells in the spleen. c, Representative gating strategy for HEK293 cells transfected with Havcr2 constructs. d, Immune checkpoint genes were quantified by RT-qPCR in microglia in the brain (live CD45intCD11b+), and peripheral immune cells in the spleen (n = 3/condition, independent mice). e, The expression of immune checkpoint genes, including Havcr2, Lag3, and Vsir, is shown in mouse microglia and other cell populations in the central and peripheral nervous system using a public scRNAseq dataset19. f, The previous dataset19 was reanalyzed to detect a dissociation signature103 in microglia and perivascular macrophages. Unsupervised clustering divided microglia into 3 populations (MGL1, 2, and 3). Though genes induced by tissue dissociation were highly expressed in two of the three mouse microglial clusters103, Havcr2, Lag3, and Vsir showed high and specific expression in microglia (e). g, The expression of immune checkpoint genes and TGFβ pathway-related genes is shown in human microglia and other cell populations in the brain20. h, Immune checkpoint receptor expression on microglia was analyzed by flow cytometry during the early postnatal period (n = 9, 3, 5 for day 7, 13, 20, respectively, independent mice). i, Developmental alterations of MGnD and homeostasis-associated genes in microglia were analyzed using a published dataset21. MGnD and homeostasis-associated genes include the top 10 DEGs in Clec7a+ plaque-associated microglia10 and several selected genes. j, Primary microglia (live CD45intCD11b+) were cultured with either TGFβ (2, 10, or 50 ng/mL), M-CSF (10 ng/mL), GM-CSF (10 ng/mL), IL-27 (10 ng/mL), or LPS (10 ng/mL) for 24 h. RT-qPCR quantified the immune checkpoint gene expression (n = 3/condition, independent samples). k, TPM value of Havcr2 in the RNAseq analysis of sorted microglia (live CD45intCD11b+) from one-month-old Havcr2cKO mice (n = 4 (3 males, 1 female)), and Havcr2flox/flox mice (n = 5 (4 males, 1 female)). Avg Expr, average expression; Pct Expr, percent expressed; MGL, microglia; PVM, perivascular macrophage. Results are shown from one experiment, representing at least two independent experiments for h. Data are mean ± s.e.m. One-way ANOVA with Dunnett’s multiple comparisons test.
Extended Data Fig. 2 TGFβ pathway-related genes are not altered in Havcr2cKO microglia.
a, The structure of Dox-inducible (Tet-on system) Flag-conjugated HAVCR2 plasmid, which was used for CoIP-MS screening for binding partners of Tim-3. b, Tim-3 expression was evaluated by western blot in Jurkat cells transduced with Dox-inducible Flag-conjugated HAVCR2, after treatment with Dox. Supernatant samples were from the supernatant solution after incubation of input cell lysate with anti-Flag antibody-conjugated dynabeads. c, d, Proximity ligation assay for Tim-3, Smad2, and Tgfbr2 in primary microglia from the brain (c) and the spinal cord (d) of WT mice. e, pSmad2, Smad2, and Gapdh were quantified in Havcr2KO primary microglia by western blotting. The samples for the control Gapdh were run on a separate gel as sample processing controls (Supplementary Fig. 1 for gel source data). f, pSmad3 and Smad3 expression was quantified in Havcr2cKO microglia by flow cytometry (n = 5 control, 4 Havcr2cKO, independent mice). g, Microglia (live CD45intCD11b+) were sorted from control and Havcr2cKO male mice and cultured in the presence of M-CSF (10 ng/mL), GM-CSF (10 ng/mL), or TGFβ (10 ng/mL) for 16 h. TGFβ pathway-related gene expressions were quantified by RT-qPCR (n = 3/condition, 2 for Havcr2cKO-GM-CSF group, independent samples. h, HEK293 cells were transfected with WT-Havcr2 plasmid. Smad3 and pSmad3 expression was quantified by flow cytometry (n = 4/condition, independent samples). i, Tgfbr2KO BV2 cells were stimulated with TGFβ, and evaluated for pSmad2 expression by flow cytometry. j, Smad2 was quantified in HEK293 cells by flow cytometry after transfection with WT and mutant Havcr2 constructs (n = 3/condition, independent samples). k, HEK293 cells were transfected with WT-Havcr2 plasmid and either non-phosphorylated Smad2 mimetic (3SA) or phosphorylated Smad2 mimetic (2 SD) plasmid. The cell lysate was coimmunoprecipitated for HA-Tim-3 and blotted for Smad2 mimetics. The samples for the control β actin were run on the same gel as loading controls (Supplementary Fig. 1 for gel source data) l, Microglia (live CD45intCD11b+) were sorted and cultured in the presence of Galectin 9 (3 ug/mL), or CEACAM1 (3 ug/mL), combined with TGFβ (1 ng/mL) for 16 h. pSmad2 expression was quantified by flow cytometry (n = 3/condition, independent samples). m, HEK293 cells were transfected with WT-Havcr2 plasmid and either one of three plasmids with major Smad2 domains: MH1 (Amino acid sequences (AA) 1-181), and MH2 (AA 274-467). The cell lysate was co-immunoprecipitated for HA-Tim-3 and blotted for Smad2 domains. The samples for the control β actin were run on the same gel as sample processing controls (Supplementary Fig. 1 for gel source data). Dox, doxycycline; Sup, supernatant; CoIP, coimmunoprecipitation; MS, mass spectrometry. Results are shown from one experiment, representing at least two independent experiments for h-m. Data are mean ± s.e.m.
