Abstract
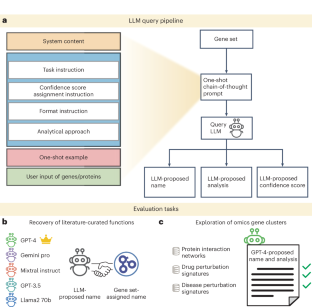
Gene set enrichment is a mainstay of functional genomics, but it relies on gene function databases that are incomplete. Here we evaluate five large language models (LLMs) for their ability to discover the common functions represented by a gene set, supported by molecular rationale and a self-confidence assessment. For curated gene sets from Gene Ontology, GPT-4 suggests functions similar to the curated name in 73% of cases, with higher self-confidence predicting higher similarity. Conversely, random gene sets correctly yield zero confidence in 87% of cases. Other LLMs (GPT-3.5, Gemini Pro, Mixtral Instruct and Llama2 70b) vary in function recovery but are falsely confident for random sets. In gene clusters from omics data, GPT-4 identifies common functions for 45% of cases, fewer than functional enrichment but with higher specificity and gene coverage. Manual review of supporting rationale and citations finds these functions are largely verifiable. These results position LLMs as valuable omics assistants.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data used in this paper are publicly available. The full GO (2023-11-15 release) is downloaded from http://release.geneontology.org/2023-11-15/ontology/index.html. The selected NeST gene set is available to download from https://github.com/idekerlab/llm_evaluation_for_gene_set_interpretation/blob/main/data/Omics_data/NeST__IAS_clixo_hidef_Nov17.edges. The L1000 data used in this study are available at https://maayanlab.cloud/static/hdfs/harmonizome/data/lincscmapchemical/gene_attribute_edges.txt.gz. The viral infection data are available at https://maayanlab.cloud/static/hdfs/harmonizome/data/geovirus/gene_attribute_matrix.txt.gz. Detailed information on data download and parsing procedures, along with all datasets used in this paper, are available in our GitHub repository at https://github.com/idekerlab/llm_evaluation_for_gene_set_interpretation.
Code availability
The code to run the LLM gene set analysis pipeline and to reproduce results for the evaluation tasks is available via GitHub at https://github.com/idekerlab/llm_evaluation_for_gene_set_interpretation or Code Ocean77 (https://doi.org/10.24433/CO.7045777.v1) with the MIT License. Note that LLM outputs are inherently stochastic and the precise names and analysis text produced by the models are not guaranteed to be the same from run to run. We minimized the variability of the outputs as described in ‘Controlling the variability of LLM responses’ section in Methods.
References
Zeeberg, B. R. et al. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 4, R28 (2003).
Breitling, R., Amtmann, A. & Herzyk, P. Iterative group analysis (iGA): a simple tool to enhance sensitivity and facilitate interpretation of microarray experiments. BMC Bioinf. 5, 34 (2004).
Beissbarth, T. & Speed, T. P. GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics 20, 1464–1465 (2004).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Al-Shahrour, F. et al. From genes to functional classes in the study of biological systems. BMC Bioinf. 8, 114 (2007).
Backes, C. et al. GeneTrail—advanced gene set enrichment analysis. Nucleic Acids Res. 35, W186–W192 (2007).
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 14, 128 (2013).
Pomaznoy, M., Ha, B. & Peters, B. GOnet: a tool for interactive Gene Ontology analysis. BMC Bioinf. 19, 470 (2018).
Cerami, E. G. et al. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res. 39, D685–D690 (2011).
Fabregat, A. et al. The Reactome pathway Knowledgebase. Nucleic Acids Res. 44, D481–D487 (2015).
Pico, A. R. et al. WikiPathways: pathway editing for the people. PLoS Biol. 6, e184 (2008).
Kanehisa, M., Goto, S., Sato, Y., Furumichi, M. & Tanabe, M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 40, D109–D114 (2012).
Pillich, R. T. et al. NDEx IQuery: a multi-method network gene set analysis leveraging the Network Data Exchange. Bioinformatics 39, btad118 (2023).
