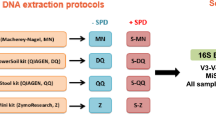
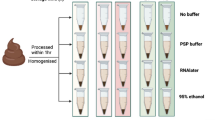
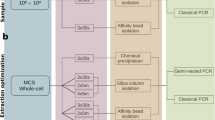
Abstract
Measurements of prokaryotic absolute abundance can provide important insights into human gut microbiome biology and correct misinterpretations of relative abundance data. Despite the existence of several relatively well-established methods for making these measurements, most microbiome studies do not report absolute abundance. To enable researchers equipped with standard molecular biology capabilities to incorporate absolute quantification into their microbiome studies, we present a detailed, step-by-step protocol for rigorous and reproducible quantification of prokaryotic concentration in stool samples. We include methods for measuring stool sample moisture content, quantifying the concentration of the 16S rRNA prokaryotic marker gene by qPCR or digital droplet PCR (ddPCR) and analyzing the resulting data. We also highlight and provide strategies to overcome common pitfalls of the quantification method, such as 16S rRNA gene contamination. The final output of this approach is 16S rRNA copies per wet or dry gram of stool. In cases where samples have matched metagenomic sequencing information, data can be converted into absolute concentration of prokaryotes and taxon-specific absolute concentrations. To enable researchers to choose the appropriate method for their specific applications, we also compare and contrast our qPCR and ddPCR methods. In 4 days, ~80 samples can be taken from frozen stool to absolute concentration by using qPCR or ddPCR without the need for resequencing. Overall, this protocol provides a sensitive and straightforward way to measure the absolute concentration of prokaryotes in human gut microbiome samples stored with or without preservative.
Key points
-
This protocol quantifies prokaryotic concentration in stool samples by measuring 16S rRNA gene concentration with qPCR or ddPCR and correcting for stool sample moisture content.
-
Absolute prokaryotic quantification can provide further insights into microbiome biology and correct misinterpretations of the relative abundance data most commonly reported in microbiome studies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Source data for figures are available at https://github.com/bhattlab/absolute-abundance-16s.
Code availability
Data analysis scripts and notebooks are available at https://github.com/bhattlab/absolute-abundance-16s.
References
Hagan, T. et al. Antibiotics-driven gut microbiome perturbation alters immunity to vaccines in humans. Cell 178, 1313–1328.e13 (2019).
Jian, C., Luukkonen, P., Yki-Järvinen, H., Salonen, A. & Korpela, K. Quantitative PCR provides a simple and accessible method for quantitative microbiota profiling. PLoS ONE 15, e0227285 (2020).
Rao, C. et al. Multi-kingdom ecological drivers of microbiota assembly in preterm infants. Nature 591, 633–638 (2021).
Rolling, T. et al. Haematopoietic cell transplantation outcomes are linked to intestinal mycobiota dynamics and an expansion of Candida parapsilosis complex species. Nat. Microbiol. 6, 1505–1515 (2021).
Vandeputte, D. et al. Quantitative microbiome profiling links gut community variation to microbial load. Nature 551, 507–511 (2017).
Barlow, J. T., Bogatyrev, S. R. & Ismagilov, R. F. A quantitative sequencing framework for absolute abundance measurements of mucosal and lumenal microbial communities. Nat. Commun. 11, 2590 (2020).
Nadkarni, M. A., Martin, F. E., Jacques, N. A. & Hunter, N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 148, 257–266 (2002).
Langenfeld, K., Chin, K., Roy, A., Wigginton, K. & Duhaime, M. B. Comparison of ultrafiltration and iron chloride flocculation in the preparation of aquatic viromes from contrasting sample types. PeerJ 9, e11111 (2021).
Tkacz, A., Hortala, M. & Poole, P. S. Absolute quantitation of microbiota abundance in environmental samples. Microbiome 6, 110 (2018).
Harris, K. A. & Brown, J. R. Diagnostic yield of broad-range 16s rRNA gene PCR varies by sample type and is improved by the addition of qPCR panels targeting the most common causative organisms. J. Med. Microbiol. 71, 001633 (2022).
