Abstract
The transcription factors nuclear factor of activated T cells (NFAT) and activator protein 1 (AP-1; Fos–Jun) cooperate to promote the effector functions of T cells, but NFAT in the absence of AP-1 imposes a negative feedback program of T cell hyporesponsiveness (exhaustion). Here, we show that basic leucine zipper ATF-like transcription factor (BATF) and interferon regulatory factor 4 (IRF4) cooperate to counter T cell exhaustion in mouse tumor models. Overexpression of BATF in CD8+ T cells expressing a chimeric antigen receptor (CAR) promoted the survival and expansion of tumor-infiltrating CAR T cells, increased the production of effector cytokines, decreased the expression of inhibitory receptors and the exhaustion-associated transcription factor TOX and supported the generation of long-lived memory T cells that controlled tumor recurrence. These responses were dependent on BATF–IRF interaction, since cells expressing a BATF variant unable to interact with IRF4 did not survive in tumors and did not effectively delay tumor growth. BATF may improve the antitumor responses of CAR T cells by skewing their phenotypes and transcriptional profiles away from exhaustion and towards increased effector function.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The RNA-seq, ATAC-seq and ChIP-seq data have been deposited at the Gene Expression Omnibus (GSE154747). Source data are provided with this paper. Other data that support the findings of this study are available from the corresponding authors upon request.
Code availability
The base code used to preprocess the ATAC-seq, RNA-seq and ChIP-seq data and the code used for the analysis of differential gene expression have been deposited in GitHub at https://github.com/Edahi/NGSDataAnalysis.
References
Blank, C. U. et al. Defining ‘T cell exhaustion’. Nat. Rev. Immunol. 19, 665–674 (2019).
McLane, L. M., Abdel-Hakeem, M. S. & Wherry, E. J. CD8 T cell exhaustion during chronic viral infection and cancer. Annu. Rev. Immunol. 37, 457–495 (2015).
Pereira, R. M., Hogan, P. G., Rao, A. & Martinez, G. J. Transcriptional and epigenetic regulation of T cell hyporesponsiveness. J. Leukoc. Biol. 102, 601–615 (2017).
Liu, X. et al. Genome-wide analysis identifies NR4A1 as a key mediator of T cell dysfunction. Nature 567, 525–529 (2019).
Chen, J. et al. NR4A transcription factors limit CAR T cell function in solid tumours. Nature 567, 530–534 (2019).
Seo, H. et al. TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8+ T cell exhaustion. Proc. Natl Acad. Sci. USA 116, 12410–12415 (2019).
Scott, A. C. et al. TOX is a critical regulator of tumour-specific T cell differentiation. Nature 571, 270–274 (2019).
Wang, X. et al. TOX promotes the exhaustion of antitumor CD8+ T cells by preventing PD1 degradation in hepatocellular carcinoma. J. Hepatol. 71, 731–741 (2019).
Alfei, F. et al. TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection. Nature 571, 265–269 (2019).
Khan, O. et al. TOX transcriptionally and epigenetically programs CD8+ T cell exhaustion. Nature 571, 211–218 (2019).
Macián, F. et al. Transcriptional mechanisms underlying lymphocyte tolerance. Cell 109, 719–731 (2002).
Martinez, G. J. et al. The transcription factor NFAT promotes exhaustion of activated CD8+ T cells. Immunity 42, 265–278 (2015).
Hogan, P. G., Chen, L., Nardone, J. & Rao, A. Transcriptional regulation by calcium, calcineurin, and NFAT. Gene Dev. 17, 2205–2232 (2003).
Karin, M., Liu, Z. & Zandi, E. AP-1 function and regulation. Curr. Opin. Cell Biol. 9, 240–246 (1997).
Oh-hora, M. et al. Dual functions for the endoplasmic reticulum calcium sensors STIM1 and STIM2 in T cell activation and tolerance. Nat. Immunol. 9, 432–443 (2008).
Scott-Browne, J. P. et al. Dynamic changes in chromatin accessibility occur in CD8+ T cells responding to viral infection. Immunity 45, 1327–1340 (2016).
Hogan, P. G. Calcium–NFAT transcriptional signalling in T cell activation and T cell exhaustion. Cell Calcium 63, 66–69 (2017).