Wang, S. et al. Typing tumors using pathways selected by somatic evolution. Nat. Commun. 9, 4159 (2018).
Ashburner, M. et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 (2000).
Gene Ontology Consortiumet al. The Gene Ontology knowledgebase in 2023. Genetics 224, iyad031 (2023).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951 (2019).
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51, D587–D592 (2023).
Croft, D. Reactome: a database of biological pathways. Nat. Preced. https://doi.org/10.1038/npre.2010.5025.1 (2010).
Jassal, B. et al. The reactome pathway knowledgebase. Nucleic Acids Res. 48, D498–D503 (2020).
Sollis, E. et al. The NHGRI-EBI GWAS Catalog: knowledgebase and deposition resource. Nucleic Acids Res. 51, D977–D985 (2023).
Blake, J. A. et al. The Mouse Genome Database genotypes::phenotypes. Nucleic Acids Res. 37, D712–D719 (2009).
Weng, M.-P. & Liao, B.-Y. MamPhEA: a web tool for mammalian phenotype enrichment analysis. Bioinformatics 26, 2212–2213 (2010).
Keenan, A. B. et al. ChEA3: transcription factor enrichment analysis by orthogonal omics integration. Nucleic Acids Res. 47, W212–W224 (2019).
Rubin, J. D. et al. Transcription factor enrichment analysis (TFEA) quantifies the activity of multiple transcription factors from a single experiment. Commun. Biol. 4, 661 (2021).
Franzén, O., Gan, L.-M. & Björkegren, J. L. M. PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database 2019, baz046 (2019).
Zhang, X. et al. CellMarker: a manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 47, D721–D728 (2019).
Hu, C. et al. CellMarker 2.0: an updated database of manually curated cell markers in human/mouse and web tools based on scRNA-seq data. Nucleic Acids Res. 51, D870–D876 (2023).
Devlin, J., Chang, M.-W., Lee, K. & Toutanova, K. BERT: pre-training of deep bidirectional transformers for language understanding. In Proc. 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers) (eds Burstein, J. et al.) 4171–4186 (Association for Computational Linguistics, 2019).
Brown, T. B. et al. Language models are few-shot learners. In Advances in Neural Information Processing Systems (eds H. Larochelle, et al.) 1877–190 (NeurIPS, 2020).
Vaswani, A. et al. Attention is all you need. Neural Inf. Process Syst. 30, 5998–6008 (2017).
OpenAI. GPT-4 technical report. Preprint at https://arxiv.org/abs/2303.08774 (2023).
Touvron, H. et al. Llama 2: open foundation and fine-tuned chat models. Preprint at https://arxiv.org/abs/2307.09288 (2023).
Jiang, A. Q. et al. Mixtral of experts. Preprint at https://arxiv.org/abs/2401.04088 (2024).
Gemini Team et al. Gemini: a family of highly capable multimodal models. Preprint at https://arxiv.org/abs/2312.11805 (2023).
Joachimiak, M. P., Harry Caufield, J., Harris, N. L., Kim, H. & Mungall, C. J. Gene set summarization using large language models. Preprint at https://arxiv.org/abs/2305.13338 (2023).
Moghaddam, S. R. & Honey, C. J. Boosting theory-of-mind performance in large language models via prompting. Preprint at https://arxiv.org/abs/2304.11490 (2023).
Hebenstreit, K., Praas, R., Kiesewetter, L. P. & Samwald, M. An automatically discovered chain-of-thought prompt generalizes to novel models and datasets. Preprint at https://arxiv.org/abs/2305.02897 (2023).
Wei, J. et al. Chain-of-thought prompting elicits reasoning in large language models. In Advances in Neural Information Processing Systems (eds Koyejo, S. et al.) 24824–24837 (NeurIPS, 2022).
Caufield, J. H. et al. Structured prompt interrogation and recursive extraction of semantics (SPIRES): a method for populating knowledge bases using zero-shot learning. Bioinformatics 40, btae104 (2024).
Miller, G. A. & Charles, W. G. Contextual correlates of semantic similarity. Lang. Cogn. Process. 6, 1–28 (1991).
