Abstract
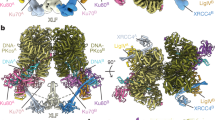
Non-homologous end joining (NHEJ) is the main repair pathway of double-strand DNA breaks in higher eukaryotes1,2. Here we report reconstitution of the final steps of NHEJ and structures of DNA polymerase μ and ligase IV (LIG4) engaged in gap filling and end joining. These reactions take place in a flexible ω-shaped framework composed of XRCC4 and XLF. Two broken DNA ends, each encircled by Ku70–Ku80 internally, are docked onto the ω frame, mediated by LIG4. DNA polymerase and ligase attached to each ω arm repair only one broken strand of a defined polarity; the final steps of NHEJ requires coordination and toggling of a pair of such enzymes. The facilitators XLF and PAXX additively stimulate NHEJ reactions. As DNA-end sensor and protector, LIG4 replaces DNA-PKcs for end joining and bridges the two DNA ends for polymerase to fill remaining gaps. These assemblies present new targets for NHEJ inhibition to enhance efficacy of radiotherapy and accuracy of gene editing.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Cryo-EM maps and structure coordinates of full-length XLF complexes have been deposited with the Electron Microscopy Data Bank (EMDB) and Protein Data Bank (PDB) with the following accession codes: gap-filling complex (EMD-49108, 9N81); ligation complex (EMD-49110, 9N83) and AMP–Lys complex (EMD-49109, 9N82). We also deposited maps associated with gap filling complex, EMD-49246 (global consensus map), EMD-49115 (focused Pol μ), EMD-49132 (focused active KU), EMD-49134 (focused active L4X4), EMD-49133 (focused supportive KU), EMD-49135 (focused supportive L4X4), EMD-49136 (focused XLF), EMD-49137 (focused LIG4 NTD and DBD), ligation complex and AMP–Lys complex, EMD-49244/49245 (global consensus maps), EMD-49144/49143 (focused LIG4cat), EMD-49138 (focused active KU), EMD-49140 (focused active L4X4), EMD-49139 (focused supportive KU), EMD-49141 (focused supportive L4X4) and EMD-49142 (focused XLF). Cryo-EM maps and structure coordinates of the truncated XLF complexes have been deposited with EMDB and PDB with the following accession codes: gap-filling complex (EMD-45807, 9CQ3); ligation complex (EMD-45809, 9CQ6) and ligation-like complex (EMD-45813, 9CQC). We also deposited maps associated with gap filling complex, EMD-45838 (global consensus map), EMD-45839 (focused Pol μ), EMD-45840 (focused active KU), EMD-45841 (focused active L4X4), EMD-45842 (focused supportive KU), EMD-45843 (focused supportive L4X4), EMD-45844 (focused XLF), EMD-45845 (focused LIG4 NTD and DBD), EMD-45846 (focused PAXX), ligation and ligation-like complexes, EMD-45847/45855 (global consensus maps), EMD-45848/45857 (focused LIG4cat), EMD-45849/45858 (focused active KU), EMD-45850/45859 (focused active L4X4), EMD-45851/45860 (focused supportive KU), EMD-45852/45861 (focused supportive L4X4) and EMD-45853/45862 (focused XLF). The above listed data are available at rcsb.org and www.ebi.ac.uk/emdb for download. Coordinates with PDB codes 1JEY, 1T2V, 2DUN, 2R9A, 3II6, 3RWR, 3Q4F, 3SR2, 3WTD, 4M0M, 6BKG, 7LSY, 7SGL, 7ZYG and 8ASC are also available at rcsb.org.
References
Shibata, A. & Jeggo, P. A historical reflection on our understanding of radiation-induced DNA double strand break repair in somatic mammalian cells; interfacing the past with the present. Int. J. Radiat. Biol. 95, 945–956 (2019).
Stinson, B. M. & Loparo, J. J. Repair of DNA double-strand breaks by the nonhomologous end joining pathway. Annu. Rev. Biochem. 90, 137–164 (2021).
Arter, M. & Keeney, S. Divergence and conservation of the meiotic recombination machinery. Nat. Rev. Genet. 25, 309–325 (2024).
