Abstract
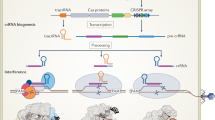
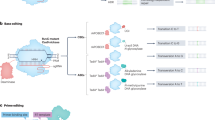
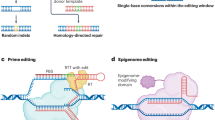
Genome editing with CRISPR–Cas systems is revolutionizing medicine, molecular biology and biotechnology. In this Review, we discuss the contributions of deep learning-based structure prediction algorithms, physics-based simulations, neural networks, graph neural networks and generative models, including diffusion and large language models, in engineering and optimizing CRISPR systems and in understanding their mechanistic basis. We highlight the challenges and limitations to the transformative effects of computational modeling and tools in the context of the development of programmable genome editors for biomedicine and biotechnology.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Karplus, M. & McCammon, J. A. Molecular dynamics simulations of biomolecules. Nat. Struct. Biol. 9, 646–652 (2002).
van den Bedem, H. & Fraser, J. S. Integrative, dynamic structural biology at atomic resolution—it’s about time. Nat. Methods 12, 307–318 (2015).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Wang, J. Y., Pausch, P. & Doudna, J. A. Structural biology of CRISPR–Cas immunity and genome editing enzymes. Nat. Rev. Microbiol. 11, 641–656 (2022).
Sinha, S., Pindi, C., Ahsan, M., Arantes, P. R. & Palermo, G. Machines on genes through the computational microscope. J. Chem. Theory Comput. 19, 1945–1964 (2023).
Palermo, G., Miao, Y., Walker, R. C., Jinek, M. & McCammon, J. A. Striking plasticity of CRISPR–Cas9 and key role of non-target DNA, as revealed by molecular simulations. ACS Cent. Sci. 2, 756–763 (2016).
Palermo, G., Miao, Y., Walker, R. C., Jinek, M. & McCammon, J. A. CRISPR–Cas9 conformational activation as elucidated from enhanced molecular simulations. Proc. Natl Acad. Sci. USA 114, 7260–7265 (2017).
Saha, A. et al. An alpha-helical lid guides the target DNA toward catalysis in CRISPR–Cas12a. Nat. Commun. 15, 1473 (2024).
Zuo, Z. et al. Structural and functional insights into the bona fide catalytic state of Streptococcus pyogenes Cas9 HNH nuclease domain. eLife 8, e46500 (2019).
Nierzwicki et al. Principles of target DNA cleavage and the role of Mg2+ in the catalysis of CRISPR–Cas9. Nat. Catal. 5, 912–922 (2022).
Casalino, L., Nierzwicki, Ł, Jinek, M. & Palermo, G. Catalytic mechanism of non-target DNA cleavage in CRISPR–Cas9 revealed by ab initio molecular dynamics. ACS Catal. 10, 13596–13605 (2020).
Van, R. et al. Exploring CRISPR–Cas9 HNH-domain-catalyzed DNA cleavage using accelerated quantum mechanical molecular mechanical free energy simulation. Biochemistry 64, 289–299 (2024).
Yoon, H., Zhao, L. N. & Warshel, A. Exploring the catalytic mechanism of Cas9 using information inferred from endonuclease VII. ACS Catal. 9, 1329–1336 (2019).
Skeens, E. et al. High-fidelity, hyper-accurate, and evolved mutants rewire atomic level communication in CRISPR–Cas9. Sci. Adv. 10, eadl1045 (2024).
Nierzwicki, L. et al. Enhanced specificity mutations perturb allosteric signaling in CRISPR–Cas9. eLife 10, e73601 (2021).
Babu, K. et al. Bridge helix of Cas9 modulates target DNA cleavage and mismatch tolerance. Biochemistry 58, 1905–1917 (2019).
Sinha, S. et al. Unveiling the RNA-mediated allosteric activation discloses functional hotspots in CRISPR–as13a. Nucleic Acids Res. 52, 906–920 (2024).
Molina Vargas, A. M. et al. New design strategies for ultra-specific CRISPR–Cas13a-based RNA detection with single-nucleotide mismatch sensitivity. Nucleic Acids Res. 52, 921–939 (2024).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Iturralde, A. B., Weller, C. A., Giovanetti, S. M. & Sadhu, M. J. Comprehensive deletion scan of anti-CRISPR AcrIIA4 reveals essential and dispensable domains for Cas9 inhibition. Proc. Natl Acad. Sci. USA 121, e2413743121 (2024).