Lynn, R. C. et al. c-Jun overexpression in CAR T cells induces exhaustion resistance. Nature 576, 293–300 (2019).
Kurachi, M. et al. The transcription factor BATF operates as an essential differentiation checkpoint in early effector CD8+ T cells. Nat. Immunol. 15, 373–383 (2014).
Murphy, T. L., Tussiwand, R. & Murphy, K. M. Specificity through cooperation: BATF–IRF interactions control immune-regulatory networks. Nat. Rev. Immunol. 13, 499–509 (2013).
Li, P. et al. BATF–JUN is critical for IRF4-mediated transcription in T cells. Nature 490, 543–546 (2012).
Ciofani, M. et al. A validated regulatory network for TH17 cell specification. Cell 151, 289–303 (2012).
Glasmacher, E. et al. A genomic regulatory element that directs assembly and function of immune-specific AP-1–IRF complexes. Science 338, 975–980 (2012).
Tussiwand, R. et al. Compensatory dendritic cell development mediated by BATF–IRF interactions. Nature 490, 502–507 (2012).
Man, K. et al. Transcription factor IRF4 promotes CD8+ T cell exhaustion and limits the development of memory-like T cells during chronic infection. Immunity 47, 1129–1141.e5 (2017).
Quigley, M. et al. Transcriptional analysis of HIV-specific CD8+ T cells shows that PD-1 inhibits T cell function by upregulating BATF. Nat. Med. 16, 1147–1151 (2010).
Im, S. J. et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537, 417–421 (2016).
Kurtulus, S. et al. Checkpoint blockade immunotherapy induces dynamic changes in PD-1−CD8+ tumor-infiltrating T cells. Immunity 50, 181–194.e6 (2019).
Siddiqui, I. et al. Intratumoral Tcf1+PD-1+CD8+ T cells with stem-like properties promote tumor control in response to vaccination and checkpoint blockade immunotherapy. Immunity 50, 195–211.e10 (2019).
Miller, B. C. et al. Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nat. Immunol. 20, 326–336 (2019).
Utzschneider, D. T. et al. T cell factor 1-expressing memory-like CD8+ T cells sustain the immune response to chronic viral infections. Immunity 45, 415–427 (2016).
Brogdon, J., June, C. H., Loew, A., Maus, M. & Scholler, J. Treatment of cancer using humanized anti-CD19 chimeric antigen receptor. WO patent WO2014153270A1 (2014).
Iwata, A. et al. Quality of TCR signaling determined by differential affinities of enhancers for the composite BATF–IRF4 transcription factor complex. Nat. Immunol. 18, 563–572 (2017).
Xiao, Z., Casey, K. A., Jameson, S. C., Curtsinger, J. M. & Mescher, M. F. Programming for CD8 T cell memory development requires IL-12 or type I IFN. J. Immunol. 182, 2786–2794 (2009).
Huber, J. P. & Farrar, J. D. Regulation of effector and memory T‐cell functions by type I interferon. Immunology 132, 466–474 (2011).
Mognol, G. P. et al. Exhaustion-associated regulatory regions in CD8+ tumor-infiltrating T cells. Proc. Natl Acad. Sci. USA 114, E2776–E2785 (2017).
Jadhav, R. R. et al. Epigenetic signature of PD-1+ TCF1+ CD8 T cells that act as resource cells during chronic viral infection and respond to PD-1 blockade. Proc. Natl Acad. Sci. USA 116, 14113–14118 (2019).
Muthusamy, N., Barton, K. & Leiden, J. M. Defective activation and survival of T cells lacking the Ets-1 transcription factor. Nature 377, 639–642 (1995).
Grenningloh, R. et al. Ets-1 maintains IL-7 receptor expression in peripheral T cells. J. Immunol. 186, 969–976 (2011).
Wherry, E. J. T cell exhaustion. Nat. Immunol. 12, 492–499 (2011).
Wherry, E. J. & Kurachi, M. Molecular and cellular insights into T cell exhaustion. Nat. Rev. Immunol. 15, 486–499 (2015).
Li, J., He, Y., Hao, J., Ni, L. & Dong, C. High levels of Eomes promote exhaustion of anti-tumor CD8+ T cells. Front. Immunol. 9, 2981 (2018).
Li, P. & Leonard, W. J. Chromatin accessibility and interactions in the transcriptional regulation of T cells. Front. Immunol. 9, 2738 (2018).
