Abstract
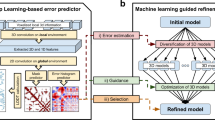
The trRosetta (transform-restrained Rosetta) server is a web-based platform for fast and accurate protein structure prediction, powered by deep learning and Rosetta. With the input of a protein’s amino acid sequence, a deep neural network is first used to predict the inter-residue geometries, including distance and orientations. The predicted geometries are then transformed as restraints to guide the structure prediction on the basis of direct energy minimization, which is implemented under the framework of Rosetta. The trRosetta server distinguishes itself from other similar structure prediction servers in terms of rapid and accurate de novo structure prediction. As an illustration, trRosetta was applied to two Pfam families with unknown structures, for which the predicted de novo models were estimated to have high accuracy. Nevertheless, to take advantage of homology modeling, homologous templates are used as additional inputs to the network automatically. In general, it takes ~1 h to predict the final structure for a typical protein with ~300 amino acids, using a maximum of 10 CPU cores in parallel in our cluster system. To enable large-scale structure modeling, a downloadable package of trRosetta with open-source codes is available as well. A detailed guidance for using the package is also available in this protocol. The server and the package are available at https://yanglab.nankai.edu.cn/trRosetta/ and https://yanglab.nankai.edu.cn/trRosetta/download/, respectively.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The example input and output files can be downloaded from https://yanglab.nankai.edu.cn/trRosetta.
Code availability
The trRosetta server and the standalone package are freely available at https://yanglab.nankai.edu.cn/trRosetta.
References
Moult, J., Fidelis, K., Kryshtafovych, A., Schwede, T. & Tramontano, A. Critical assessment of methods of protein structure prediction (CASP)-Round XII. Proteins 86, 7–15 (2018).
Kryshtafovych, A., Schwede, T., Topf, M., Fidelis, K. & Moult, J. Critical assessment of methods of protein structure prediction (CASP)-Round XIII. Proteins 87, 1011–1020 (2019).
Wang, S., Sun, S., Li, Z., Zhang, R. & Xu, J. Accurate de novo prediction of protein contact map by ultra-deep learning model. PLoS Comput. Biol. 13, e1005324 (2017).
Schaarschmidt, J., Monastyrskyy, B., Kryshtafovych, A. & Bonvin, A. Assessment of contact predictions in CASP12: co-evolution and deep learning coming of age. Proteins 86, 51–66 (2018).
Xu, J. Distance-based protein folding powered by deep learning. Proc. Natl Acad. Sci. USA 116, 16856 (2019).
Yang, J. et al. Improved protein structure prediction using predicted interresidue orientations. Proc. Natl Acad. Sci. USA 117, 1496–1503 (2020).
Senior, A. W. et al. Improved protein structure prediction using potentials from deep learning. Nature 577, 706–710 (2020).
Zhang, Y. & Skolnick, J. Scoring function for automated assessment of protein structure template quality. Proteins 57, 702–710 (2004).
Zemla, A. LGA: a method for finding 3D similarities in protein structures. Nucleic Acids Res. 31, 3370–3374 (2003).
Callaway, E. ‘It will change everything’: DeepMind’s AI makes gigantic leap in solving protein structures. Nature 588, 203–204 (2020).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Baek, M. et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science 373, 871–876 (2021).
Remmert, M., Biegert, A., Hauser, A. & Söding, J. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat. Methods 9, 173–175 (2012).
Söding, J. Protein homology detection by HMM–HMM comparison. Bioinformatics 21, 951–960 (2004).
Rohl, C. A., Strauss, C. E., Misura, K. M. & Baker, D. Protein structure prediction using Rosetta. Methods Enzymol. 383, 66–93 (2004).
Mistry, J. et al. Pfam: the protein families database in 2021. Nucleic Acids Res. 49, D412–D419 (2021).
Gao, S. H. et al. Res2Net: a new multi-scale backbone architecture. IEEE Trans. Pattern Anal. Mach. Intell. 43, 652–662 (2021).
Berman, H. M. et al. The Protein Data Bank. Nucleic Acids Res. 28, 235–242 (2000).
Pleiner, T. et al. Structural basis for membrane insertion by the human ER membrane protein complex. Science 369, 433–436 (2020).
O’Donnell, J. P. et al. The architecture of EMC reveals a path for membrane protein insertion. Elife 9, e57887 (2020).
Mashtalir, N. et al. A structural model of the endogenous human BAF complex informs disease mechanisms. Cell 183, 802–817.e24 (2020).
Banerjee, A. K. et al. SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses. Cell 183, 1325–1339.e21 (2020).
Gordon, D. E. et al. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science 370, eabe9403 (2020).
Anishchenko, I., Chidyausiku, T. M., Ovchinnikov, S., Pellock, S. J. & Baker, D. De novo protein design by deep network hallucination. Preprint at https://doi.org/10.1101/2020.07.22.211482 (2020).
Xu, J. & Zhang, Y. How significant is a protein structure similarity with TM-score = 0.5? Bioinformatics 26, 889–895 (2010).
Eddy, S. R. Profile hidden Markov models. Bioinformatics 14, 755–763 (1998).
Ovchinnikov, S. et al. Protein structure determination using metagenome sequence data. Science 355, 294–298 (2017).
Wu, Q. et al. Protein contact prediction using metagenome sequence data and residual neural networks. Bioinformatics 36, 41–48 (2020).