Christie, S. M., Fijen, C. & Rothenberg, E. V(D)J recombination: recent insights in formation of the recombinase complex and recruitment of DNA repair machinery. Front. Cell Dev. Biol. 10, 886718 (2022).
Wang, X. S., Lee, B. J. & Zha, S. The recent advances in non-homologous end-joining through the lens of lymphocyte development. DNA Repair 94, 102874 (2020).
Wang, J. Y. & Doudna, J. A. CRISPR technology: a decade of genome editing is only the beginning. Science 379, eadd8643 (2023).
Jasin, M. & Haber, J. E. The democratization of gene editing: insights from site-specific cleavage and double-strand break repair. DNA Repair 44, 6–16 (2016).
Bossaert, M. et al. Identification of the main barriers to Ku accumulation in chromatin. Cell Rep. 43, 114538 (2024).
Williams, G. J. et al. Structural insights into NHEJ: building up an integrated picture of the dynamic DSB repair super complex, one component and interaction at a time. DNA Repair 17, 110–120 (2014).
Woodbine, L., Gennery, A. R. & Jeggo, P. A. The clinical impact of deficiency in DNA non-homologous end-joining. DNA Repair 16, 84–96 (2014).
Pryor, J. M. et al. Essential role for polymerase specialization in cellular nonhomologous end joining. Proc. Natl Acad. Sci. USA 112, E4537–E4545 (2015).
Sallmyr, A., Rashid, I., Bhandari, S. K., Naila, T. & Tomkinson, A. E. Human DNA ligases in replication and repair. DNA Repair 93, 102908 (2020).
Grawunder, U. et al. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature 388, 492–495 (1997).
Critchlow, S. E., Bowater, R. P. & Jackson, S. P. Mammalian DNA double-strand break repair protein XRCC4 interacts with DNA ligase IV. Curr. Biol. 7, 588–598 (1997).
Junop, M. S. et al. Crystal structure of the Xrcc4 DNA repair protein and implications for end joining. EMBO J. 19, 5962–5970 (2000).
Sibanda, B. L. et al. Crystal structure of an Xrcc4-DNA ligase IV complex. Nat. Struct. Biol. 8, 1015–1019 (2001).
Wu, P. Y. et al. Structural and functional interaction between the human DNA repair proteins DNA ligase IV and XRCC4. Mol. Cell. Biol. 29, 3163–3172 (2009).
Walker, J. R., Corpina, R. A. & Goldberg, J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 412, 607–614 (2001).
Graham, T. G. W., Carney, S. M., Walter, J. C. & Loparo, J. J. A single XLF dimer bridges DNA ends during nonhomologous end joining. Nat. Struct. Mol. Biol. 25, 877–884 (2018).
Nemoz, C. et al. XLF and APLF bind Ku80 at two remote sites to ensure DNA repair by non-homologous end joining. Nat. Struct. Mol. Biol. 25, 971–980 (2018).
Ochi, T. et al. DNA repair. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair. Science 347, 185–188 (2015).
Seif-El-Dahan, M. et al. PAXX binding to the NHEJ machinery explains functional redundancy with XLF. Sci. Adv. 9, eadg2834 (2023).
Chen, S. et al. Structural basis of long-range to short-range synaptic transition in NHEJ. Nature 593, 294–298 (2021).
Buck, D. et al. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell 124, 287–299 (2006).
Balmus, G. et al. Synthetic lethality between PAXX and XLF in mammalian development. Genes Dev. 30, 2152–2157 (2016).
Lescale, C. et al. Specific roles of XRCC4 paralogs PAXX and XLF during V(D)J recombination. Cell Rep. 16, 2967–2979 (2016).
Liu, X., Shao, Z., Jiang, W., Lee, B. J. & Zha, S. PAXX promotes KU accumulation at DNA breaks and is essential for end-joining in XLF-deficient mice. Nat. Commun. 8, 13816 (2017).
Moon, A. F. et al. Sustained active site rigidity during synthesis by human DNA polymerase mu. Nat. Struct. Mol. Biol. 21, 253–260 (2014).