Pham, D. et al. Batf pioneers the reorganization of chromatin in developing effector T cells via Ets1-dependent recruitment of Ctcf. Cell Rep. 29, 1203–1220.e7 (2019).
Chang, Y. K., Zuo, Z. & Stormo, G. D. Quantitative profiling of BATF family proteins/JUNB/IRF hetero-trimers using Spec-seq. BMC Mol. Biol. 19, 5 (2018).
Man, K. et al. The transcription factor IRF4 is essential for TCR affinity-mediated metabolic programming and clonal expansion of T cells. Nat. Immunol. 14, 1155–1165 (2013).
Wei, J. et al. Targeting REGNASE-1 programs long-lived effector T cells for cancer therapy. Nature 576, 471–476 (2019).
Xin, G. et al. A critical role of IL-21-induced BATF in sustaining CD8-T-cell-mediated chronic viral control. Cell Rep. 13, 1118–1124 (2015).
Nicholson, I. C. et al. Construction and characterisation of a functional CD19 specific single chain Fv fragment for immunotherapy of B lineage leukaemia and lymphoma. Mol. Immunol. 34, 1157–1165 (1997).
Roybal, K. T. et al. Precision tumor recognition by T cells with combinatorial antigen-sensing circuits. Cell 164, 770–779 (2016).
Hooijberg, E., Bakker, A. Q., Ruizendaal, J. J. & Spits, H. NFAT-controlled expression of GFP permits visualization and isolation of antigen-stimulated primary human T cells. Blood 96, 459–466 (2000).
Corces, M. R. et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 (2017).
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Krueger, F. Trim galore: a wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files. (2015); https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. ENBnet J. 17, 10–12 (2011).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Broad Institute. Picard Toolkit (2018); http://broadinstitute.github.io/picard/
Amemiya, H. M., Kundaje, A. & Boyle, A. P. The ENCODE blacklist: identification of problematic regions of the genome. Sci. Rep. 9, 9354 (2019).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Zhang, Y. et al. Model-based analysis of ChIP-seq (MACS). Genome Biol. 9, R137 (2008).
Law, C. W., Chen, Y., Shi, W. & Smyth, G. K. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29 (2014).
R Development Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2018).
Kluyver, T., Angerer, P. & Schulz, J. IRdisplay: ‘Jupyter’ Display Machinery. R package v0.7.0 (2016); https://CRAN.R-project.org/package=IRdisplay
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Su, S. et al. Glimma: interactive graphics for gene expression analysis. Bioinformatics 33, btx094 (2017).
Bioconductor Core Team. Mus.musculus: Annotation package for the Mus.musculus object. R package v1.3.1 (2015).
Neuwirth, E. RColorBrewer: ColorBrewer palettes. R package v1.1-2 (2014); https://CRAN.R-project.org/package=RColorBrewer
Wickham, H. in ggplot2: Elegant Graphics for Data Analysis (Use R!) 147–168 (Springer, 2016).
Lawrence, M. et al. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 9, e1003118 (2013).
Kolde, R. pheatmap: Pretty heatmaps. R package v1.0.12 (2019); https://CRAN.R-project.org/package=pheatmap
MATLAB v7.10.0 (R2010a) (The MathWorks, 2010).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Warnes, G. R. et al. gplots: Various R programming tools for plotting data. R package v3.0.1.1 (2009); https://CRAN.R-project.org/package=gplots
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Chen, H. VennDiagram: Generate high-resolution Venn and Euler plots. R package v1.6.20 (2018); https://CRAN.R-project.org/package=VennDiagram
Chen, H. & Boutros, P. C. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics 12, 35 (2011).
Garnier, S. Colorblind-friendly color maps for R. R package v0.6.1. (2018); https://CRAN.R-project.org/package=viridis
Ramírez, F., Dündar, F., Diehl, S., Grüning, B. A. & Manke, T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–W191 (2014).