Zemb, O. et al. Absolute quantitation of microbes using 16S rRNA gene metabarcoding: a rapid normalization of relative abundances by quantitative PCR targeting a 16S rRNA gene spike-in standard. Microbiologyopen 9, e977 (2020).
Liao, C. et al. Oral bacteria relative abundance in faeces increases due to gut microbiota depletion and is linked with patient outcomes. Nat. Microbiol. 9, 1555–1565 (2024).
Yao, B. et al. Quantification and characterization of mouse and human tissue-resident microbiota by qPCR and 16S sequencing. STAR Protoc. 3, 101765 (2022).
Sharpton, T. J. An introduction to the analysis of shotgun metagenomic data. Front. Plant Sci. 5, 209 (2014).
Saheb Kashaf, S., Almeida, A., Segre, J. A. & Finn, R. D. Recovering prokaryotic genomes from host-associated, short-read shotgun metagenomic sequencing data. Nat. Protoc. 16, 2520–2541 (2021).
Quince, C., Walker, A. W., Simpson, J. T., Loman, N. J. & Segata, N. Shotgun metagenomics, from sampling to analysis. Nat. Biotechnol. 35, 833–844 (2017).
Jin, J., Yamamoto, R. & Shiroguchi, K. High-throughput identification and quantification of bacterial cells in the microbiota based on 16S rRNA sequencing with single-base accuracy using BarBIQ. Nat. Protoc. 19, 207–239 (2024).
Tourlousse, D. M. et al. Synthetic spike-in standards for high-throughput 16S rRNA gene amplicon sequencing. Nucleic Acids Res. 45, e23 (2017).
Smets, W. et al. A method for simultaneous measurement of soil bacterial abundances and community composition via 16S rRNA gene sequencing. Soil Biol. Biochem. 96, 145–151 (2016).
Prest, E. I., Hammes, F., Kötzsch, S., van Loosdrecht, M. C. M. & Vrouwenvelder, J. S. Monitoring microbiological changes in drinking water systems using a fast and reproducible flow cytometric method. Water Res. 47, 7131–7142 (2013).
Bezirtzoglou, E., Tsiotsias, A. & Welling, G. W. Microbiota profile in feces of breast- and formula-fed newborns by using fluorescence in situ hybridization (FISH). Anaerobe 17, 478–482 (2011).
Vieira-Silva, S. et al. Quantitative microbiome profiling disentangles inflammation- and bile duct obstruction-associated microbiota alterations across PSC/IBD diagnoses. Nat. Microbiol. 4, 1826–1831 (2019).
van de Velde, C. C. et al. Fast quantification of gut bacterial species in cocultures using flow cytometry and supervised classification. ISME Commun. 2, 40 (2022).
Batani, G., Bayer, K., Böge, J., Hentschel, U. & Thomas, T. Fluorescence in situ hybridization (FISH) and cell sorting of living bacteria. Sci. Rep. 9, 18618 (2019).
Christensen, H., Hansen, M. & Sorensen, J. Counting and size classification of active soil bacteria by fluorescence in situ hybridization with an rRNA oligonucleotide probe. Appl. Environ. Microbiol. 65, 1753–1761 (1999).
Sze, M. A., Abbasi, M., Hogg, J. C. & Sin, D. D. A comparison between droplet digital and quantitative PCR in the analysis of bacterial 16S load in lung tissue samples from control and COPD GOLD 2. PLoS ONE 9, e110351 (2014).
Brankatschk, R., Bodenhausen, N., Zeyer, J. & Bürgmann, H. Simple absolute quantification method correcting for quantitative PCR efficiency variations for microbial community samples. Appl. Environ. Microbiol. 78, 4481–4489 (2012).
Gobert, G. et al. Droplet digital PCR improves absolute quantification of viable lactic acid bacteria in faecal samples. J. Microbiol. Methods 148, 64–73 (2018).