Kang, J. et al. Structural investigation of the anti-CRISPR protein AcrIE7. Proteins 93, 1645–1656 (2025).
Belato, H. B. et al. Structural and dynamic insights into the HNH nuclease of divergent Cas9 species. J. Struct. Biol. 214, 107814 (2022).
Halpin-Healy, T. S., Klompe, S. E., Sternberg, S. H. & Fernández, I. S. Structural basis of DNA targeting by a transposon-encoded CRISPR–Cas system. Nature 577, 271–274 (2020).
Chaudhury, S., Lyskov, S. & Gray, J. J. PyRosetta: a script-based interface for implementing molecular modeling algorithms using Rosetta. Bioinformatics 26, 689–691 (2010).
Patel, A. C., Sinha, S., Arantes, P. R. & Palermo, G. Unveiling Cas8 dynamics and regulation within a transposon-encoded Cascade–TniQ complex. Proc. Natl Acad. Sci. USA 122, e2422895122 (2025).
Zhao, F. et al. A strategy for Cas13 miniaturization based on the structure and AlphaFold. Nat. Commun. 14, 5545 (2023).
Yoon, P. H. et al. Structure-guided discovery of ancestral CRISPR–Cas13 ribonucleases. Science 385, 538–543 (2024).
Varadi, M. et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, 439–444 (2022).
Holm, L. Benchmarking fold detection by DaliLite v.5. Bioinformatics 35, 5326–5327 (2019).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Pan, L. et al. Optimization of CRISPR/Cas12f1 guide RNAs using AlphaFold 3 for enhanced nucleic acid detection. Microchem. J. 212, 113194 (2025).
Schneider, B. et al. When will RNA get its AlphaFold moment? Nucleic Acids Res. 51, 9522–9532 (2023).
McDonnell, R. T., Henderson, A. N. & Elcock, A. H. Structure prediction of large RNAs with AlphaFold3 highlights its capabilities and limitations. J. Mol. Biol. 436, 168816 (2024).
LeCun, Y., Bengio, Y. & Hinton, G. Deep learning. Nature 521, 436–444 (2015).
Gallego, V. & Ríos Insua, D. Current advances in neural networks. Annu. Rev. Stat. Appl. 9, 197–222 (2022).
Alzubaidi, L. et al. Review of deep learning: concepts, CNN architectures, challenges, applications, future directions. J. Big Data 8, 53 (2021).
Jurtz, V. I. et al. An introduction to deep learning on biological sequence data: examples and solutions. Bioinformatics 33, 3685–3690 (2017).
Doench, J. G. et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR–Cas9. Nat. Biotechnol. 34, 184–191 (2016).
Chuai, G. et al. DeepCRISPR: optimized CRISPR guide RNA design by deep learning. Genome Biol. 19, 80 (2018).
Lin, J., Zhang, Z., Zhang, S., Chen, J. & Wong, K. CRISPR-Net: a recurrent convolutional network quantifies CRISPR off-target activities with mismatches and indels. Adv. Sci. 7, 1903562 (2020).
Kim, H. K. et al. SpCas9 activity prediction by DeepSpCas9, a deep learning-based model with high generalization performance. Sci. Adv. 5, eaax9249 (2019).
Xue, L., Tang, B., Chen, W. & Luo, J. Prediction of CRISPR sgRNA activity using a deep convolutional neural network. J. Chem. Inf. Model. 59, 615–624 (2019).
Wang, D. et al. Optimized CRISPR guide RNA design for two high-fidelity Cas9 variants by deep learning. Nat. Commun. 10, 4284 (2019).
Xiao, L.-M., Wan, Y.-Q. & Jiang, Z.-R. AttCRISPR: a spacetime interpretable model for prediction of sgRNA on-target activity. BMC Bioinformatics 22, 589 (2021).
Li, C., Zou, Q., Li, J. & Feng, H. Prediction of CRISPR–Cas9 on-target activity based on a hybrid neural network. Comput. Struct. Biotechnol. J. 27, 2098–2106 (2025).
Anthon, C., Corsi, G. I. & Gorodkin, J. CRISPRon/off: CRISPR/Cas9 on- and off-target gRNA design. Bioinformatics 38, 5437–5439 (2022).
Sun, J., Guo, J. & Liu, J. CRISPR-M: predicting sgRNA off-target effect using a multi-view deep learning network. PLoS Comput. Biol. 20, e1011972 (2024).