Acknowledgements
We thank C. Kim, D. Hinz and C. Dillingham of the LJI Flow Cytometry Core Facility for the CyTOF experiments; D. Hinz, C. Dillingham, S. Ellis, M. Haynes, S. Sehic and C. Kim for cell sorting; J. Day, C. Kim and K. Tanguay of the LJI Next Generation Sequencing Facility for sequencing; the LJI DLAC and the animal facility for excellent support; and R. Nowak for excellent laboratory management. This work was funded in part by NIH R01 grants AI109842 and AI040127 (to P.G.H. and A.R.); U01 grant DE028227 (to A.R.); NIH S10 instrumentation grants RR027366 and OD018499 (to the LJI); AACR-Genentech Immuno-oncology Research Fellowship 18-40-18-SEO and Donald J. Gogel Cancer Research Institute Irvington Fellowship (to H.S.); University of California Institute for Mexico and the United States (UC MEXUS) and El Consejo Nacional de Ciencia y Tecnología (UC MEXUS/CONACYT) predoctoral fellowship (to E.G.-A.); NIH T32 predoctoral training grant in the University of California, San Diego Cardiovascular Bioengineering Training Program (to W.Z.); Cancer Research Institute Irvington Fellowship (to C.Y.); an Independent Investigator Fund (LJI/Kyowa Kirin) and a Career Transition Award from the National Cancer Institute (K22CA241290 to C.-W.J. L.).
Author information
Authors and Affiliations
Contributions
H.S. designed and performed the experiments, analyzed the data and prepared the sequencing libraries. E.G.-A. performed the computational analyses of the genome-wide sequencing data. W.Z. cloned the plasmid vectors, performed the NFAT–AP-1 reporter screening assays and assisted with the in vivo and in vitro experiments. C.-W.J.L. provided the pMIG-BATF and IRF4 vectors. P.R. and C.Y. assisted with the in vitro experiments. A.R. and P.G.H. supervised the study. H.S., E.G.-A., A.R. and P.G.H. interpreted the data and wrote the manuscript. All authors contributed to writing and providing feedback.
Corresponding authors
Ethics declarations
Competing interests
A.R. and P.G.H. were recipients of a grant from Lyell Immunopharma for a separate research project related to cancer immunotherapy. The La Jolla Institute for Immunology has filed a patent application, PCT/62/976,706, covering the production and use of engineered immune cells expressing BATF and IRF4. H.S., A.R. and P.G.H. are listed as inventors. The other authors have no competing interests.
Additional information
Peer review information Nature Immunology thanks Rahul Roychoudhuri, Harinder Singh and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available. L. A. Dempsey was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Identification of bZIP transcription factors capable of increasing NFAT:AP-1 reporter activity.
a, MA plots of basic region-leucine zipper (bZIP) transcription factor gene expression in TOX-depleted (TOX DKO, left) or NR4A-depleted (Nr4a TKO, right) CAR TILs5,6— which mount increased anti-tumor responses— relative to control CAR TILs. Differentially expressed genes (adjusted p-value < 0.1, log2 fold-change ≥ 0.5 or ≤ −0.5) are highlighted; selected genes are labeled. b, Basis of the experiment to identify bZIP transcription factors that activate an NFAT:AP-1 reporter through a positive feedback loop, either directly by binding adjacent to NFAT on the NFAT:AP-1 composite site or indirectly by increasing the expression or activity of NFAT or AP-1. Mouse CD8+ T cells were transduced with retroviral expression vectors encoding a Thy1.1 reporter, separated by a P2A sequence from a co-expressed bZIP transcription factor, and under the control of six tandem NFAT:AP1 sites upstream of a minimal IL-2 promoter. Transcription factors for testing were chosen based on the data analysis in a. c, Gating strategy for the experiments. d, g, Histograms of Thy1.1 expression after CD8+ T cell transduction. No Thy1.1, transduced with empty retrovirus without Thy1.1 or bZIP transcription factor; No bZIP, transduced with retroviral vector encoding Thy1.1 but no bZIP transcription factor, a condition that assesses the background induction of Thy1.1 by endogenous NFAT and AP-1; Jun, Maff, Batf, and Batf3 (d) and JunD, Fosl2, and Nfil3 (g), transduced with vector encoding the indicated bZIP transcription factor. e, h, Percentage of Thy1.1+ cells in three replicate experiments. f, i, MFI of Thy1.1 expression in these experiments. j, k, Results for each sample, normalized to those of the No bZIP control from the same experiment. Each circle in e, f, h, I, j and k represents one mouse. Data are representative of (c, d, g) or obtained from (e, f, h-k) three biological replicate experiments. Data in e and h were analyzed by one-way ANOVA. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001.
