Abstract
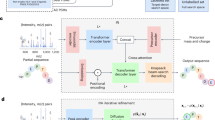
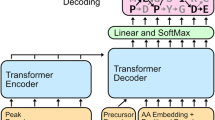
A core computational challenge in the analysis of mass spectrometry data is the de novo sequencing problem, in which the generating amino acid sequence is inferred directly from an observed fragmentation spectrum without the use of a sequence database. Recently, deep learning models have made substantial advances in de novo sequencing by learning from massive datasets of high-confidence labeled mass spectra. However, these methods are designed primarily for data-dependent acquisition experiments. Over the past decade, the field of mass spectrometry has been moving toward using data-independent acquisition (DIA) protocols for the analysis of complex proteomic samples owing to their superior specificity and reproducibility. Hence, we present a de novo sequencing model called Cascadia, which uses a transformer architecture to handle the more complex data generated by DIA protocols. In comparisons with existing approaches for de novo sequencing of DIA data, Cascadia achieves substantially improved performance across a range of instruments and experimental protocols.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The MassIVE-KB DDA data used for pretraining are available at https://noble.gs.washington.edu/~melih/mskb_casanovo_splits.zip. The wide-window DIA data from the CPTAC consortium used for training Cascadia are available through Proteomic Data Commons with study identifiers PDC000341 (ref. 35) and PDC000200 (ref. 36). The original DIA test set used by DeepNovo-DIA is available on the MassIVE repository with accession number MSV000082368 (ref. 11). The Astral mouse training data and the data used in our variant prediction experiments are available at https://panoramaweb.org/cascadia.url with ProteomeXchange ID PXD053291, and the HeLa and human plasma EV astral test sets are available at https://panoramaweb.org/AstralBenchmarking.url with ProteomeXchange ID PXD042704 (ref. 17). The yeast DIA data were downloaded from https://panoramaweb.org/Carafe.url with ProteomeXchange ID PXD056793 (ref. 7). The reference proteomes for Homo sapiens, Mus musculus and Saccharomyces cerevisiae are available on UniProt with identifiers UP000005640, UP000000589 and UP000002311 respectively37.
Code availability
The open-source Cascadia software and associated model weights are available with an Apache license via GitHub at https://github.com/Noble-Lab/cascadia. A dockerized version of Cascadia was also added as an option in the Skyline DIA Nextflow workflow available at https://nf-skyline-dia-ms.readthedocs.io/, allowing proteomics researchers with less technical expertise to more easily integrate de novo sequencing into their data analysis. This workflow outputs a Skyline document that can be loaded into the commonly used DIA analysis software Skyline, enabling easy visualization of results38. As such, Cascadia can be directly incorporated into existing DIA protomics analysis pipeline either in place of or alongside a traditional search engine.
References
Bittremieux, W. et al. Deep learning methods for de novo peptide sequencing. Mass Spectrom. Rev. https://doi.org/10.1002/mas.21919 (2024).
Vaswani, A. et al. Attention is all you need. Adv. Neural Inf. Process. Syst. 30, 5998–6008 (2017).
Venable, J. D., Dong, M. Q., Wohlsclegel, J., Dillin, A. & Yates III, J. R. Automated approach for quantitative analysis of complex peptide mixtures from tandem mass spectra. Nat. Methods 1, 39–45 (2004).
Tsou, C.-C. et al. DIA-Umpire: a comprehensive computational framework for data-independent acquisition proteomics. Nat. Methods 12, 258–264 (2015).
Liu, K., Ye, Y., Li, S. & Tang, H. Accurate de novo peptide sequencing using fully convolutional neural networks. Nat. Commun. 14, 7974 (2023).
Wu, S., Luan, Z., Fu, Z., Wang, Q. & Guo, T. BiATNovo: a self-attention based bidirectional peptide sequencing method. Preprint at bioRxiv https://doi.org/10.1101/2023.05.11.540352 (2023).
Wen, B. et al. Carafe enables high quality in silico spectral library generation for data-independent acquisition proteomics. Preprint at bioRxiv https://doi.org/10.1101/2024.10.15.618504 (2024).
Searle, B. C. et al. Chromatogram libraries improve peptide detection and quantification by data independent acquisition mass spectrometry. Nat. Commun. 9, 5128 (2018).
Pino, L. K., Just, S. C., MacCoss, M. J. & Searle, B. C. Acquiring and analyzing data independent acquisition proteomics experiments without spectrum libraries. Mol. Cell. Proteomics 19, 1088–1103 (2020).
Isaksson, M., Karlsson, C., Laurell, T., Kirkeby, A. & Heusel, M. MSLibrarian: optimized predicted spectral libraries for data-independent acquisition proteomics. J. Proteome Res. 21, 535–546 (2022).
Tran, N. H. et al. Deep learning enables de novo peptide sequencing from data-independent-acquisition mass spectrometry. Nat. Methods 16, 63–66 (2019).
Ebrahimi, S. & Guo, X. Transformer-based de novo peptide sequencing for data-independent acquisition mass spectrometry. In Proc. IEEE 23rd International Conference on Bioinformatics and Bioengineering 28–35 (2024).
Yilmaz, M., Fondrie, W. E., Bittremieux, W., Oh, S. & Noble, W. S. De novo mass spectrometry peptide sequencing with a transformer model. In Proc. 39th International Conference on Machine Learning Vol. 62, 25514–25522 (PMLR, 2022).
Edwards, N. et al. The CPTAC data portal: a resource for cancer proteomics research. J. Proteome Res. 14, 2707–2713 (2015).
