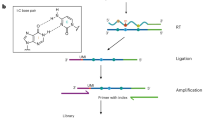
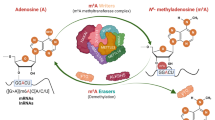
Abstract
Methods for absolute quantification of N6-methyladenosine (m6A) have emerged as powerful tools in epitranscriptomics. We previously reported GLORI, a chemical-assisted approach to achieve unbiased and precise m6A measurement. However, its lengthy reaction time and severe RNA degradation have limited its applicability, particularly for low-input samples. Here, we present two updated GLORI approaches that are ultrafast, mild and enable absolute m6A quantification from one to two orders of magnitude less than the RNA starting material: GLORI 2.0 is compatible with RNA from ~10,000 cells and enhances sensitivity for both transcriptome-wide and locus-specific m6A detection; GLORI 3.0 further utilizes a reverse transcription-silent carrier RNA to achieve m6A quantification from as low as 500–1,000 cells. Using limited RNA from mouse dorsal hippocampus, we reveal a high modification level in synapse-related gene sets. We envision that the updated GLORI methods will greatly expand the applicability of absolute quantification of m6A in biology.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The sequencing data generated in this study have been deposited in the NCBI Gene Expression Omnibus (GEO), under accession code GSE270643. The public GLORI 1.0 datasets were downloaded from the GEO under accession code GSE210563. Source data are provided with this paper.
Code availability
The scripts used for processing the raw data are deposited on Zenodo (https://doi.org/10.5281/zenodo.14233421)59.
References
Moshitch-Moshkovitz, S., Dominissini, D. & Rechavi, G. The epitranscriptome toolbox. Cell 185, 764–776 (2022).
Roundtree, I. A., Evans, M. E., Pan, T. & He, C. Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200 (2017).
Frye, M., Jaffrey, S. R., Pan, T., Rechavi, G. & Suzuki, T. RNA modifications: what have we learned and where are we headed? Nat. Rev. Genet. 17, 365–372 (2016).
Sun, H., Li, K., Liu, C. & Yi, C. Regulation and functions of non-m6A mRNA modifications. Nat. Rev. Mol. Cell Biol. 24, 714–731 (2023).
Dominissini, D. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012).
Meyer, K. D. et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646 (2012).
Li, X., Xiong, X. & Yi, C. Epitranscriptome sequencing technologies: decoding RNA modifications. Nat. Methods 14, 23–31 (2016).
Harcourt, E. M., Kietrys, A. M. & Kool, E. T. Chemical and structural effects of base modifications in messenger RNA. Nature 541, 339–346 (2017).
Liu, C. et al. Absolute quantification of single-base m6A methylation in the mammalian transcriptome using GLORI. Nat. Biotechnol. 41, 355–366 (2023).
Xiao, Y. L. et al. Transcriptome-wide profiling and quantification of N6-methyladenosine by enzyme-assisted adenosine deamination. Nat. Biotechnol. 41, 993–1003 (2023).
Shen, W. et al. GLORI for absolute quantification of transcriptome-wide m6A at single-base resolution. Nat. Protoc. 19, 1252–1287 (2024).
Shapiro, R. & Pohl, S. H. The reaction of ribonucleosides with nitrous acid. Side products and kinetics. Biochemistry 7, 448–455 (1968).
Schuster, H. & Wilhelm, R. C. Reaction differences between tobacco mosaic virus and its free ribonucleic acid with nitrous acid. Biochim. Biophys. Acta 68, 554–560 (1963).
Shapiro, R. & Hachmann, J. The reaction of guanine derivatives with 1,2-dicarbonyl compounds. Biochemistry 5, 2799–2807 (1966).
Nakaya, K., Takenaka, O., Horinishi, H. & Shibata, K. Reactions of glyoxal with nucleic acids. Nucleotides and their component bases. Biochim. Biophys. Acta 161, 23–31 (1968).
Knutson, S. D. et al. Thermoreversible control of nucleic acid structure and function with glyoxal caging. J. Am. Chem. Soc. 142, 17766–17781 (2020).
Karloff, D. B. et al. Glyoxal caging of nucleoside antivirals toward self-activating, extended-release prodrugs. J. Am. Chem. Soc. 146, 29402–29406 (2024).
Zhang, Z. et al. Systematic calibration of epitranscriptomic maps using a synthetic modification-free RNA library. Nat. Methods 18, 1213–1222 (2021).
Liu, N. et al. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA 19, 1848–1856 (2013).
Xiao, Y. et al. An elongation- and ligation-based qPCR amplification method for the radiolabeling-free detection of locus-specific N6-ethyladenosine modification. Angew. Chem. Int. Ed. Engl. 57, 15995–16000 (2018).