Zhang, Z., Lamson, A. R., Shelley, M. & Troyanskaya, O. Interpretable neural architecture search and transfer learning for understanding CRISPR–Cas9 off-target enzymatic reactions. Nat. Comput. Sci. 3, 1056–1066 (2023).
Anzalone, A. V., Koblan, L. W. & Liu, D. R. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 38, 824–844 (2020).
Park, J. & Kim, H. K. Prediction of base editing efficiencies and outcomes using DeepABE and DeepCBE. Methods Mol. Biol. 2606, 23–32 (2023).
Kim, N. et al. Deep learning models to predict the editing efficiencies and outcomes of diverse base editors. Nat. Biotechnol. 42, 484–497 (2024).
Silverstein, R. A. et al. Custom CRISPR–Cas9 PAM variants via scalable engineering and machine learning. Nature 643, 539–550 (2025).
Vieyra, F., Pindi, C., Lisi, G. P., Morzan, U. N., & Palermo, G. Design rules for expanding PAM compatibility in CRISPR-Cas9 from the VQR, VRER and EQR variants. J. Phys. Chem. B 129, 11949–11958 (2025).
Kleinstiver, B. P. et al. Engineered CRISPR–Cas9 nucleases with altered PAM specificities. Nature 523, 481–485 (2015).
Wang, Y., Li, Z. & Farimani, A. B. Graph neural networks for molecules. Preprint at arXiv https://doi.org/10.48550/arXiv.2209.05582 (2022).
Veličković, P. Everything is connected: graph neural networks. Curr. Opin. Struct. Biol. 79, 102538 (2023).
Park, J.-U. et al. Structures of the holo CRISPR RNA-guided transposon integration complex. Nature 613, 775–782 (2023).
Veličković, P. et al. Graph attention networks. Preprint at arXiv https://doi.org/10.48550/arXiv.1710.10903 (2017).
Pindi, C., Ahsan, M., Sinha, S. & Palermo, G. Graph attention neural networks reveal TnsC filament assembly in a CRISPR-associated transposon. Preprint at bioRxiv https://doi.org/10.1101/2025.06.17.659969 (2025).
Patel, A. C., Sinha, S. & Palermo, G. Graph theory approaches for molecular dynamics simulations. Q. Rev. Biophys. 57, e15 (2024).
Liu, H., Jian, Y., Zeng, C. & Zhao, Y. RNA–protein interaction prediction using network-guided deep learning. Commun. Biol. 8, 247 (2025).
Jiang, Y., Li, B., Xiong, J. & Liu, X. Graph-CRISPR: a gene editing efficiency prediction model based on graph neural network with integrated sequence and secondary structure feature extraction. Brief. Bioinform. 26, bbaf410 (2025).
Chen, G., Hou, L., Li, Z., Xie, B. & Liu, Y. A new strategy for Cas protein recognition based on graph neural networks and SMILES encoding. Sci. Rep. 15, 15236 (2025).
Abudayyeh, O. O. et al. RNA targeting with CRISPR–Cas13. Nature 550, 280–284 (2017).
East-Seletsky, A. et al. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 538, 270–273 (2016).
Knott, G. J. et al. Guide-bound structures of an RNA-targeting A-cleaving CRISPR–Cas13a enzyme. Nat. Struct. Mol. Biol. 24, 825–833 (2017).
Liu, L. et al. Two distant catalytic sites are responsible for C2c2 RNase activities. Cell 168, 121–134 (2017).
Tambe, A., East-Seletsky, A., Knott, G. J., Doudna, J. A. & O’Connell, M. R. RNA binding and HEPN-nuclease activation are decoupled in CRISPR–Cas13a. Cell Rep. 24, 1025–1036 (2018).
Fei, H. et al. Advancing protein evolution with inverse folding models integrating structural and evolutionary constraints. Cell 188, 4674–4692 (2025).
Dauparas, J. et al. Robust deep learning-based protein sequence design using ProteinMPNN. Science 378, 49–56 (2022).
Hsu, C. et al. Learning inverse folding from millions of predicted structures. In Proc. 39th International Conference on Machine Learning (eds Chaudhuri, K. et al.) 8946–8970 (PMLR, 2022).
Ruffolo, J. A. & Madani, A. Designing proteins with language models. Nat. Biotechnol. 42, 200–202 (2024).