Kaminski, A. M. et al. Structures of DNA-bound human ligase IV catalytic core reveal insights into substrate binding and catalysis. Nat. Commun. 9, 2642 (2018).
Stinson, B. M., Carney, S. M., Walter, J. C. & Loparo, J. J. Structural role for DNA ligase IV in promoting the fidelity of non-homologous end joining. Nat. Commun. 15, 1250 (2024).
Zhao, B., Rothenberg, E., Ramsden, D. A. & Lieber, M. R. The molecular basis and disease relevance of non-homologous DNA end joining. Nat. Rev. Mol. Cell Biol. 21, 765–781 (2020).
Riballo, E. et al. XLF-Cernunnos promotes DNA ligase IV-XRCC4 re-adenylation following ligation. Nucleic Acids Res. 37, 482–492 (2009).
Ropars, V. et al. Structural characterization of filaments formed by human Xrcc4-Cernunnos/XLF complex involved in nonhomologous DNA end-joining. Proc. Natl Acad. Sci. USA 108, 12663–12668 (2011).
Hammel, M. et al. XRCC4 protein interactions with XRCC4-like factor (XLF) create an extended grooved scaffold for DNA ligation and double strand break repair. J. Biol. Chem. 286, 32638–32650 (2011).
Chen, S. et al. Cryo-EM visualization of DNA-PKcs structural intermediates in NHEJ. Sci. Adv. 9, eadg2838 (2023).
Cottarel, J. et al. A noncatalytic function of the ligation complex during nonhomologous end joining. J. Cell Biol. 200, 173–186 (2013).
Graham, T. G., Walter, J. C. & Loparo, J. J. Two-stage synapsis of DNA ends during non-homologous end joining. Mol. Cell 61, 850–858 (2016).
Goff, N. J. et al. Catalytically inactive DNA ligase IV promotes DNA repair in living cells. Nucleic Acids Res. 50, 11058–11071 (2022).
Waters, C. A. et al. The fidelity of the ligation step determines how ends are resolved during nonhomologous end joining. Nat. Commun. 5, 4286 (2014).
Kaminski, A. M. et al. DNA polymerase lambda Loop1 variant yields unexpected gain-of-function capabilities in nonhomologous end-joining. DNA Repair 136, 103645 (2024).
Loc’h, J. & Delarue, M. Terminal deoxynucleotidyltransferase: the story of an untemplated DNA polymerase capable of DNA bridging and templated synthesis across strands. Curr. Opin. Struct. Biol. 53, 22–31 (2018).
Ochi, T., Gu, X. & Blundell, T. L. Structure of the catalytic region of DNA ligase IV in complex with an Artemis fragment sheds light on double-strand break repair. Structure 21, 672–679 (2013).
Pascal, J. M., O’Brien, P. J., Tomkinson, A. E. & Ellenberger, T. Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature 432, 473–478 (2004).
Cotner-Gohara, E. et al. Human DNA ligase III recognizes DNA ends by dynamic switching between two DNA-bound states. Biochemistry 49, 6165–6176 (2010).
Conlin, M. P. et al. DNA ligase IV guides end-processing choice during nonhomologous end joining. Cell Rep 20, 2810–2819 (2017).
De Ioannes, P., Malu, S., Cortes, P. & Aggarwal, A. K. Structural basis of DNA ligase IV–Artemis interaction in nonhomologous end-joining. Cell Rep. 2, 1505–1512 (2012).
Malu, S. et al. Artemis C-terminal region facilitates V(D)J recombination through its interactions with DNA ligase IV and DNA-PKcs. J. Exp. Med. 209, 955–963 (2012).
DeRose, E. F. et al. Solution structure of polymerase mu’s BRCT domain reveals an element essential for its role in nonhomologous end joining. Biochemistry 46, 12100–12110 (2007).
Mueller, G. A. et al. A comparison of BRCT domains involved in nonhomologous end-joining: introducing the solution structure of the BRCT domain of polymerase lambda. DNA Repair 7, 1340–1351 (2008).
