Abstract
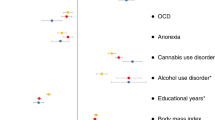
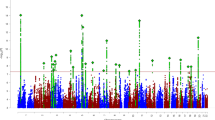
We performed a genome-wide association meta-analysis (GWAMA) of 290,134 attention-deficit/hyperactivity disorder (ADHD) symptom measures of 70,953 unique individuals from multiple raters, ages and instruments (ADHDSYMP). Next, we meta-analyzed the results with a study of ADHD diagnosis (ADHDOVERALL). ADHDSYMP returned no genome-wide significant variants. We show that the combined ADHDOVERALL GWAMA identified 39 independent loci, of which 17 were new. Using a recently developed gene-mapping method, Fine-mapped Locus Assessment Model of Effector genes, we identified 22 potential ADHD effector genes implicating several new biological processes and pathways. Moderate negative genetic correlations (rg < −0.40) were observed with multiple cognitive traits. In three cohorts, polygenic scores (PGSs) based on ADHDOVERALL outperformed PGSs based on ADHD symptoms and diagnosis alone. Our findings support the notion that clinical ADHD is at the extreme end of a continuous liability that is indexed by ADHD symptoms. We show that including ADHD symptom counts helps to identify new genes implicated in ADHD.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Summary statistics for ADHDSYMP and ADHDOVERALL are available for download through GWAS Catalog (ebi.ac.uk/gwas/studies/GCST90568440 and ebi.ac.uk/gwas/studies/GCST90568441). ADHDDIAG summary statistics are available for download at the Psychiatric Genomics Consortium (PGC) website (https://www.med.unc.edu/pgc/download-results/). Raw data are available upon request through the individual participating cohorts. Individual cohort GWAS summary statistics are available upon request through the corresponding author. Datasets used for gene mapping and hypergeometric gene-set tests in FUMA are listed in Supplementary Methods.
Code availability
The complete analysis plan is available for download at https://www.action-euproject.eu/sites/default/files/Action%20AGG%20AP%20SOP.pdf. The N-weighted GWAMA code is available via GitHub at https://github.com/baselmans/multivariate_GWAMA and via Zenodo at https://doi.org/10.5281/zenodo.15862079 (ref. 44. For a list of software and versions used, see Supplementary Methods.
Change history
29 September 2025
A Correction to this paper has been published: https://doi.org/10.1038/s41588-025-02383-z
References
Faraone, S. V., Biederman, J. & Mick, E. The age-dependent decline of attention deficit hyperactivity disorder: a meta-analysis of follow-up studies. Psychol. Med. 36, 159–165 (2006).
Kan, K.-J. et al. Genetic and environmental stability in attention problems across the lifespan: evidence from the Netherlands twin register. J. Am. Acad. Child Adolesc. Psychiatry 52, 12–25 (2013).
American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition (American Psychiatric Association Publishing, 2013).
Faraone, S. V. et al. Attention-deficit/hyperactivity disorder. Nat. Rev. Dis. Primers 1, 15020 (2015).
Caci, H. et al. Daily life impairments associated with self-reported childhood/adolescent attention-deficit/hyperactivity disorder and experiences of diagnosis and treatment: results from the European Lifetime Impairment Survey. Eur. Psychiatry 29, 316–323 (2014).
Caci, H. et al. Daily life impairments associated with childhood/adolescent attention-deficit/hyperactivity disorder as recalled by adults: results from the European Lifetime Impairment Survey. CNS Spectr. 20, 112–121 (2015).
Faraone, S. V. et al. Attention-deficit/hyperactivity disorder. Nat. Rev. Dis. Primers 10, 11 (2024).
Faraone, S. V. & Larsson, H. Genetics of attention deficit hyperactivity disorder. Mol. Psychiatry 24, 562–575 (2019).
Kan, K.-J., van Beijsterveldt, C. E. M., Bartels, M. & Boomsma, D. I. Assessing genetic influences on behavior: informant and context dependency as illustrated by the analysis of attention problems. Behav. Genet. 44, 326–336 (2014).