Xiong, M. et al. Can LLMs express their uncertainty? An empirical evaluation of confidence elicitation in LLMs. In The Twelfth International Conference on Learning Representations (ICLR, 20234).
Fu, J., Ng, S.-K., Jiang, Z. & Liu, P. GPTScore: evaluate as you desire. In Proc. 2024 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1: Long Papers (eds Duh, K. et al.) 6556–6576 (Association for Computational Linguistics, 2024).
Kolberg, L. et al. g:Profiler-interoperable web service for functional enrichment analysis and gene identifier mapping (2023 update). Nucleic Acids Res. 51, W207–W212 (2023).
Duan, Q. et al. LINCS Canvas Browser: interactive web app to query, browse and interrogate LINCS L1000 gene expression signatures. Nucleic Acids Res. 42, W449–W460 (2014).
Subramanian, A. et al. A next generation connectivity map: L1000 platform and the first 1,000,000. Profiles Cell 171, 1437–1452.e17 (2017).
Barrett, T. et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 41, D991–D995 (2013).
Zheng, F. et al. Interpretation of cancer mutations using a multiscale map of protein systems. Science 374, eabf3067 (2021).
Pinkas, D. M. et al. Structural complexity in the KCTD family of Cullin3-dependent E3 ubiquitin ligases. Biochem. J. 474, 3747–3761 (2017).
Dhanoa, B. S., Cogliati, T., Satish, A. G., Bruford, E. A. & Friedman, J. S. Update on the Kelch-like (KLHL) gene family. Hum. Genomics 7, 13 (2013).
Pleiner, T. et al. WNK1 is an assembly factor for the human ER membrane protein complex. Mol. Cell 81, 2693–2704.e12 (2021).
Berthold, J. et al. Characterization of RhoBTB-dependent Cul3 ubiquitin ligase complexes—evidence for an autoregulatory mechanism. Exp. Cell. Res. 314, 3453–3465 (2008).
McCormick, J. A. et al. Hyperkalemic hypertension-associated cullin 3 promotes WNK signaling by degrading KLHL3. J. Clin. Invest. 124, 4723–4736 (2014).
Sohara, E. & Uchida, S. Kelch-like 3/Cullin 3 ubiquitin ligase complex and WNK signaling in salt-sensitive hypertension and electrolyte disorder. Nephrol. Dial. Transpl. 31, 1417–1424 (2016).
Tang, H., Finn, R. D. & Thomas, P. D. TreeGrafter: phylogenetic tree-based annotation of proteins with Gene Ontology terms and other annotations. Bioinformatics 35, 518–520 (2019).
Groh, B. S. et al. The antiobesity factor WDTC1 suppresses adipogenesis via the CRL4WDTC1 E3 ligase. EMBO Rep. 17, 638–647 (2016).
Ji, W. & Rivero, F. Atypical rho GTPases of the RhoBTB subfamily: roles in vesicle trafficking and tumorigenesis. Cells 5, 28 (2016).
Nori, H., King, N., McKinney, S. M., Carignan, D. & Horvitz, E. Capabilities of GPT-4 on medical challenge problems. Preprint at https://arxiv.org/abs/2303.13375 (2023).
López Espejel, J., Ettifouri, E. H., Yahaya Alassan, M. S., Chouham, E. M. & Dahhane, W. GPT-3.5, GPT-4, or BARD? Evaluating LLMs reasoning ability in zero-shot learning and performance boosting through prompts. Nat. Lang. Process. J. 5, 100032 (2023).
Yu, H. et al. Evaluation of retrieval-augmented generation: a survey. Preprint at https://arxiv.org/abs/2405.07437 (2024).
Yao, S. et al. ReAct: synergizing reasoning and acting in language models. In The Eleventh International Conference on Learning Representations (ICLR, 2022).
Nair, V., Schumacher, E., Tso, G. & Kannan, A. DERA: enhancing large language model completions with dialog-enabled resolving agents. In Proc. 6th Clinical Natural Language Processing Workshop (eds Naumann, T. et al.) 122–161 (2023).