Extended Data Fig. 3 Havcr2cKO microglia share gene expression signature with Tgfbr2cKO microglia, phagocytosing microglia, and MGnD.
a, Circos plot comparison of DEGs up- and down-regulated in Havcr2cKO, phagocytosing, Tgfbr2cKO, and Clec7a+ microglia compared to control microglia. Only islands with DEGs shared by at least 3 of the 4 comparisons were included. Permutation test p-values were displayed as ns P > 0.025, *P ≤ 0.025, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001, and *****P ≤ 0.00001. b, Heatmap visualization of DEGs shared by at least 3 of the 4 comparisons. c, Heatmap representation of the correlation between vectors of the gene expression levels of all DEGs of (1) Havcr2cKO microglia compared to control microglia, and those shared by comparison (2) to (4) (phagocytosing, Tgfbr2cKO, Clec7a+ microglia compared to their corresponding control groups).
Extended Data Fig. 4 Aβ plaque load is not reduced in Havcr2icKO:5xFAD mice at five months.
a, Tim-3 expression was quantified in microglia and infiltrating monocytes by flow cytometry at 4 and 7 months (n = 19, 13, 14, 20 (4 M), 8, 9, 9, and 6 (7 M), from the left). b, Tim-3 expression was quantified in macrophages and Ly6Chigh and Ly6Clow monocytes in the spleen by flow cytometry at 7 months (n = 9, 8, 3, and 10, from the left). c, Havcr2 gene expression was quantified in microglia from 5xFAD and Havcr2icKO:5xFAD mice at the age of 4 and 7 months by qPCR (n = 16, 13, 14, 20 (4 M), 5, 13, 14, and 14 (7 M), from the left). d, Locomotor activity measures (including total arm entries, distance moved, movement duration, and velocity) were assessed by spontaneous alternation Y-maze test (n = 37, 33, 30, and 35, from the left). No significant difference was observed among the groups regarding the locomotor activity. e, Representative tile images of Aβ (HJ3.4b antibody) and Clec7a staining in male 5xFAD and Havcr2icKO:5xFAD mice at the age of 6 months. f, Confocal images of Tmem119, Clec7a and HJ3.4b staining at the age of 6 months. g, Aβ plaque load and size were quantified in the cortices at 7 months (n = 17, 17, 12, and 15, from the left). The difference was still significant after separating sexes, both for plaque load and size. h, Aβ plaque load and size were quantified in the cortices at 5 months (n = 4, 7, 8, and 4, from the left). i, Gene expressions of molecules involved in Aβ production were quantified in brain homogenate at 7 months (n = 6 and 8, from the left). Results are shown using three, one, one, and six independent cohorts for 4, 5, 6, and 7 months, respectively. Data are mean ± s.e.m. Kruskal-Wallis test adjusted by the false discovery rate (FDR) for d, student’s two-tailed t-test for g and h.