Merwood, A. et al. Different heritabilities but shared etiological influences for parent, teacher and self-ratings of ADHD symptoms: an adolescent twin study. Psychol. Med. 43, 1973–1984 (2013).
Demontis, D. et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat. Genet. 51, 63–75 (2019).
Middeldorp, C. M. et al. A genome-wide association meta-analysis of attention-deficit/hyperactivity disorder symptoms in population-based pediatric cohorts. J. Am. Acad. Child Adolesc. Psychiatry 55, 896–905 (2016).
Demontis, D. et al. Genome-wide analyses of ADHD identify 27 risk loci, refine the genetic architecture and implicate several cognitive domains. Nat. Genet. 55, 198–208 (2023).
Wang, X. et al. Polygenic risk prediction: why and when out-of-sample prediction R2 can exceed SNP-based heritability. Am. J. Hum. Genet. 110, 1207–1215 (2023).
Larsson, H., Anckarsater, H., Råstam, M., Chang, Z. & Lichtenstein, P. Childhood attention‐deficit hyperactivity disorder as an extreme of a continuous trait: a quantitative genetic study of 8,500 twin pairs. J. Child Psychol. Psychiatry 53, 73–80 (2012).
Levy, F., Hay, D. A., McStephen, M., Wood, C. & Waldman, I. Attention-deficit hyperactivity disorder: a category or a continuum? Genetic analysis of a large-scale twin study. J. Am. Acad. Child Adolesc. Psychiatry 36, 737–744 (1997).
Derks, E. M. et al. Genetic and environmental influences on the relation between attention problems and attention deficit hyperactivity disorder. Behav. Genet. 38, 11–23 (2008).
Rönnegård, L. et al. Increasing the power of genome wide association studies in natural populations using repeated measures – evaluation and implementation. Methods Ecol. Evol. 7, 792–799 (2016).
Ip, H. F. et al. Genetic association study of childhood aggression across raters, instruments, and age. Transl. Psychiatry 11, 413 (2021).
Watanabe, K., Taskesen, E., van Bochoven, A. & Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 8, 1826 (2017).
Hormozdiari, F., Kostem, E., Kang, E. Y., Pasaniuc, B. & Eskin, E. Identifying causal variants at loci with multiple signals of association. Genetics 198, 497–508 (2014).
Benner, C. et al. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics 32, 1493–1501 (2016).
Kichaev, G. et al. Integrating functional data to prioritize causal variants in statistical fine-mapping studies. PLoS Genet. 10, e1004722 (2014).
De Leeuw, C. A., Mooij, J. M., Heskes, T. & Posthuma, D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 11, e1004219 (2015).
Schipper, M. et al. Prioritizing effector genes at trait-associated loci using multimodal evidence. Nat. Genet. 57, 323–333 (2025).
Marees, A. T. et al. Potential influence of socioeconomic status on genetic correlations between alcohol consumption measures and mental health. Psychol. Med. 50, 484–498 (2020).
Groen-Blokhuis, M. M., Middeldorp, C. M., van Beijsterveldt, C. E. M. & Boomsma, D. I. Evidence for a causal association of low birth weight and attention problems. J. Am. Acad. Child Adolesc. Psychiatry 50, 1247–1254 (2011).
Bartels, M. et al. Childhood aggression and the co-occurrence of behavioural and emotional problems: results across ages 3–16 years from multiple raters in six cohorts in the EU-ACTION project. Eur. Child Adolesc. Psychiatry 27, 1105–1121 (2018).
Carey, G. Inference about genetic correlations. Behav. Genet. 18, 329–338 (1988).
Carlson, C. S. et al. Generalization and dilution of association results from European GWAS in populations of non-European ancestry: the PAGE study. PLoS Biol. 11, e1001661 (2013).