Harcourt, E. M., Ehrenschwender, T., Batista, P. J., Chang, H. Y. & Kool, E. T. Identification of a selective polymerase enables detection of N6-methyladenosine in RNA. J. Am. Chem. Soc. 135, 19079–19082 (2013).
Hong, T. et al. Precise antibody-independent m6A identification via 4SedTTP-involved and FTO-assisted strategy at single-nucleotide resolution. J. Am. Chem. Soc. 140, 5886–5889 (2018).
Bai, D. et al. Simultaneous single-cell analysis of 5mC and 5hmC with SIMPLE-seq. Nat. Biotechnol. 43, 85–96 (2024).
Guo, H. et al. The DNA methylation landscape of human early embryos. Nature 511, 606–610 (2014).
Yoshida, M. & Ukita, T. Modification of nucleosides and nucleotides. VII. Selective cyanoethylation of inosine and pseudouridine in yeast transfer ribonucleic acid. Biochim. Biophys. Acta 157, 455–465 (1968).
Ofengand, J. The function of pseudouridylic acid in transfer ribonucleic acid. I. The specific cyanoethylation of pseudouridine, inosine, and 4-thiouridine by acrylonitrile. J. Biol. Chem. 242, 5034–5045 (1967).
Sakurai, M., Yano, T., Kawabata, H., Ueda, H. & Suzuki, T. Inosine cyanoethylation identifies A-to-I RNA editing sites in the human transcriptome. Nat. Chem. Biol. 6, 733–740 (2010).
Li, Y. et al. Single-cell m6A mapping in vivo using picoMeRIP-seq. Nat. Biotechnol. 42, 591–596 (2024).
Hamashima, K. et al. Single-nucleus multiomic mapping of m6A methylomes and transcriptomes in native populations of cells with sn-m6A-CT. Mol. Cell https://doi.org/10.1016/j.molcel.2023.08.010 (2023).
Yao, H. et al. scm6A-seq reveals single-cell landscapes of the dynamic m6A during oocyte maturation and early embryonic development. Nat. Commun. 14, 315 (2023).
Tegowski, M., Flamand, M. N. & Meyer, K. D. scDART-seq reveals distinct m6A signatures and mRNA methylation heterogeneity in single cells. Mol. Cell 82, 868–878 (2022).
Martin, K. C. & Ephrussi, A. mRNA localization: gene expression in the spatial dimension. Cell 136, 719–730 (2009).
Jung, H., Yoon, B. C. & Holt, C. E. Axonal mRNA localization and local protein synthesis in nervous system assembly, maintenance and repair. Nat. Rev. Neurosci. 13, 308–324 (2012).
Shi, H. et al. m6A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature 563, 249–253 (2018).
Zhang, Z. et al. METTL3-mediated N6-methyladenosine mRNA modification enhances long-term memory consolidation. Cell Res. 28, 1050–1061 (2018).
Merkurjev, D. et al. Synaptic N6-methyladenosine (m6A) epitranscriptome reveals functional partitioning of localized transcripts. Nat. Neurosci. 21, 1004–1014 (2018).
Flamand, M. N. & Meyer, K. D. m6A and YTHDF proteins contribute to the localization of select neuronal mRNAs. Nucleic Acids Res. 50, 4464–4483 (2022).
Livneh, I., Moshitch-Moshkovitz, S., Amariglio, N., Rechavi, G. & Dominissini, D. The m6A epitranscriptome: transcriptome plasticity in brain development and function. Nat. Rev. Neurosci. 21, 36–51 (2020).
Yu, Y. et al. Bi-functionality of glyoxal caged nucleic acid coupled with CRISPR/Cas12a system for Hg2+ determination. Mikrochim. Acta 191, 120 (2024).
Knutson, S. D., Arthur, R. A., Johnston, H. R. & Heemstra, J. M. Selective enrichment of A-to-I edited transcripts from cellular RNA using endonuclease V. J. Am. Chem. Soc. 142, 5241–5251 (2020).
Xiao, Z. et al. Holo-Seq: single-cell sequencing of holo-transcriptome. Genome Biol. 19, 163 (2018).
Taryma-Lesniak, O., Sokolowska, K. E. & Wojdacz, T. K. Current status of development of methylation biomarkers for in vitro diagnostic IVD applications. Clin. Epigenetics 12, 100 (2020).
Barbieri, I. & Kouzarides, T. Role of RNA modifications in cancer. Nat. Rev. Cancer 20, 303–322 (2020).
Delaunay, S. & Frye, M. RNA modifications regulating cell fate in cancer. Nat. Cell Biol. 21, 552–559 (2019).
Suzuki, T., Ueda, H., Okada, S. & Sakurai, M. Transcriptome-wide identification of adenosine-to-inosine editing using the ICE-seq method. Nat. Protoc. 10, 715–732 (2015).
Wang, Y. et al. LEAD-m6A-seq for locus-specific detection of N6-methyladenosine and quantification of differential methylation. Angew. Chem. Int. Ed. Engl. 60, 873–880 (2021).