Shinn, N. et al. Reflexion: language agents with verbal reinforcement learning. In Advances in Neural Information Processing Systems (eds Oh, A. et al.) 8634–8652 (NeurIPS, 2023).
Li, G., Al Kader Hammoud, H. A., Itani, H., Khizbullin, D. & Ghanem, B. CAMEL: Communicative Agents for ‘Mind’ Exploration of Large Scale Language Model Society. In Advances in Neural Information Processing Systems (eds Oh, A. et al.) 36, 51991–52008 (NeurIPS, 2023).
Schick, T. et al. Toolformer: language models can teach themselves to use tools. In Advances in Neural Information Processing Systems (eds Oh, A. et al.) 68539–68551 (NeurIPS, 2023).
Shen, Y. et al. HuggingGPT: solving AI tasks with ChatGPT and its friends in Hugging Face. In Advances in Neural Information Processing Systems (eds Oh, A. et al.) 36, 38154–38180 (NeurIPS, 2023).
Keskar, N. S., McCann, B., Varshney, L. R., Xiong, C. & Socher, R. CTRL: a conditional transformer language model for controllable generation. Preprint at https://arxiv.org/abs/1909.05858 (2019).
Holtzman, A., Buys, J., Du, L., Forbes, M. & Choi, Y. The curious case of neural text degeneration. In The Eighth International Conference on Learning Representations (ICLR, 2020).
Smith, B. et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat. Biotechnol. 25, 1251–1255 (2007).
Tirmizi, S. H. et al. Mapping between the OBO and OWL ontology languages. J. Biomed. Semant. 2, S3 (2011).
Liu, F., Shareghi, E., Meng, Z., Basaldella, M. & Collier, N. Self-alignment pretraining for biomedical entity representations. In Proc. 2021 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies (eds Toutanova, K. et al.) 4228–4238 (Association for Computational Linguistics, 2021).
Wolf, T. et al. Transformers: state-of-the-art natural language processing. In Proc. 2020 Conference on Empirical Methods in Natural Language Processing: System Demonstrations (eds Liu, Q. & Schlangen, D.) 38–45 (Association for Computational Linguistics, 2020).
Rouillard, A. D. et al. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database 2016, baw100 (2016).
Seal, R. L. et al. Genenames.org: the HGNC resources in 2023. Nucleic Acids Res. 51, D1003–D1009 (2023).
Hu, M. et al. Evaluation of Large Language Models for Discovery of Gene Set Function (Code Ocean, 2024); https://doi.org/10.24433/CO.7045777.V1
Acknowledgements
This work was supported by National Institutes of Health grants U24 CA269436 (R.T.P., D.F., K.S., T.I. and D.P.), OT2 OD032742 (M.H., T.I. and D.P.), U24 HG012107 (D.F., T.I. and D.P.) and U01 MH115747 (S.A. and T.I.). Additional support was received from Schmidt Futures (M.H. and T.I.). We thank X. Zhao and A. Singhal for insightful comments, M. R. Kelly for providing the NeST data raw files and C. Churas and J. Lenkiewicz for helping to improve the GitHub repository.
Author information
Authors and Affiliations
Contributions
M.H., S.A., T.I. and D.P. designed the study. M.H. and S.A. developed and implemented the automated LLM-based gene set interpretation pipeline, performed the data analysis and organized the GitHub repository. S.A. developed and assessed the semantic similarity calculation. I.L. and M.H. contributed to the development of the citation search and validation pipeline. D.P. contributed to the coding and the evaluation of the analysis. R.T.P. assisted in the study design, prompt engineering and the evaluation of the analysis. M.H., R.T.P., R.B. and D.P. conducted the scientific review of the LLM output. M.H. and D.P. contributed to the user interface design for the GSAI tool. D.F. built the web interface for the GSAI tool, and K.S. set up the server for accessing open-source LLMs. M.H., S.A., T.I. and D.P. wrote the paper with input from all authors. All authors approved the final version of this paper.