Boomsma, D. I. Aggression in children: unravelling the interplay of genes and environment through (epi)genetics and metabolomics. J. Pediatr. Neonatal Individ. Med. 4, e040251 (2015).
Middeldorp, C. M., Felix, J. F., Mahajan, A. & McCarthy, M. I. The Early Growth Genetics (EGG) and Early Genetics and Lifecourse Epidemiology (EAGLE) consortia: design, results and future prospects. Eur. J. Epidemiol. 34, 279–300 (2019).
Achenbach, T. M., Ivanova, M. Y. & Rescorla, L. A. Empirically based assessment and taxonomy of psychopathology for ages 1½–90+ years: developmental, multi-informant, and multicultural findings. Compr. Psychiatry 79, 4–18 (2017).
Goodman, R. Psychometric properties of the strengths and difficulties questionnaire. J. Am. Acad. Child Adolesc. Psychiatry 40, 1337–1345 (2001).
Tucker, G. et al. Two-variance-component model improves genetic prediction in family datasets. Am. J. Hum. Genet. 97, 677–690 (2015).
Minică, C. C., Dolan, C. V., Kampert, M. M. D., Boomsma, D. I. & Vink, J. M. Sandwich corrected standard errors in family-based genome-wide association studies. Eur. J. Hum. Genet. 23, 388–394 (2015).
Liu, Q. et al. Systematic assessment of imputation performance using the 1000 Genomes reference panels. Brief. Bioinform. 16, 549–562 (2015).
Bulik-Sullivan, B. K. et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Baselmans, B. M. L. et al. Multivariate genome-wide analyses of the well-being spectrum. Nat. Genet. 51, 445–451 (2019).
Grotzinger, A. D. et al. Genomic structural equation modelling provides insights into the multivariate genetic architecture of complex traits. Nat. Hum. Behav. 3, 513–525 (2019).
Weeks, E. M. et al. Leveraging polygenic enrichments of gene features to predict genes underlying complex traits and diseases. Nat. Genet. 55, 1267–1276 (2023).
Koopmans, F. et al. SynGO: an evidence-based, expert-curated knowledge base for the synapse. Neuron 103, 217–234 (2019).
Bulik-Sullivan, B. K. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 47, 1236–1241 (2015).
van der Laan, C. N_weighted_GWAMA. Zenodo https://doi.org/10.5281/zenodo.15862079 (2025).
Acknowledgements
We would like to thank all participants, their parents and teachers for making this study possible. This project was supported by the ACTION project. ACTION received funding from the European Union Seventh Framework Program (FP7/2007-2013) under grant agreement 602768. Cohort-specific acknowledgments and funding information are included in the Supplementary Note.
Author information
Authors and Affiliations
Contributions
C.M.v.d.L. conducted the central analyses and wrote the manuscript. M. Schipper performed the FLAMES analyses. H.F.I., B.St.P., T.Z., R. Pool, E.M.L.K., I.B., M.S.A., J.C.-D., S.A., I.M.N., K.B., T.P., H.Z., S.G., F.A., C.A.W., G.S., V.K., D.E.A., R. Border, R.E.P., J.A.P., E.T., N.V.-T., T.K., E.V., A.K.H., S. Llop, M.-J.L.-E., C.J., D.M.D., T.S.A., A.A., K.R., Q.C., Y.L., J.M., R. Bosch, N.L., A.N., J.E., K.L.G., J.J.M., X.T., S.M., J.G.S., A.A.S., L.M.E., K.S., L.S., R.V., C.J., Q.L., J.P., J. Horwood and W.E.C. performed cohort-specific analyses. J.-J.H., R.C., S.J., M.S.A., J.C.-D., S.A., R. Bosch, N.L., S.A.B., C.C., J. Haavik, A.K.H., A.S., S. Llop, M.-J.L.-E., L.A., M.M., N.V.-T., E.A.E., K.K., M. Stallings and M.R. coordinated genotyping. B.St.P., S.A., J.S., H.L., S. Lundström, R.J.R., A.G.U., F.A.H., H.S., Ø.H., A.R., A.K.H., S. Llop, M.