Watson, J. L. et al. De novo design of protein structure and function with RFdiffusion. Nature 620, 1089–1100 (2023).
Ruffolo, J. A. et al. Design of highly functional genome editors by modelling CRISPR–Cas sequences. Nature 645, 518–525 (2025).
Madani, A. et al. Large language models generate functional protein sequences across diverse families. Nat. Biotechnol. 41, 1099–1106 (2023).
Qu, Y. et al. CRISPR-GPT for agentic automation of gene-editing experiments. Nat. Biomed. Eng. https://doi.org/10.1038/s41551-025-01463-z (2025).
Feng, Y. et al. Discovery of CRISPR–Cas12a clades using a large language model. Nat. Commun. 16, 7877 (2025).
Jiang, K. et al. Rapid in silico directed evolution by a protein language model with EVOLVEpro. Science 387, eadr6006 (2025).
Nguyen, E. et al. Sequence modeling and design from molecular to genome scale with Evo. Science 386, eado9336 (2024).
Taveneau, C. et al. De novo design of potent CRISPR–Cas13 inhibitors. Nat. Chem. Biol. https://doi.org/10.1038/s41589-025-02136-3 (2026).
Park, J.-C. et al. AI-generated MLH1 small binder improves prime editing efficiency. Cell 188, 5831–5846 (2025).
Pacesa, M. et al. One-shot design of functional protein binders with BindCraft. Nature 464, 483–492 (2025).
Lauko, A. et al. Computational design of serine hydrolases. Science 388, eadu2454 (2025).
O’Brien, A. R., Burgio, G. & Bauer, D. C. Domain-specific introduction to machine learning terminology, pitfalls and opportunities in CRISPR-based gene editing. Brief. Bioinform. 22, 308–314 (2021).
Fong, J. H. C. & Wong, A. S. L. Advancing CRISPR/Cas gene editing with machine learning. Curr. Opin. Biomed. Eng. 28, 100477 (2023).
Abbaszadeh, A. & Shahlai, A. Artificial intelligence for CRISPR guide RNA design: explainable models and off-target safety. Preprint at arXiv https://doi.org/10.48550/arXiv.2508.20130 (2025).
Xiang, X. et al. Enhancing CRISPR–Cas9 gRNA efficiency prediction by data integration and deep learning. Nat. Commun. 12, 3238 (2021).
Murdoch, W. J., Singh, C., Kumbier, K., Abbasi-Asl, R. & Yu, B. Definitions, methods, and applications in interpretable machine learning. Proc. Natl Acad. Sci. USA 116, 22071–22080 (2019).
Rudin, C. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nat. Mach. Intell. 1, 206–215 (2019).
Kim, M., Go, M., Kang, S.-H., Jeong, S. & Lim, K. Revolutionizing CRISPR technology with artificial intelligence. Exp. Mol. Med. 57, 1419–1431 (2025).
Dixit, S., Kumar, A., Srinivasan, K., Vincent, P. M. D. R. & Ramu Krishnan, N. Advancing genome editing with artificial intelligence: opportunities, challenges, and future directions. Front. Bioeng. Biotechnol. 11, 1335901 (2024).
Abbasi, A. F., Asim, M. N. & Dengel, A. Transitioning from wet lab to artificial intelligence: a systematic review of AI predictors in CRISPR. J. Transl. Med. 23, 153 (2025).
Liu, L. et al. The molecular architecture for RNA-guided RNA cleavage by Cas13a. Cell 170, 714–726 (2017).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996).
Acknowledgements
This material is based upon work supported by the National Institutes of Health (R01GM141329) and the National Science Foundation (CHE-2144823). G.P. also acknowledges support from the Sloan Foundation (FG-2023-20431) and the Camille and Henry Dreyfus Foundation (TC-24-063).
Author information
Authors and Affiliations
Contributions
G.P. conceptualized and drafted the review article. C.P. contributed to the writing. Both authors jointly developed the graphical representations.
Corresponding author
Ethics declarations
Competing interests
C.P. and G.P. are coinventors on patent applications filed by the University of California, Riverside.
Peer review
Peer review information
Nature Structural & Molecular Biology thanks Qi Liu, Li Yang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pindi, C., Palermo, G. Computation and deep-learning-driven advances in CRISPR genome editing. Nat Struct Mol Biol 33, 203–214 (2026). https://doi.org/10.1038/s41594-025-01739-7
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41594-025-01739-7