Williams, R. S., Lee, M. S., Hau, D. D. & Glover, J. N. Structural basis of phosphopeptide recognition by the BRCT domain of BRCA1. Nat. Struct. Mol. Biol. 11, 519–525 (2004).
Chen, X. et al. Structure of an activated DNA-PK and its implications for NHEJ. Mol. Cell 81, 801–810.e803 (2021).
Liu, L. et al. Autophosphorylation transforms DNA-PK from protecting to processing DNA ends. Mol. Cell 82, 177–189.e174 (2022).
Tadi, S. K. et al. PAXX is an accessory c-NHEJ factor that associates with Ku70 and has overlapping functions with XLF. Cell Rep 17, 541–555 (2016).
Xing, M. et al. Interactome analysis identifies a new paralogue of XRCC4 in non-homologous end joining DNA repair pathway. Nat. Commun. 6, 6233 (2015).
Cisneros-Aguirre, M., Lopezcolorado, F. W., Ping, X., Chen, R. & Stark, J. M. Distinct functions of PAXX and MRI during chromosomal end joining. iScience https://doi.org/10.1016/j.isci.2025.112722 (2025).
Buehl, C. J. et al. Two distinct long-range synaptic complexes promote different aspects of end processing prior to repair of DNA breaks by non-homologous end joining. Mol. Cell 83, 698–714 e694 (2023).
Matsumoto, Y. et al. Development and evolution of DNA-dependent protein kinase inhibitors toward cancer therapy. Int. J. Mol. Sci. 23, 4264 (2022).
Denes, C. E. et al. Approaches to enhance precise CRISPR/Cas9-mediated genome editing. Int. J. Mol. Sci. 22, 8571 (2021).
Shao, Z. et al. DNA-PKcs has KU-dependent function in rRNA processing and haematopoiesis. Nature 579, 291–296 (2020).
Greco, G. E. et al. SCR7 is neither a selective nor a potent inhibitor of human DNA ligase IV. DNA Repair 43, 18–23 (2016).
Yang, W. & Gao, Y. Translesion and repair DNA polymerases: diverse structure and mechanism. Annu. Rev. Biochem. 87, 239–261 (2018).
Cisneros-Aguirre, M., Lopezcolorado, F. W., Tsai, L. J., Bhargava, R. & Stark, J. M. The importance of DNAPKcs for blunt DNA end joining is magnified when XLF is weakened. Nat. Commun. 13, 3662 (2022).
Schorb, M., Haberbosch, I., Hagen, W. J. H., Schwab, Y. & Mastronarde, D. N. Software tools for automated transmission electron microscopy. Nat. Methods 16, 471–477 (2019).
Fernandez-Leiro, R. & Scheres, S. H. W. A pipeline approach to single-particle processing in RELION. Acta Crystallogr. D Struct. Biol. 73, 496–502 (2017).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Sanchez-Garcia, R. et al. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun. Biol. 4, 874 (2021).
Rosenthal, P. B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Acknowledgements
We thank D. Leahy for critical reading of the manuscript. This work used the Cryo-Electron Microscopy Core facility, NIDDK and the NIH Multi-Institute Cryo-EM Facility (MICEF). This research was supported by National Institute of Diabetes, Digestive and Kidney Disease to M.G. (DK036167) and W.Y. (DK036147); the National Cancer Institute of the National Institutes of Health: R01CA256989, R01CA240392 (J.M.S.); P30CA33572 (City of Hope Core Facilities) and F99CA284248 (M.C.-A.).
Author information
Authors and Affiliations
Contributions
L.L. carried out biochemical experiments, cryo-EM grid preparation and data processing. J.L. collected cryo-EM data and built models. M.C.-A., A.M. and J.M.S. performed the cell-based NHEJ assay. W.Y. and M.G. designed and supervised this study. All authors participated in manuscript preparation.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Jean-Baptiste Charbonnier and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 End-joining and gap-filling reaction.
a. The presence of DNA-PKcs reduces the ligation efficiency of fully complementary DNA ends. b. Efficiency of gap filling and end joining on DNA substrate with different combinations of NHEJ factors (same as in Fig. 1c). The raw data are shown in a denaturing DNA gel. All assays were performed in triplicate, and the means and standard deviations were presented in Fig. 1d,e. For gel source data, see SI Fig. 1d,e.