Extended Data Fig. 2 Phenotypic analyses of pMIG- and BATF-transduced CAR TILs.
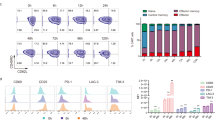
a, Retroviral transduction efficiencies for CAR and MSCV-IRES-eGFP retroviral expression plasmids, assessed as expression of Thy1.1 and GFP respectively. FMO, fluorescence-minus-one control. b, Histograms showing BATF (left), JUN (middle), and MAFF (right) expression after retroviral transduction of CD8+ T cells with the corresponding retroviral expression plasmids or pMIG empty-vector control, assessed by flow cytometry with antibodies to the endogenous proteins. c, Histograms showing CAR expression (assessed by staining for the Myc tag) in pMIG- (grey), BATF- (red), JUN- (sky blue) and MAFF- (orange) transduced CAR T cells. d-e, Replicate tumor growth experiments using B16F0-hCD19 (d) and MC38-hCD19 (e) tumor cells. 1×105 tumor cells were injected subcutaneously into the left flank of C57BL/6 mice at day 0 (D0) in 100 μl phosphate-buffered saline (PBS); 3×106 control pMIG-, JUN-, MAFF-, or BATF-transduced CAR T cells were adoptively transferred by retro-orbital injection at day 7. Tumor sizes were measured by caliper. f, Histograms showing expression of the indicated markers by each group of CAR TILs, assessed by flow cytometry. g, Left panels, Histograms showing expression of CD44, CD62L, CD127 and KLRG1. Middle panels, Overlaid contour plots of CD44 and CD62L (top) and CD127 and KLRG1 (bottom) in pMIG- (grey) and BATF- (red) transduced CAR TILs. Right panel, expression of the markers quantitated as MFI. h, Left panels, Histograms showing expression of TNF, IFNγ, granzyme B, and CD107a after resting in T cell media or after stimulation with PMA and ionomycin for 4 h. Right panel, expression of the markers quantitated as MFI. i, pMIG (n = 6) and BATF (n = 6). Quantitation of TCF1+ and TCF1– CAR TILs. Each circle in g, h, and i represents one mouse, and the bar graphs represent the mean ± standard error of mean (s.e.m.). Data in d-i were obtained from two independent biological experiments. Data in g, h, and i were analyzed by two-tailed unpaired Student’s t-test.
Extended Data Fig. 3 Mass cytometric analyses.
a, Gating strategy for mass cytometry of tumor-infiltrating lymphocytes (TILs). b, UMAP plots of data from TILs of tumour-bearing mice, showing expression of CD8α (top) as a marker for all CD8+ T cells (including endogenous T cells) and Thy1.1 (bottom) as a marker for adoptively transferred pMIG- or BATF-transduced CAR T cells. c, Mass cytometric analysis of CD8+ T cells from draining lymph nodes (left) and spleens (right) of mice in the tumor rechallenge experiments (Fig. 3). Endogenous CD8+ T cells and BATF-transduced CAR T cells are clearly distinguishable in the UMAP views. d, Expression of selected surface markers on lymphocytes obtained from three groups of age-matched C57BL/6 mice: inguinal lymph nodes of completely unmanipulated, non-tumor-bearing mice (left panels); draining (inguinal) lymph nodes of mice inoculated with tumors 14 days previously (middle panels), and draining (inguinal) lymph nodes of rechallenged mice. The UMAP plots show that each marker is expressed by all or by a large fraction of lymph node CAR T cells of the rechallenged mice. Each group of samples in c and d was pooled from 5 mice. The data are representative of two independent biological experiments. e, Replicate of the experiment in Fig. 3e. 1×105 B16F0-hCD19 tumor cells were injected subcutaneously into the right flank of age-matched C57BL/6 mice (n = 3) to yield the ‘tumor-naïve’ control group, or into the right flank of the surviving tumor-free mice (n = 3) from the experiment of Extended Data Fig. 2d. In this replicate B16-hCD19 rechallenge experiment, CAR T cells accounted for ~10% of CD8+ T cells in spleen and LN of the mice, and exhibited a memory phenotype (CD44high, CD62Lhigh, TCF1high) as in the first experiment.