-J.L.-E., L.A., M.K., M.M., J.R.H., G.P.S.K., P.R.N., A. Mamun, J.M.N., S.B., C.H., C.A.R., M. Stallings, S.W., T.L.W., L.E., J.L.S., A. Miller, A.H., K.B., J.S., M. Standl, J. Heinrich, J.B., J. Horwood, R. Pool, H.H.M., W.E.C., C.M.M, N.W., M.-R.J., W.I., A.C., T.E.M., A.J.O.W., C.E.P., K.L.K., D.M.D., M.S.A., J.C.-D., S.A., R. Bosch, N.L., S.A.B., J.A.R.-Q., R.C., A.M.W., T.K., E.V., T.R.-K., N.G.M., S.E.M., T.V., J.K., H.T., C.A.H., A.J.O., M.C., P.L., R. Pool, M.B., M.G.N. and D.I.B. collected samples and conducted phenotyping. B.St.P., J.S., H.L., S. Lundström, S.A.B., R.J.R., A.G.U., J.R.H., G.P.S.K., P.R.N., J.M.N., S.B., C.H., J. Hewitt, M. Stallings, S.W., L.E., J.L.S., H.H.M., W.E.C., C.M.M., N.W., M.-R.J., W.I., A.C., T.E.M., A.J.O.W., C.E.P., KL.K., D.M.D., J.A.R.-Q., H.S., A.S., S. Llop, M.-J.L.-E., L.A., M.K., G.M.W., M.M., G.M.W., T.R.-K., N.G.M., S.E.M., T.V., J.K., H.T., G.D.S., C.A.T., A.J.O., M.C., M.R., P.L., R. Plomin, M.B., M.G.N. and D.I.B. led the study design and principal investigator oversight. All above mentioned authors and E.M.D. contributed to critical revisions of the manuscript and approved the final version for submission.
Corresponding author
Ethics declarations
Competing interests
J.A.R.-Q. was on the speakers’ bureau and/or acted as a consultant for Biogen, Idorsia, Casen-Recordati, Janssen-Cilag, Novartis, Takeda, Bial, Sincrolab, Neuraxpharm, BMS, Medice, Rubió, Uriach, Technofarma and Raffo in the last 3 years. He also received travel awards (air tickets and hotel) for taking part in psychiatric meetings from Idorsia, Janssen-Cilag, Rubió, Takeda, Bial and Medice. The Department of Psychiatry, chaired by him, received unrestricted educational and research support from the following companies in the last 3 years: Exeltis, Idorsia, Janssen-Cilag, Neuraxpharm, Oryzon, Roche, Probitas and Rubió. M.C. has received fees to give talks for TAKEDA and Laboratorios RUBIO. The other authors declare no competing interests.
Peer review
Peer review information
Nature Genetics thanks Joshua Gray, Andrew McQuillin and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–14, Notes and Methods.
Supplementary Data 1
Forest plots of cohort-specific GWAS regression estimates for all independent significant loci. Dots indicate regression estimates; bars indicate 95% confidence intervals. The red line represents the regression estimate from the overall meta-analysis. Sample sizes for the individual cohorts are shown in Supplementary Table 2.
Supplementary Data 2
Forest plots of stratified GWAS regression estimates for all independent significant loci. Dots indicate regression estimates; bars indicate 95% confidence intervals. The red line represents the regression estimate from the overall meta-analysis.
Supplementary Data 3
Locus plots of independent significant loci. More information for these loci is available in Supplementary Table 13.
Supplementary Tables 1–20
Supplementary Tables 1–20.
Rights and permissions
About this article
Cite this article
van der Laan, C.M., Ip, H.F., Schipper, M. et al. Genome-wide association meta-analysis of childhood ADHD symptoms and diagnosis identifies new loci and potential effector genes. Nat Genet 57, 2427–2435 (2025). https://doi.org/10.1038/s41588-025-02295-y
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41588-025-02295-y