Extended Data Fig. 2 Data processing and structure refinement of ligation Complex.
a. SDS gel of proteins used in this study. b. A representative cryoEM micrograph of 10,133 collected. c. Images of 2D classification. d. Workflow of image processing. e. FSC analysis of the data quality and map resolution. The map-model FSC (M) and the half-map FSC (H) are indicated. f. The composite map is colored according to the resolution scale bar on the side. g. Angular distribution of particles used in the final map calculation. h-l. The cryoEM map of specific regions.
Extended Data Fig. 3 The ω framework and its flexibility.
a. The cryoEM structure of end-joining complexes. The AMP-DNA (multi-color) and AMP-Lys (blue gray) complexes are superimposed, and the mobile domains are encircled. b. The NHEJ ligation (cartoon) and the SR complex (PDB: 7LSY, as semi-transparency yellow surface) are shown after superimposing the LIG4 catalytic core. The different regions between the cartoon and surface are boxed in red. The DNA in SR complex is marked by a red open arrow. c. A front view of the ω frame consisting of XLF (yellow and copper), XRCC4 (slate and orange) and two BRCT domains of LIG4 (green). Dimensions of the ω frame are marked. d. Two DNAs are suspended between L4X4 arms without touching the protein frame. e. A top view of panel d. The top (DNAs) and bottom (XRCC4-XLF-XRCC4) lines of the trapezoid are at an angle of ~75°. f. Superposition of one XRCC4 subunit (blue ovals) of the active and supportive half of the gap-filling complex. Structural changes are marked by red arrows, whose sizes are related to degrees of changes. The flexible joint in L4X4 is boxed in pink. g. Superposition of the XLF head domain (blue circle) between the two halves of the ω framework reveals subtle structural changes in dimeric interface of XRCC4 and XLF as well as between XRCC4 and XLF (boxed in pink).
Extended Data Fig. 4 Data processing and structure refinement of the gap-filling complex.
a. A representative cryoEM micrograph of 11,180 collected. b. Images of 2D classification. c. Workflow of image processing. d. Comparison of Ligation-like complex with Ligation complex from Extended Data Fig. 2. e. FSC analysis of the quality and map resolution. The map-model FSC (M) and the half-map FSC (H) are indicated. f. The composite map is colored according to the resolution scale bar on the side. g. Angular distribution of particles used in the final map calculation. h-l. The cryoEM map of specific regions.
Extended Data Fig. 5 Comparison of XLF-XRCC4 interface in crystal structures and NHEJ complexes.
a. Superposition of three crystal structures of XLF-XRCC4 complexes (PDB: 3SR2, 3RWR and 3Q4F) colored teal, pink and gray) with an XLR head domain in the ligation complex (olive). The different positions of the interacting XRCC4 head domain among crystal structures are obvious, and 3Q4F (gray) is most similar to the XRCC4 in the ligation complex (blue). b. Superposition of the four XLF-XRCC4 interfaces among ligation and gap-filling complexes. The supportive and active side of the two complexes are nearly the same. The changes between the two sides of NHEJ complexes are smaller than the differences among the crystal structures of XLF-XRCC4.
Extended Data Fig. 6 Binding partner and structure modulation of Ku70 and Ku80.
a. Binding of the N-terminal peptide of Ku70 to DBD of LIG4. Binding of FAM-labeled Ku70 and Artemis peptides to DBD domain of LIG4 (amino acids 1–240) were measured by fluorescence polarization changes. Kds were determined based on triplicate measurements. Error bars represent standard deviations. b. Binding of X-KBM (yellow) to Ku80 (cyan) displaces a self-peptide of Ku80 (magenta), which links vWA with the rest of Ku80 (pink), and causes the relative shift of vWA. c. The shifts of vWA domain of Ku80 and associated DNA in the presence (chocolate and yellow) are coordinated relative to their positions in the absence of X-KBM (gray), which is revealed after superposition of Ku70. The DNA is bent 20° when the vWA domain of Ku80 is opened by X-KBM, while the Ku80-DNA interface is maintained the same. The DNA axes are displayed as yellow (with X-KBM) and gray balls (without X-KBM). d. PAXX dimerization domain is docked into the cryoEM map volume in the XLF-truncated gap-filling complex (see SI).