Extended Data Fig. 4 BATF overexpression improves effector function in human CAR T cells.
a, Histograms showing expression of human BATF (hBATF, left panel) and human CAR (hCAR [stained by Goat Anti-Armenian Hamster IgG (H + L)], right panel) in the corresponding lentivirally transduced human CD8+ T cells. FMO, fluorescence-minus-one control; TIG, control cells transduced with the empty vector. b, Left, Histograms showing CellTrace Violet (CTV) dilution in lentivirally transduced human CD8+ CAR T cells. Right, Proliferation index calculated as (Day 0 CTV MFI) / (Day 4 CTV MFI). c, Histograms showing expression of the indicated cytokines after resting in X-Vivo media or after stimulation with PMA and ionomycin for 4 h. d-e, Human CAR T cells were labeled with CellTrace Violet and co-cultured with NALM6 cells for 5 h. d, Gating strategy for in vitro cytotoxicity assay of control (TIG) or hBATF-transduced human CAR T cells. e, Histograms showing ratio between target cells (NALM6) and effector cells (human CAR T cells). Percent cytotoxicity was calculated as (1- (R5/R0)) x 100, where R5 = (target cells as % of total at 5 h) / (effector cells as % of total at 5 h), R0 = (target cells as % of total at 0 h) / (effector cells as % of total at 0 h). Each dot in b-e represents an individual donor. Data were obtained from four biological experiments and analyzed by one-tailed unpaired Student’s t-test.
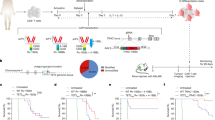
Extended Data Fig. 5 Tumor growth and TIL expansion/survival in mice receiving BATF-transduced, BATF-HKE-transduced, or BATF-deficient CD8+ T cells.
a, Tumor growth curves for the individual mice from Fig. 4b,c (PBS (n = 12), pMIG (n = 16), BATF (n = 25) and HKE (n = 12)). b-f, 1×105 B16F0-hCD19 tumor cells were subcutaneously injected into the left flank of C57BL/6 mice at day 0 (D0). 100 μl of PBS, without cells or containing 1.5×106 CAR T cells transduced with retroviral expression plasmids encoding either pMIG control, BATF, or BATF HKE-mutant, were adoptively transferred into C57BL/6 recipient mice by retro-orbital injection on day 12. TILs were isolated on days 13, 16, 19, and 22. c, Expression of CAR T cell marker Thy1.1 on CD8+ TILs on the indicated days. d-f, Frequencies and MFIs of the indicated PD-1- and TIM3-expressing populations from Fig. 4j,k. g-m, 1×105 B16F0-hCD19 tumor cells were injected subcutaneously into the left flank of C57BL/6 mice at day 0 (D0). 1.5×106 wild-type (WT, n = 4) or BATF-deficient (BATF KO, n = 4) CAR T cells were adoptively transferred at day 12. Tumor-infiltrating lymphocytes were isolated at day 20. h, Tumor growth curves for individual mice (dashed lines) and average (bold lines) of all tumor growth curves in a group. i-k, Contour plots of Thy1.1 expression in CD8+ TILs (i), percentages of Thy1.1+ CAR TILs (j) and numbers of Thy1.1+ CAR TILs normalized to tumor size (k) in tumor-bearing BATF WT or BATF KO mice. l, Contour plots of PD-1 and TIM3 expression (left) and percentage of PD-1hiTIM3+ cells (right) in WT or BATF KO CAR TILs. m, Contour plots of PD-1 and TOX expression (top left), TIM3 and TCF1 expression (bottom left), and percentages of the indicated populations (right) in WT or BATF KO CAR TILs. Data in a were obtained from three independent experiments. Each circle in d-f and j-m represents one mouse, and the bar graphs represent the mean ± standard error of mean (s.e.m.). Data in d-m are representative of two independent experiments. Data in j-m were analyzed by two-tailed unpaired Student’s t-test.