Extended Data Fig. 5 Reduction of Aβ plaque burden is observed for all the plaque phenotypes in Havcr2icKO:5xFAD mice.
a, The plaques in the cortices at 7 months were classified into filamentous, compact, and inert areas by double staining with Methoxy-X04 and HJ3.4b. The percentage of each area was calculated per plaque depending on its size (at least 100 um2). #: significant only in females. b, Aβ plaque number with more than 100 um2 of size was quantified per 1×105 um2 of the cortices at 7 months. The plaque area was calculated for total, filamentous, compact, and inert phenotypes. The inert area difference was insignificant when assessed only for female mice. c, The percentage area of each phenotype was calculated per each plaque with more than 100 um2 of size at 5 months. As plaque groups, plaques with ≥ 50% area of any phenotype were classified into the category of that phenotype. Results are shown using one and six independent cohorts for 5 and 7 months, respectively. d, Confocal images of Thioflavin-S+ and HJ3.4b+ plaques in the cortex from 7-month-old 5xFAD and Havcr2icKO:5xFAD mice. e, Quantification of the areas of filamentous plaques (HJ3.4b+Thioflavin-S–), compact plaques (HJ3.4b+Thioflavin-S+), and inert plaques (HJ3.4b–Thioflavin-S+) from 5xFAD and Havcr2icKO:5xFAD mice at the age of 7 months (n = 13-15 ROIs/group for filamentous area, n = 14 ROIs/group for compact area, n = 14 ROIs/group for inert area; 2-3 ROIs per mouse). f, Quantification of the areas of filamentous plaques, compact plaques, and inert plaques from 5-month-old 5xFAD and Havcr2icKO:5xFAD mice (n = 11-14 ROIs/group for filamentous area, n = 11-14 ROIs/group for compact area, n = 11-14 ROIs/group for inert area; 2-3 ROIs per mouse). g, The number of synapses detected by colocalization of Homer1 and Vglut2 was quantified in the cortices at 7 months (n = 6, 12, 6, and 17, from the left). The difference was significant when both sexes were combined. Data are mean ± s.e.m. Student’s two-tailed t-test for a-c and e. One-way ANOVA with Holm-Sidak’s multiple comparisons test for g.
Extended Data Fig. 6 Galectin-3-positive area is reduced in Havcr2icKO:5xFAD mice.
a, Representative tile images of galectin-3 staining in 5xFAD and Havcr2icKO:5xFAD mice of both sexes at the age of 6 months. b, Confocal images of galectin-3 staining in females at the age of 6 months. c, Quantification of galectin-3-positive area and fluorescence intensity in cortical regions in females at the age of 6 months (n = 4 and 7 for 5xFAD and Havcr2icKO:5xFAD mice, respectively). d, Confocal images of galectin-3 staining in males at the age of 6 months. e, Quantification of galectin-3-positive area and fluorescence intensity in cortical regions in males at the age of 6 months (n = 6/each group). f, g, The frequencies of immune cell populations were quantified among live CD45+ cells (except for DC1 and DC2, among DC) in the spleen at 7 months. Results are shown using two independent cohorts. Data are mean ± s.e.m. Student’s two-tailed t-test.
Extended Data Fig. 7 Microglial clusters found in snRNAseq of the 5xFAD cohort.
a, b, RNAseq analysis of sorted microglia (live CD45intCD11b+Tmem119+Gr1−) from 5xFAD cohort at 4 and 7 months old females. Gene expression signatures of Havcr2icKO microglia (DEGs between Havcr2icKO and control microglia) and 5xFAD microglia (DEGs between 5xFAD and control microglia) were compared. c, d, Boxplots showing the gene expression levels (log2 TPM) of Havcr2 at 4 and 7 months (n = 4, 2, 2, 4 (4 M female), 2, 3, 6, 2 (4 M male), 8, 8, 7, 6 (7 M female), 8, 5, 6, and 11 (7 M male) from the left). e, The frequency of Clec7a+ MGnD-type microglia was evaluated by flow cytometry at 4 months. f, UMAP visualization of 63,119 nuclei across all genotypes (n = 10,063, 12,531, 18,734, and 21,791 nuclei from control, Havcr2icKO, 5xFAD, and Havcr2icKO:5xFAD mice, respectively) with 26 transcriptionally distinct populations (color in the figure). g, DotPlot visualization of Havcr2 expression by genotype in 26 populations. Size of the dot is the percentage of cells in the cluster (column) expressing the marker gene (row). Color represents the average scaled expression of the gene among expressing cells. h-j, Barplot visualization of proportions (Y-axis) of individual mouse (a), and genotype (b) per microglial cluster (X-axis); and cluster per mouse (X-axis) (c). k, Dotplot visualization of top genes distinguishing each microglial cluster from snRNAseq (Fig. 5a). Size of the dot is the percentage of cells in the cluster (column) expressing the marker gene (row). Color represents the average scaled expression of the gene among expressing cells. l, m, Visualization of the MGnD (g) and homeostasis (h) signature score (Y-axis) across microglia clusters (X-axis) and genotypes (color). Each point represents a microglial cell. n, Proportions of microglial cluster 0-4 in 5xFAD and Havcr2icKO:5xFAD mice (n = 3 for each). o, Violin plot of scores for Tgfbr2cKO signature genes among genotypes in cluster 0. Results are shown using three independent cohorts for e. Data are mean ± s.e.m. Permutation test p-values were used in a and b. Student’s two-tailed t-test with Bonferroni correction for n. Student’s two-tailed t-test for o.