Extended Data Fig. 7 Different DNA conformations in complexes with KU.
a. The KU-DNA on the supportive side of the gap-filling complex. An overwound and tilted DNA structure is supported by the cryoEM map. b. Superposition of KU-DNAs in the gap-filling and ligation complex. The DNA sequences are identical. c. The KU-DNA in ligation complex (as shown in panel b). The B-form DNA agrees with the corresponding cryoEM map. d. The B-like DNA is observed in all previously reported KU-DNA structures. Structures of KU-DNA of resolutions better than 3.4 Å (cryoEM) or 2.5 Å (crystal) are superimposed with the KU-DNA in ligation complex (as in panel c).
Extended Data Fig. 8 DNA is bent at the protein interface of Ku70.
a. DNA-PK complexed with Artemis and hairpin DNA (PDB: 7SGL). A black arrow marks where DNA is bent. b-c. NHEJ complex (this study) in two views. The catalytic domain of LIG4 (green) encircles two DNA ends. DNA is bent near the interface between Ku70 and LIG4 (DBD). d-e. DNA is always bent at the interface between Ku70 and DNA-PK, either in the presence of P-KBM (PDB:8EZA) (d) or absence of PAXX (PDB: 7K0Y) (e).
Supplementary information
Supplementary Information
Supplementary Figs, Table 1, Methods and legends to videos.
Supplementary Video 1
Comparison of AMP-Lys and AMP-DNA complexes. With one XLF of each complex superimposed, NTD of LIG4 and the adjacent active side KU are slightly opened in the AMP-Lys compared with the AMP-DNA (the ligation) complex. The OBD domain of LIG4 is disordered in the AMP-Lys complex, whereas the ω framework remains nearly the same.
Supplementary Video 2
Transition from the gap-filling to end-joining structure. Both LIG4 and Pol μ are attached to the same active side of the ω framework. After superimposing Ku70 on the supportive side motions of the active side allow the two broken DNA ends to translocate and adjust from bent 80° in the active site of Pol μ to 35° bent for LIG4 to join them. DBD of LIG4 bridges the template and primer strands on two DNA pieces for Pol μ to act on.
Supplementary Video 3
Structural differences between two halves of the gap-filling complex. Superposition of one XRCC4 head domain (orange) of the active and supportive half reveals large movements of BRCT1 and the linker between two BRCT domains of LIG4, which leads to the change of KU and DNA position, and small shifts of dimeric interfaces of XRCC4 and XLF as well as the interface between them. Similar structural changes are observed between gap filling and end joining complexes.
Supplementary Video 4
The DNAs bound to KU in the ligation and gap-filling complexes show two different conformations. Toggling of the B-form DNA in the ligation complex, which is the same as observed in all previously reported KU-DNA complexes, to the DNA in the gap-filling complex (both of supportive side), reveals the overwinding of two strands and a shift of one base pair in the gap-filling complex. An orthogonal view reveals expansion of the DNA diameter and the DNA binding channel in KU. The change of DNA seems to be correlated with the binding of BRCT domain of Pol μ to KU.
Supplementary Video 5
The opening of the vWA domains of Ku70 and Ku80. Binding of X-KBM to Ku80 leads to the opening of Ku80. Binding of P-KBM to Ku70 or interaction of Ku70 with DNA-PKcs, Artemis or LIG4 in the functional complexes of NHEJ results in the opening of Ku70.
Rights and permissions
About this article
Cite this article
Liu, L., Li, J., Cisneros-Aguirre, M. et al. Dynamic assemblies and coordinated reactions of non-homologous end joining. Nature 643, 847–854 (2025). https://doi.org/10.1038/s41586-025-09078-9
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41586-025-09078-9