Extended Data Fig. 6 Tumor growth rates, survival curves, and phenotypic analysis of CAR TILs.
a-e, 2.5×105 B16F10-OVA tumor cells were injected subcutaneously into the left flank of C57BL/6 mice at day 0 (D0). 100 μl of PBS (n = 10), without cells or containing 1×106 OT-I T cells transduced with retroviral expression plasmids encoding pMIG control (n = 10), BATF (n = 10), IRF4 (n = 10), or BATF + IRF4 (n = 10), were adoptively transferred by retro-orbital injection at day 7. b, Averaged tumor growth curves for all mice in the indicated groups. c, Tumor sizes measured in individual mice at day 18. d, Mouse survival curves. e, Tumor growth curves in individual mice. f-m, 2.5×105 B16F10-OVA tumor cells were injected subcutaneously into the left flank of C57BL/6 mice at day 0. 1×106 pMIG control- (n = 4), BATF- (n = 5), IRF4- (n = 5), or BATF + IRF4-transduced (n = 5) OT-I cells were adoptively transferred at day 12. TILs were isolated at day 20. g, Gating strategy for flow cytometric analysis of OT-I TILs. h, Averaged tumor growth curves for all mice in the indicated groups. i, Left, Contour plots of CD8α and CD45.1 expression in OT-I TILs. Middle, Percentage of OT-I TILs in CD8+ TILs. Right, Number of OT-I TILs normalized to tumor size. j, Left, Contour plots of PD-1 and TIM3 expression in each group of OT-I TILs. Right, Percentages of the indicated PD-1- and TIM3-expressing cell populations. k, Left, Contour plots of PD-1 and TOX expression in the indicated OT-I TILs. Right, Percentage of PD-1+TOX+ cells in OT-I TILs. l, m, Left, Contour plots of expression of granzyme B (l) and the indicated cytokines (m) under resting conditions or after PMA/ionomycin stimulation for 4 h. Right, Percentages of OT-I TILs expressing granzyme B (l) or the indicated cytokines (m) under resting conditions or after PMA/ionomycin stimulation for 4 h. Data obtained from two biological experiments. n, Histograms showing JUN and BATF expression in the indicated groups of transduced OT-I T cells. o, Tumor growth curves for individual mice given pMIG control-, BATF-, JUN-, or BATF + JUN-transduced OT-1 cells (top) and averaged tumor growth curves for all mice in each group (bottom). Experimental scheme as in a. Each circle in i, j, k, l, and m represents one mouse, and the bar graphs represent the mean ± standard error of mean (s.e.m.). Data were obtained from two independent biological experiments. Data in h, j, l, and m were analyzed by two-way ANOVA test; data from i and k, by one-way ANOVA test. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001.
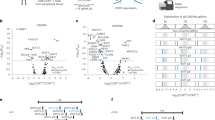
Extended Data Fig. 7 Differentially accessible regions in BATF- compared to pMIG-transduced CAR TILs.
a, Left, Venn diagrams showing the overlap of the 351 regions more accessible in BATF- versus pMIG-transduced TILs (Fig. 5c) with the exhaustion-related (top) or activation-related (bottom) regions from Mognol et al36. Right, Histograms illustrate the significance calculation by one-tailed Fisher’s exact test. b, Genomic annotation of the commonly and differentially accessible regions in CAR TILs. c, Enrichment for transcription factor binding motifs in regions differentially accessible in BATF CAR TILs. d, Heatmap of ATAC-seq signal (z-score) from BATF- and pMIG-transduced CD8+ T cells or CAR TILs, in the 640 regions more accessible in BATF-transduced compared to pMIG-transduced CD8+ T cells (left; see Fig. 5b) and in the 351 regions more accessible in BATF-transduced compared to pMIG CAR TILs (right; see Fig. 5c). Each column represents a biological replicate. Data obtained from two biological experiments.
Extended Data Fig. 8 BATF and IRF4 binding and gene expression changes in pMIG-, HKE-, and BATF-transduced cells.