Extended Data Fig. 8 Havcr2icKO mice have unique microglial states in an Alzheimer’s disease model.
a, b, Heatmap of top DEGs distinguishing P1 (a) and P2 (b) subpopulations in Cluster 2/MGnD. c, d, UMAP Visualization of microglial snRNA-seq clusters colored by P1 (c) and P2 (d) signature scores.
Extended Data Fig. 9 Signature scores of different cell types in each microglial cluster.
a-h, Violin plots of microglia, border-associated macrophages (BAM), perivascular macrophages (PVM), infiltrating monocyte, and monocyte/monocyte-derived antigen-presenting cells (APC) signature scores (Y-axis) across microglial clusters (X-axis) and genotypes (color). Each point represents a microglial cell. i, j, Violin plot of signature scores (Y-axis) of P1 and P2 populations from Havcr2icKO:5xFAD mice across all genotypes and microglial clusters (X-axis).
Extended Data Fig. 10 Tim-3 expression is inversely correlated with P2 marker expressions in MGnD cluster.
a, Boxplot of pairwise spearman correlation coefficient (Y-axis) of whole gene expression pseudobulk microglial profiles between conditions (X-axis). Each point represents the spearman correlation coefficient between profiles of two biological replicates. Havcr2icKO:5xFAD;P1 vs 5xFAD: n = 9 spearman correlations derived from 34 cells from 3 biologically independent Havcr2icKO:5xFAD samples and 201 cells from 3 biologically independent 5xFAD samples. Havcr2icKO:5xFAD;P2 vs 5xFAD: n = 9 spearman correlations derived from 23 cells from 3 biologically independent Havcr2icKO:5xFAD samples and 201 cells from 3 biologically independent 5xFAD samples. Havcr2icKO:5xFAD;P1 vs Havcr2icKO:5xFAD;P2: n = 9 spearman correlations derived from 57 cells (34 cells, 23 cells, respectively) from 3 biologically independent Havcr2icKO:5xFAD samples. b, Twelve human microglial signatures were curated from a previous paper using samples from patients with AD58. These signature scores were plotted for cluster 2 in our snRNAseq data. A significant difference was found only in human MG8 signature annotated as inflammatory61. c, d, Scatterplot of P1(Y-axis, a) or P2 (Y-axis, b), and Hallmark TGFB signature scores among C2 5xFAD and Havcr2icKO microglia in current study. e-g, Scatterplot of P1 (Y-axis) or P2 (Y-axis), and Hallmark TGFB signature scores among 5xFAD microglia in MGnD clusters in public datasets (GSE9896957 cluster 2 (e), GSE9896957 cluster 3 (f), GSE14051059 (g)). h, i, The cells isolated from the brain were analyzed by flow cytometry, and identified clusters were annotated according to their markers, with cluster 12 annotated as MGnD. The proportion of cluster 12 among all the cells was compared between control, 5xFAD, and Havcr2icKO:5xFAD mice at the age of 4.5−5 months (n = 3, 3, 3 (female), 2, 4, 3 (male), from the left). j, Phenograph clustering of microglia from 4-month-old control, 5xFAD, and Havcr2icKO:5xFAD mice using flow cytometry data. Cluster 12 represents Clec7a+ MGnD among the microglia. k, Pearson’s correlation between MFI of paired markers of interest within combined Clec7a+ MGnD from 4-month-old control, 5xFAD, and Havcr2icKO:5xFAD was analyzed. The coefficient indicates the strength and direction of the correlation. Data are mean ± s.e.m. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 and ****P ≤ 0.0001; Student’s two-tailed t-test for a. One-way ANOVA with Tukey’s HSD correction for b. Spearman correlation test was used for c-g. One-way ANOVA with Holm-Sidak’s multiple comparisons test for i. Pearson’s correlation analysis was adjusted by the false discovery rate (FDR) method for k.