Ziegler, I., Lindström, S., Källgren, M., Strålin, K. & Mölling, P. 16S rDNA droplet digital PCR for monitoring bacterial DNAemia in bloodstream infections. PLoS ONE 14, e0224656 (2019).
Galazzo, G. et al. How to count our microbes? The effect of different quantitative microbiome profiling approaches. Front. Cell. Infect. Microbiol. 10, 403 (2020).
Wang, X., Howe, S., Deng, F. & Zhao, J. Current applications of absolute bacterial quantification in microbiome studies and decision-making regarding different biological questions. Microorganisms 9, 1797 (2021).
Bustin, S. A. et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622 (2009).
dMIQE Group, Huggett, J. F. The digital MIQE guidelines update: minimum information for publication of quantitative digital PCR experiments for 2020. Clin. Chem. 66, 1012–1029 (2020).
Biorad. Droplet DigitalTM PCR Applications Guide https://www.bio-rad.com/webroot/web/pdf/lsr/literature/Bulletin_6407.pdf (2018).
Yuan, S., Cohen, D. B., Ravel, J., Abdo, Z. & Forney, L. J. Evaluation of methods for the extraction and purification of DNA from the human microbiome. PLoS ONE 7, e33865 (2012).
Brooks, J. P. et al. The truth about metagenomics: quantifying and counteracting bias in 16S rRNA studies. BMC Microbiol. 15, 66 (2015).
Wesolowska-Andersen, A. et al. Choice of bacterial DNA extraction method from fecal material influences community structure as evaluated by metagenomic analysis. Microbiome 2, 19 (2014).
Greathouse, K. L., Sinha, R. & Vogtmann, E. DNA extraction for human microbiome studies: the issue of standardization. Genome Biol. 20, 212 (2019).
Maghini, D. G. et al. Quantifying bias introduced by sample collection in relative and absolute microbiome measurements. Nat. Biotechnol. 42, 328–338 (2024).
Kralj, J. et al. Reference Material 8376 Microbial Pathogen DNA Standards for Detection and Identification https://nvlpubs.nist.gov/nistpubs/SpecialPublications/NIST.SP.260-225.pdf (2022).
Costea, P. I. et al. Towards standards for human fecal sample processing in metagenomic studies. Nat. Biotechnol. 35, 1069–1076 (2017).
Stoddard, S. F., Smith, B. J., Hein, R., Roller, B. R. K. & Schmidt, T. M. rrnDB: improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Res. 43, D593–D598 (2015).
Li, F. et al. Highly accurate and sensitive absolute quantification of bacterial strains in human fecal samples. Microbiome 12, 168 (2024).
Carra, E. et al. A probe-based qPCR method, targeting 16S rRNA gene, for the quantification of Paenibacillus larvae spores in powdered sugar samples. Appl. Sci. 12, 9895 (2022).
Yang, Y.-W. et al. Use of 16S rRNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in mouse feces. Appl. Environ. Microbiol. 81, 6749–6756 (2015).
Lloyd-Price, J. et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569, 655–662 (2019).
Ning, L. et al. Microbiome and metabolome features in inflammatory bowel disease via multi-omics integration analyses across cohorts. Nat. Commun. 14, 7135 (2023).
Konstantinidis, T. et al. Effects of antibiotics upon the gut microbiome: a review of the literature. Biomedicines 8, 502 (2020).
Willing, B. P., Russell, S. L. & Finlay, B. B. Shifting the balance: antibiotic effects on host-microbiota mutualism. Nat. Rev. Microbiol. 9, 233–243 (2011).
Papanicolas, L. E. et al. Conventional myelosuppressive chemotherapy for non-haematological malignancy disrupts the intestinal microbiome. BMC Cancer 21, 591 (2021).
Contijoch, E. J. et al. Gut microbiota density influences host physiology and is shaped by host and microbial factors. Elife 8, e40553 (2019).
Paone, P. et al. Prebiotic oligofructose protects against high-fat diet-induced obesity by changing the gut microbiota, intestinal mucus production, glycosylation and secretion. Gut Microbes 14, 2152307 (2022).