a, Contour plot relating BATF ChIP-seq signals (log2(CPM)) in BATF-transduced CD8+ T cells to signals from the corresponding peaks in pMIG-transduced cells. b, Left, Distribution of BATF ChIP-seq reads in peaks in BATF-transduced cells (red) versus pMIG-transduced cells (black). Right, BATF ChIP-seq signal distribution in the ‘BATF and pMIG’ peaks common to the two conditions (blue) and in ‘BATF-new’ peaks observed only in BATF-transduced cells (red). The BATF-new peaks defined here are largely the same as the BATF-only peaks in Fig. 6a. c, Heatmaps of the IRF4 ChIP-seq signal in BATF-transduced, BATF-HKE-transduced, and pMIG-transduced cells, at IRF4 peak locations called in BATF-transduced cells. Curves at the top show the average signal taken over all peaks in the respective heatmap. d, Heatmaps of the BATF ChIP-seq signal in BATF-transduced, BATF-HKE-transduced and pMIG-transduced cells, at IRF4 peak locations called in pMIG-transduced cells. Curves at the top show the average signal taken over all peaks in the respective heatmap. e, Genome browser view of Ctla4 gene locus showing BATF and IRF4 ChIP-seq signals from pMIG-, BATF-, and HKE-transduced CD8+ T cells. f, Heatmap of normalized RNA-seq reads (as z-scores) under the indicated conditions, for the top 100 genes differentially expressed after αCD3/αCD28 stimulation of pMIG-transduced cells. Data obtained from two or three biological experiments.
Extended Data Fig. 9 Redistribution of IRF4 binding in BATF-overexpressing cells.
a, The redistribution of the normalized IRF4 ChIP-seq signal in BATF-overexpressing cells is most evident when the median deviation of the y-coordinate (αIRF4 ChIP-seq signal in BATF-overexpressing cells) from the diagonal in Fig. 7c, left, is plotted as a function of position on the x-axis (αIRF4 ChIP-seq signal in control cells). The median for all peaks in each slice of 0.1 log2(CPM) units on the x-axis was determined. The inset replicates Fig. 7c, left, with a red rectangle indicating the slice between log2(CPM) = 0.5 and log2(CPM) = 0.6 on the x-axis. b, Spurious ‘αIRF4’ ChIP-seq regions (as defined in Methods), incorporated into the graph of panel a. c, IRF4 does not redistribute in BATF-HKE-overexpressing cells. The median deviation of the y-coordinate (αIRF4 ChIP-seq signal in HKE-overexpressing cells) from the diagonal in Fig. 7c, right, is plotted as a function of position on the x-axis (αIRF4 ChIP-seq signal in control cells), as in a. d, IRF4 (left) and IRF8 (right) expression detected by flow cytometry (MFI) in pMIG- and HKE-transduced CD8+ T cells that had been expanded in vitro, at the indicated times after restimulation with αCD3/αCD28. The black symbol on the y-axis shows expression in naïve CD8+ T cells. Values for pMIG-transduced and naïve CD8+ T cells were obtained in the same experiments and are replotted from Fig. 6f. Overexpression of BATF-HKE did not attenuate IRF4 or IRF8 induction. e, Similar to Fig. 8a. Dot plot highlighting regions of the IRF4 ChIP-seq data from Fig. 7c where IRF4 binding increases (log2FC ≥ 0.75, red dots), does not change substantially (light grey dots), or decreases (log2FC ≤ -0.75, blue dots) in BATF-overexpressing relative to pMIG-transduced cells. Peaks with very low (log2(signal) < -1.25) or high (log2(signal) > 2.5) IRF4 binding in pMIG cells were judged unlikely to be informative and were omitted from the analysis. f, ATAC-seq signal (CPM) in peaks in each of the three categories defined in e. g, Similar to f, but downsampling the subsets to match the 2312 regions with increased IRF4 binding in BATF-transfected cells, for clearer visualization. Data were obtained from (a-c, e-g), or are representative of (d), two independent biological experiments.
Supplementary information
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Rights and permissions
About this article
Cite this article
Seo, H., González-Avalos, E., Zhang, W. et al. BATF and IRF4 cooperate to counter exhaustion in tumor-infiltrating CAR T cells. Nat Immunol 22, 983–995 (2021). https://doi.org/10.1038/s41590-021-00964-8
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41590-021-00964-8
This article is cited by
-
Regulators of CD8+ T cell exhaustion
Nature Reviews Immunology (2026)
-
Single-cell RNA sequencing unveils CCDC86-driven immune modulation and antigen-presenting cell dynamics in head and neck squamous cell carcinoma
Discover Oncology (2026)
-
Strategies and mechanisms for the enhancement of chimeric antigen receptor T-cell functions
Science China Life Sciences (2026)
-
High expression of IRF4 mediated by m6A modification promotes immunosuppression in the late phase of sepsis
Anesthesiology and Perioperative Science (2026)
-
Atlas-guided discovery of transcription factors for T cell programming
Nature (2026)