Corresponding authors
Ethics declarations
Competing interests
T.I. is a cofounder and member of the advisory board and has an equity interest in Data4Cure and Serinus Biosciences. T.I. is a consultant for and has an equity interest in Ideaya Biosciences. The terms of these arrangements have been reviewed and approved by the University of California San Diego in accordance with its conflict-of-interest policies. The other authors declare no competing interests.
Peer review
Peer review information
Nature Methods thanks Qiao Jin, Zhiyong Lu, Zhizheng Wang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Lin Tang, in collaboration with the Nature Methods team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Schematic of the citation module.
a, GPT-4 is asked to provide gene symbol keywords and functional keywords separately. Multiple gene keywords and functions are combined and used to search PubMed for relevant paper titles and abstracts in the scientific literature. GPT-4 is queried to evaluate each abstract, saving supporting references. b, Prompts used to query the GPT-4 model.
Extended Data Fig. 2 Distribution of GO term gene sizes.
a, Distribution of term size (number of genes) for terms in the Biological Process branch (GO-BP). Terms with 3-100 genes shown (n = 8,910). b, Distribution of term size for the 1000 GO terms used in Task 1.
Extended Data Fig. 3 Evaluation of GPT-4 in recovery of GO-CC and GO-MF names.
a, Cumulative number of GO-CC term names recovered by GPT-4 (y-axis) at a given similarity percentile (x-axis). 0 = least similar, 100 = most similar. Blue curve: semantic similarities between GPT-4 names and assigned GO-CC term names. Grey dashed curve: semantic similarities between GPT-4 names and random GO-CC term names. The red dotted line marks that 642 of the 1000 sampled GO-CC names are recovered by GPT-4 at a similarity percentile of 95%. b, As for panel a, but for GO-MF terms rather than GO-CC. The red dotted line marks that 757 of the 1000 sampled GO-MF names are recovered by GPT-4 at a similarity percentile of 95%.
Extended Data Fig. 4 Supplemental analysis of the confidence score.
a, Distribution of confidence scores (n = 300) assigned by GPT-4 with confidence level threshold set based on the distribution pattern. “High confidence” (red): 0.87–1.00; “Medium confidence” (blue): 0.82–0.86; “Low confidence” (dark orange): 0.01–0.81; “Name not assigned” (gray): 0. b, Scatter plot of naming accuracy versus GPT-4 self-assessed confidence score for real gene sets drawn from GO (points, n = 100). Accuracy is estimated by the semantic similarity between the GPT-4 proposed name and the real GO term name. The best-fit regression line is shown in dark gray. The correlation coefficient (R) is determined by a two-sided Pearson’s correlation with p-value shown.
Extended Data Fig. 5 Distribution of ‘omics gene set sizes.
Distribution shown for all ‘omics gene sets considered in this study (n = 300).
Supplementary information
Supplementary Information
Description for Supplementary Tables 1–4.
Supplementary Tables 1–4
Supplementary Table 1. Complete analysis of GO terms, 50/50 mix and random for all models (related to task 1: Figs. 2a and 3b and Table 1). Supplementary Table 2. Complete GPT-4 analysis of GO terms (related to task 1: Fig. 2). Supplementary Table 3. Complete GPT-4 analysis of omics gene sets (related to task 2: Fig. 4 and Extended Data Table 4). Supplementary Table 4. Reviewer fact-checking of GPT-4 analysis text and citation relevance (related to task 2: Fig. 5).
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hu, M., Alkhairy, S., Lee, I. et al. Evaluation of large language models for discovery of gene set function. Nat Methods 22, 82–91 (2025). https://doi.org/10.1038/s41592-024-02525-x
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41592-024-02525-x
This article is cited by
-
AI-powered Immune Cell Knowledge Graph (ICKG) with granular immune contexts enables immune program interpretation
npj Artificial Intelligence (2026)
-
Emergence and evolution of protein-coding de novo genes
Nature Reviews Genetics (2026)
-
Natural language processing of gene descriptions for overrepresentation analysis with GeneTEA
Genome Biology (2025)
-
scDrugMap: benchmarking large foundation models for drug response prediction
Nature Communications (2025)
-
GeneAgent: self-verification language agent for gene-set analysis using domain databases
Nature Methods (2025)