Sinha, R., Abnet, C. C., White, O., Knight, R. & Huttenhower, C. The microbiome quality control project: baseline study design and future directions. Genome Biol. 16, 276 (2015).
Nearing, J. T., Comeau, A. M. & Langille, M. G. I. Identifying biases and their potential solutions in human microbiome studies. Microbiome 9, 113 (2021).
Pollock, J., Glendinning, L., Wisedchanwet, T. & Watson, M. The madness of microbiome: attempting to find consensus ‘best practice’ for 16S microbiome studies. Appl. Environ. Microbiol. 84, e02627-17 (2018).
Bartolomaeus, T. U. P. et al. Quantifying technical confounders in microbiome studies. Cardiovasc. Res. 117, 863–875 (2021).
Bahl, M. I., Bergström, A. & Licht, T. R. Freezing fecal samples prior to DNA extraction affects the Firmicutes to Bacteroidetes ratio determined by downstream quantitative PCR analysis. FEMS Microbiol. Lett. 329, 193–197 (2012).
Broussard, I. M. & Kahwaji, C. I. Universal precautions. StatPearls [Internet] https://www.ncbi.nlm.nih.gov/books/NBK470223/ (updated 29 July 2023).
Acknowledgements
We thank all members of the Bhatt Laboratory for discussions and experimental advice; K. Langenfeld for guidance on the ddPCR assay; A. Boehm for access to the ddPCR workflow equipment; D. Solow-Cordero and S. Sim for assistance in using the Stanford High-Throughput Bioscience Center and Stanford Functional Genomics Facility, which is supported by NIH Shared Instrumentation Grants S10RR019513, S10RR026338, S10OD025004 and S10OD026899 and by an anonymous donation; L. Nichols for guidance on flow cytometry microbial counting methods; and J. Axel for feedback on the manuscript, input on the figures and testing of the scripts. A.S.B. was supported by National Institutes of Health R01 AI148623 and R01 AI143757, a Distinguished Investigator Award from the Paul Allen Foundation and a Convergence grant from the Stand Up 2 Cancer Foundation. The Bhatt Laboratory is also supported by The Phil & Penny Knight Initiative for Brain Resilience at the Wu Tsai Neurosciences Institute, Stanford University. B.D. was supported by the Stanford Medical Scholars Fellowship Program, Stanford Berg Scholars Program and a Physician Scientist Institutional Award (PSIA) from the Burroughs Wellcome Fund. M.D. was supported by NIH Cellular and Molecular Biology Training Program Training Grant T32GM007276. D.G.M. was supported by the Stanford Gerald J. Lieberman Fellowship and the NIH Fogarty Global Health Equity Scholars Program (NIH FIC D43TW010540). Figures 1–4 were created with BioRender.com.
Author information
Authors and Affiliations
Contributions
B.D., G.Z.M.R., M.D., A.N. and D.G.M. refined the approach for the qPCR and ddPCR assays. A.S.B., B.D., G.Z.M.R. and M.D. conceptualized the qPCR versus ddPCR comparison experiment. B.D., G.Z.M.R. and M.D. performed extraction, qPCR and ddPCR. B.D., G.Z.M.R. and M.D. carried out analysis and generated figures. B.D., G.Z.M.R., M.D. and A.S.B. wrote the manuscript with input from A.N. and D.G.M. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key reference
Maghini, D. G. et al. Nat. Biotechnol. 42, 328–338 (2024): https://doi.org/10.1038/s41587-023-01754-3
Supplementary information
Supplementary Information
Supplementary Methods 1–4, Supplementary Figures 1–4
Supplementary Tables 1–3
Supplementary Tables 1–3
Supplementary Video 1
Video showing how to biopsy punch samples
Supplementary Video 2
Video showing how to vortex a ddPCR reaction plate before droplet generation
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Doyle, B., Reynolds, G.Z.M., Dvorak, M. et al. Absolute quantification of prokaryotes in the microbiome by 16S rRNA qPCR or ddPCR. Nat Protoc (2025). https://doi.org/10.1038/s41596-025-01165-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41596-025-01165-5