Dong, R., Pan, S., Peng, Z., Zhang, Y. & Yang, J. mTM-align: a server for fast protein structure database search and multiple protein structure alignment. Nucleic Acids Res. 46, W380–W386 (2018).
Rego, N. & Koes, D. 3Dmol.js: molecular visualization with WebGL. Bioinformatics 31, 1322–1324 (2014).
Yang, J. et al. The I-TASSER Suite: protein structure and function prediction. Nat. Methods 12, 7–8 (2015).
Ward, J. J., McGuffin, L. J., Bryson, K., Buxton, B. F. & Jones, D. T. The DISOPRED server for the prediction of protein disorder. Bioinformatics 20, 2138–2139 (2004).
Webb, B. & Sali, A. Protein structure modeling with MODELLER. in Protein Structure Prediction (ed. Kihara, D.) 1–15 (Springer, 2014).
Ju, F. et al. CopulaNet: learning residue co-evolution directly from multiple sequence alignment for protein structure prediction. Nat. Commun. 12, 2535 (2021).
Wang, Z., Eickholt, J. & Cheng, J. MULTICOM: a multi-level combination approach to protein structure prediction and its assessments in CASP8. Bioinformatics 26, 882–888 (2010).
Kim, D. E., Chivian, D. & Baker, D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 32, W526–W531 (2004).
Mao, W., Ding, W., Xing, Y. & Gong, H. AmoebaContact and GDFold as a pipeline for rapid de novo protein structure prediction. Nat. Mach. Intell. 2, 25–33 (2020).
Zheng, L. et al. Combining deep learning enhanced hybrid potential energy for template-based modelling. CASP14 Abstracts https://predictioncenter.org/casp14/doc/CASP14_Abstracts.pdf (2020).
Greener, J. G., Kandathil, S. M. & Jones, D. T. Deep learning extends de novo protein modelling coverage of genomes using iteratively predicted structural constraints. Nat. Commun. 10, 3977 (2019).
Xu, D. & Zhang, Y. Ab initio protein structure assembly using continuous structure fragments and optimized knowledge-based force field. Proteins 80, 1715–1735 (2012).
Jin, S. et al. AWSEM-Suite: a protein structure prediction server based on template-guided, coevolutionary-enhanced optimized folding landscapes. Nucleic Acids Res. 48(W1), W25–W30 (2020).
Ko, J., Park, H., Heo, L. & Seok, C. GalaxyWEB server for protein structure prediction and refinement. Nucleic Acids Res. 40, W294–W297 (2012).
Källberg, M. et al. Template-based protein structure modeling using the RaptorX web server. Nat. Protoc. 7, 1511–1522 (2012).
Yang, Y., Faraggi, E., Zhao, H. & Zhou, Y. Improving protein fold recognition and template-based modeling by employing probabilistic-based matching between predicted one-dimensional structural properties of query and corresponding native properties of templates. Bioinformatics 27, 2076–2082 (2011).
Kelley, L. A. & Sternberg, M. J. E. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371 (2009).
Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40 (2008).
Wu, S. & Zhang, Y. LOMETS: a local meta-threading-server for protein structure prediction. Nucleic Acids Res. 35, 3375–3382 (2007).
Söding, J., Biegert, A. & Lupas, A. N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248 (2005).
Guex, N. & Peitsch, M. C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–2723 (1997).
Yan, Y., Tao, H., He, J. & Huang, S.-Y. The HDOCK server for integrated protein–protein docking. Nat. Protoc. 15, 1829–1852 (2020).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (NSFC 11871290 and 61873185), Fok Ying-Tong Education Foundation (161003) and KLMDASR.
Author information
Authors and Affiliations
Contributions
J.Y. conceived and supervised the project. Z.D., H.S., W.W., L.Y., H.W., Z.P. and J.Y. designed and performed the experiments. Z.D., J.Y., I.A. and D.B. wrote the manuscript. All authors revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Julia Leman and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference using this protocol
Yang, J. et al. Proc. Natl Acad. Sci. USA 117, 1496–1503 (2020): https://www.pnas.org/content/117/3/1496
Supplementary information
Supplementary Information
Supplementary Tables 1–4 and Supplementary Figs. 1 and 2.
Rights and permissions
About this article
Cite this article
Du, Z., Su, H., Wang, W. et al. The trRosetta server for fast and accurate protein structure prediction. Nat Protoc 16, 5634–5651 (2021). https://doi.org/10.1038/s41596-021-00628-9
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41596-021-00628-9
This article is cited by
-
Structural and functional analysis of the accessory gene regulators of Staphylococcus aureus and Staphylococcus epidermidis: an in Silico approach
BMC Microbiology (2025)
-
Damaging non-synonymous mutations in the extracellular domain of HER2 potentially alter the efficacy of Herceptin-mediated breast cancer therapy
Egyptian Journal of Medical Human Genetics (2025)
-
Immunoinformatics-Based development of a Multi-Epitope vaccine candidate targeting coinfection by Klebsiella pneumoniae and Acinetobacter baumannii
BMC Infectious Diseases (2025)
-
UniAMP: enhancing AMP prediction using deep neural networks with inferred information of peptides
BMC Bioinformatics (2025)
-
MEGDTA: multi-modal drug-target affinity prediction based on protein three-dimensional structure and ensemble graph neural network
BMC Genomics (2025)