Demichev, V., Messner, C. B., Vernardis, S. I., Lilley, K. S. & Ralser, M. DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods 17, 41–44 (2020).
Wu, C. C. et al. Mag-Net: Rapid enrichment of membrane-bound particles enables high coverage quantitative analysis of the plasma proteome. Preprint at bioRxiv https://doi.org/10.1101/2023.06.10.544439 (2023).
Heil, L. R. et al. Evaluating the performance of the astral mass analyzer for quantitative proteomics using data-independent acquisition. J. Proteome Res. 22, 3290–3300 (2023).
Janeway Jr, C. A., Travers, P., Walport, M. & Shlomchik, M. J. The Generation of Diversity in Immunoglobulins (Garland Science, 2001).
Tran, N. H. et al. Discovering and validating neoantigens by mass spectrometry-based immunopeptidomics and deep learning. Preprint at bioRxiv https://doi.org/10.1101/2022.07.05.497667 (2024).
Melendez, C. et al. Accounting for digestion enzyme bias in Casanovo. J. Proteome Res. 23, 4761–4761 (2024).
Jin, Z. et al. ContraNovo: a contrastive learning approach to enhance de novo peptide sequencing. In Proc. 38th AAAI Conference on Artificial Intelligence 144–152 (2023).
Xia, J. et al. AdaNovo: adaptive de novo peptide sequencing with conditional mutual information. Adv. Neural Inf. Process. Syst. 37, 1811–1828 (2024).
Eloff, K. et al. InstaNovo enables diffusion-powered de novo peptide sequencing in large-scale proteomics experiments. Nat. Mach. Intell. 7, 565–579 (2023).
Yang, H., Chi, H., Zeng, W., Zhou, W. & He, S. pNovo 3: precise de novo peptide sequencing using a learning-to-rank framework. Bioinformatics 35, i83–i90 (2019).
Klaproth-Andrade, D. et al. Deep learning-driven fragment ion series classification enables highly precise and sensitive de novo peptide sequencing. Nat. Commun. 15, 151 (2024).
Li, K., Vaudel, M., Zhang, B., Ren, Y. & Wen, B. PDV: an integrative proteomics data viewer. Bioinformatics 35, 1249–1251 (2019).
Zeng, W.-F. et al. AlphaPeptDeep: a modular deep learning framework to predict peptide properties for proteomics. Nat. Commun. 13, 7238 (2022).
Lu, Y. Y., Bilmes, J., Rodriguez-Mias, R. A., Villén, J. & Noble, W. S. DIAmeter: matching peptides to data-independent acquisition mass spectrometry data. Bioinformatics 37, i434–i442 (2021).
Ting, Y. S. et al. PECAN: a library free peptide detection tool for data-independent acquisition tandem mass spectrometry data. Nat. Methods 14, 903–908 (2017).
Dosovitskiy, A. et al. An image is worth 16x16 words: transformers for image recognition at scale. In Proc. International Conference on Learning Representations (2021).
Dittwald, P., Claesen, J., Burzykowski, T., Valkenborg, D. & Gambin, A. BRAIN: a universal tool for high-throughput calculations of the isotopic distribution for mass spectrometry. Anal. Chem. 85, 1991–1994 (2013).
Wen, B., Li, K., Zhang, Y. & Zhang, B. Cancer neoantigen prioritization through sensitive and reliable proteogenomics analysis. Nat. Commun. 11, 1759 (2020).
Wang, M. et al. Assembling the community-scale discoverable human proteome. Cell Syst. 7, 412–421 (2018).
Yu, F. et al. Analysis of DIA proteomics data using MSFragger-DIA and FragPipe computational platform. Nat. Commun. 14, 4154 (2023).
Cao, L. et al. Clinical Proteomic Tumor Analysis Consortium proteogenomic characterization of pancreatic ductal adenocarcinoma. Cell 184, 5031–5052 (2021).
Clark, D. J. et al. Clinical Proteomic Tumor Analysis Consortium integrated proteogenomic characterization of clear cell renal cell carcinoma. Cell 179, 964–983 (2019).
UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 43, D204–212 (2014).
MacLean, B. et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 (2010).
Acknowledgements
This work is supported by National Science Foundation award number 2245300 to W.S.N., and in part by the Intelligence Advanced Research Projects Activity (IARPA) TEI-REX and PROTEOS programs under contract numbers W911NF2220059 and 2018-18041000004 to M.J.M, and the National Science Foundation Graduate Research Fellowship Program (grant no. DGE-2140004, B.W.). The views and conclusions contained should not be interpreted as necessarily representing the official policies, either expressed or implied, of ODNI, IARPA, ARO or the US Government. The funders had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
J.S., W.S.N. and S.O. conceived of the method design and computational experiments. J.S. wrote the code and performed computational analysis, except where otherwise noted. C.C.W. collected the mouse astral data used for training. R.S.J. collected the human DIA data. P.A.R. provided an initial variant analysis of the human DIA data. B.W. helped run the database searches and perform retention time analysis. M.R. implemented the Nextflow workflow. J.S. and W.S.N. wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The MacCoss Lab at the University of Washington receives funding from Agilent, Bruker, Sciex, Shimadzu, Thermo Fisher Scientific and Waters to support the development of Skyline, a quantitative analysis software tool. M.J.M. is a paid consultant for Thermo Fisher Scientific. The other authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Notes 1–5 and Figs. 1–12.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sanders, J., Wen, B., Rudnick, P.A. et al. A transformer model for de novo sequencing of data-independent acquisition mass spectrometry data. Nat Methods 22, 1447–1453 (2025). https://doi.org/10.1038/s41592-025-02718-y
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41592-025-02718-y