Shen, W., Sipos, B. & Zhao, L. Y. SeqKit2: a Swiss army knife for sequence and alignment processing. Imeta 3, e191 (2024).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Crooks, G. E., Hon, G., Chandonia, J. M. & Brenner, S. E. WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004).
Park, Y., Figueroa, M. E., Rozek, L. S. & Sartor, M. A. MethylSig: a whole genome DNA methylation analysis pipeline. Bioinformatics 30, 2414–2422 (2014).
Wu, T. et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation 2, 100141 (2021).
Supek, F., Bosnjak, M., Skunca, N. & Smuc, T. REVIGO summarizes and visualizes long lists of Gene Ontology terms. PLoS ONE 6, e21800 (2011).
Lorenz, R. et al. ViennaRNA Package 2.0. Algorithms Mol. Biol. 6, 26 (2011).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Anders, S., Pyl, P. T. & Huber, W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Lu, B. Codes for "Mild and ultrafast GLORI enables absolute quantification of m6A methylome from low-input samples". Zenodo https://doi.org/10.5281/zenodo.14233421 (2024).
Acknowledgements
We thank the National Center for Protein Sciences at Peking University for assistance with library size distribution. We thank the High Performance Computing Platform of the Center for Life Science for assistance with the analysis. We thank the Analytical Instrumentation Center in Peking University for assistance with identification of intermediates and X. Liu for help with the analysis of LC–TOF–MS. We thank H. Yao and Y. Cai from Peking University for discussion on chemistry experiments. This work was supported by the National Key R&D Program of China (2023YFC3402200 to C.Y.), the National Natural Science Foundation of China (22425071 to C.Y.), the Beijing Natural Science Foundation (Z220013 to C.Y., 5234030 to B.L.), the Beijing Municipal Science & Technology Commission (Z231100007223010 to X.-J.W.), the China National Postdoctoral Program for Innovative Talents (BX20230029 to H.S.; BX20230411 to Z. Zhang) and the China Postdoctoral Science Foundation (GZB20230026 to B.L.). This work was supported by the New Cornerstone Science Foundation through the XPLORER PRIZE.
Author information
Authors and Affiliations
Contributions
C.Y., X.-J.W., H.S. and B.L. conceived the project and designed the experiments; C.Y., H.S., B.L. and Z. Zhang wrote the manuscript with the help of X.-J.W., Z.J. and J.P.; H.S., Y.X. and Z. Zhang performed the experiments with the help of Z.L., L.X., Y.M. and J.Z.; H.S. develop GLORI 2.0 and GLORI 3.0 and identify intermediates; B.L. designed and performed the bioinformatics analysis with the help of Z. Zhou and C.L.; Z. Zhang performed mouse experiments with the help of M.W.; C.Y. and X.-J.W. supervised the project.
Corresponding authors
Ethics declarations
Competing interests
A patent application has been filed by Peking University for the technology disclosed in this publication. C.Y. and H.S. are coinventors on a patent application describing GLORI 2.0 and GLORI 3.0. The other authors declare no competing interests.
Peer review
Peer review information
Nature Methods thanks Qihan Chen, Kate Meyer and Weixin Tang for their contribution to the peer review of this work. Primary Handling Editor: Lei Tang, in collaboration with the Nature Methods team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–24 and Supplementary Tables 1 and 2.
Supplementary Data 1
m6A sites identified by GLORI 2.0 with 50 ng, 10 ng and 2 ng mRNA input (300 M raw reads).
Supplementary Data 2
m6A sites identified by GLORI 2.0 after STM2457 treatment (300 M raw reads).
Supplementary Data 3
m6A sites identified by GLORI 1.0 with 50 ng and 10 ng mRNA input (300 M raw reads).
Supplementary Data 4
m6A sites identified by GLORI 3.0 with 100 ng, 20 ng and 10 ng total RNA input (300 M raw reads).
Supplementary Data 5
m6A sites identified by GLORI 3.0 in synapse and cytoplasm (300 M raw reads).
Supplementary Data 6
Differentially methylated m6A sites between synapse and cytoplasm (300 M raw reads).
Supplementary Data 7
Library quality assessment.
Supplementary Data 8
Primer and spike-in sequences.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Original Gel Figures
Unprocessed gel images.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sun, H., Lu, B., Zhang, Z. et al. Mild and ultrafast GLORI enables absolute quantification of m6A methylome from low-input samples. Nat Methods 22, 1226–1236 (2025). https://doi.org/10.1038/s41592-025-02680-9
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41592-025-02680-9
This article is cited by
-
Chemical biology of precise RNA targeting and intervention: advances and perspectives
Science China Chemistry (2026)
-
Uli-epic: profiling RNA modifications from ultra-low input samples
Genome Biology (2025)