Extended Data Fig. 11 General appearance of scRNAseq of microglia from 5xFAD cohort.
a, UMAP visualization of signature scores for the MGnD, homeostasis, and Interferon signatures. Each dot is a microglial cell, and the color represents the value of the signature score in that cell. b, DotPlot visualization of marker genes (Y-axis) for each cluster (X-axis) determined by differential gene expression between each cluster and all other cells. The size of the dot represents the number of cells in the cluster that have a non-zero expression of the gene and the color represents the average level of expression in scaled units (scaled across clusters). c, Barplot representing proportions of microglial cells in each cluster from either condition: 5xFAD (purple) or Havcr2icKO:5xFAD (dark yellow). d, e, Scatter plot of log2 fold changes of gene expression in microglia between conditions 5XFAD and Havcr2icKO:5xFAD on the X-axis and Clec7a− vs Clec7a+ on the Y-axis for the clusters HMG_0 (a) and MGnD_0 (b). Spearman Correlation coefficients and p-values are indicated. Spearman correlation test for d and e.
Extended Data Fig. 12 Transcriptional characteristics of Havcr2icKO:5xFAD microglia revealed by scRNAseq.
a, The characteristic genes that were differentially expressed between Havcr2icKO:5xFAD and 5xFAD mice are described in each microglial cluster. The genes were sorted into several gene categories. Color is the log2 fold change comparing Havcr2icKO:5xFAD to 5xFAD. Asterisks are the significance level by Wilcoxon test adjusted with Benjamini-Hochberg method. b, c, Scatter plot of log2 fold changes of gene expression in microglia between conditions 5XFAD and Havcr2icKO:5xFAD on the X-axis and Clec7a− vs Clec7a+ on the Y-axis for the clusters HMG_0 (b) and MGnD_0 (c). Spearman Correlation coefficients and p-values are indicated. d, e, Volcano plot of differential gene expression between conditions 5XFAD and Havcr2icKO:5xFAD and Clec7a− vs Clec7a+ in clusters HMG_0 (d) and MGnD_0 (e). Each point is a gene; the X-axis represents the average log2 foldchange between conditions and the Y-axis represents the -log10(FDR). Spearman correlation test for b and c. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 and ****P ≤ 0.0001; HMG: homeostatic microglia, CC: cell-cycling.
Extended Data Fig. 13 Gene expression signatures of Havcr2icKO:5xFAD microglia.
a, Gene expression was analyzed across all the genotypes at the age of 4.5 months by qPCR (n = 9, 5, 7, 11 (male), 8, 8, 7, 9 (female), from the left). b, Microglial signature scores curated using a previous paper61 were compared between Havcr2icKO:5xFAD and 5xFAD in each microglial cluster defined in Fig. 5k using a t-test. Numbers are the test statistics (Havcr2icKO:5xFAD against 5xFAD); background color shows upregulation or downregulation; and text color is the significance of the t-test. The p-values were adjusted using the Benjamini-Hochberg method. Data are mean ± s.e.m. One-way ANOVA with Sidak’s multiple comparisons test for a. HMG: homeostatic microglia, CC: cell-cycling.
Supplementary information
Supplementary Fig. 1
Gel source data.
Supplementary Tables
Supplementary Tables 1–8. Supplementary Table 1: List of genes differentially expressed in Havcr2cKO vs control (data for 1-month-old mice). Supplementary Table 2: List of proteins detected in Co-IP–MS screening assays. Supplementary Table 3: Sequences of mutant Havcr2 constructs. Supplementary Table 4: Overlap of DEGs across four comparisons: phagocytosis vs control, Havcr2cKO vs control, Clec7a+ vs Clec7a– and Tgfbr2cKO vs control. Supplementary Table 5: Quality-control metrics and cluster markers for scRNA-seq and snRNA-seq datasets of 5×FAD and Havcr2icKO:5×FAD microglia. Supplementary Table 6: Antibodies used in the experiments. Supplementary Table 7: The annotation of in-house generated scRNA-seq data of microglia. Supplementary Table 8: Summary of signature genes.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kimura, K., Subramanian, A., Yin, Z. et al. Immune checkpoint TIM-3 regulates microglia and Alzheimer’s disease. Nature 641, 718–731 (2025). https://doi.org/10.1038/s41586-025-08852-z
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41586-025-08852-z
This article is cited by
-
TIM-3 regulates microglial function
Nature Reviews Drug Discovery (2025)
-
Into the Spotlight: Microglia in the Peripheral Nervous System
Neuroscience Bulletin (2025)
-
Beyond Neuroinflammation: Microglia at the Crossroads of Amyloid, Tau, and Neurodegeneration in Alzheimer’s Disease
Neurological Sciences (2025)
-
A spatial map of MAPK-activated immunosuppressive myeloid populations in pediatric low-grade glioma
Nature Immunology (2025)