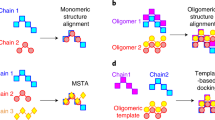
Abstract
With the success of structural biology and the advancements in deep-learning-based structure predictions, rapid and accurate structural comparisons among macromolecular structures have become increasingly important in structural bioinformatics. US-align is a highly efficient, versatile, open-source program for sequential and nonsequential structure comparisons of proteins, RNAs and DNAs in pairwise and multiple alignment forms and applicable to both monomeric and multimeric complex structures. The core algorithm of US-align is built on a highly optimized, iterative superimposition and dynamic programming alignment process, guided with a unified and sequence length-independent scoring function, TM-score. The unique design of US-align not only ensures its high accuracy and speed compared with other state-of-the-art methods designed for specific alignment tasks but also makes it the only protocol that can be applied to multiple alignment tasks and allow a structural comparison across different molecular types, the latter of which is critical for template-based heteromolecular structure prediction and function annotations. Here we describe how to install and effectively utilize US-align as a command line tool, as an online web server, and as a plugin to commonly used molecular graphic systems such as PyMOL. US-align installation takes a few minutes to setup, while the actual alignment implementation can be completed typically within 1 s.
Key points
-
US-align is a highly efficient, versatile and open-source program for the sequential and nonsequential structure comparisons of proteins, RNAs and DNAs in pairwise and multiple alignment forms. It is applicable to both monomeric and multimeric complex structures.
-
Its unique design ensures high accuracy and speed compared with other state-of-the-art methods designed for specific alignment tasks and enables it to be applied to multiple alignment tasks, allowing structural comparison across different molecular types.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
PDB files for PDB IDs 101m, 1mba, 4jhm, 4iaj, 1eh1, 1evv, 6jxm, 3am1, 1ajk and 2ayh are available through https://rcsb.org. The non-redundant PDB library (that is, the I-TASSER template library) is updated on a weekly basis and available at https://zhanggroup.org/library/PDB.tar.bz2. Files to demonstrate the ‘-TMscore’ option of US-align are available at https://zhanggroup.org/TM-score/help.zip.
Code availability
The US-align web server and source code are available at https://zhanggroup.org/US-align/. The code in this Protocol has been peer reviewed.
References
Zhang, Y. & Skolnick, J. Scoring function for automated assessment of protein structure template quality. Proteins 57, 702–710 (2004).
Xu, J. R. & Zhang, Y. How significant is a protein structure similarity with TM-score = 0.5? Bioinformatics 26, 889–895 (2010).
Gong, S., Zhang, C. & Zhang, Y. RNA-align: quick and accurate alignment of RNA 3D structures based on size-independent TM-scoreRNA. Bioinformatics 35, 4459–4461 (2019).
Kabsch, W. A solution for the best rotation to relate two sets of vectors. Acta Crystallogr. A 32, 922–923 (1976).
Zhang, Y. & Skolnick, J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 33, 2302–2309 (2005).
Holm, L. & Sander, C. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 20, 478–480 (1995).
Shindyalov, I. N. & Bourne, P. E. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 11, 739–747 (1998).
Krissinel, E. & Henrick, K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D 60, 2256–2268 (2004).
Yang, Y., Zhan, J., Zhao, H. & Zhou, Y. A new size-independent score for pairwise protein structure alignment and its application to structure classification and nucleic-acid binding prediction. Proteins 80, 2080–2088 (2012).
Ge, P. & Zhang, S. STAR3D: a stack-based RNA 3D structural alignment tool. Nucleic Acids Res. 43, e137 (2015).
Dror, O., Nussinov, R. & Wolfson, H. J. The ARTS web server for aligning RNA tertiary structures. Nucleic Acids Res. 34, W412–W415 (2006).
Zheng, J., Xie, J., Hong, X. & Liu, S. RMalign: an RNA structural alignment tool based on a novel scoring function RMscore. BMC Genomics 20, 276 (2019).
Nguyen, M. N., Sim, A. Y., Wan, Y., Madhusudhan, M. S. & Verma, C. Topology independent comparison of RNA 3D structures using the CLICK algorithm. Nucleic Acids Res. 45, e5 (2017).
Mukherjee, S. & Zhang, Y. MM-align: a quick algorithm for aligning multiple-chain protein complex structures using iterative dynamic programming. Nucleic Acids Res. 37, e83 (2009).
Minami, S., Sawada, K. & Chikenji, G. MICAN: a protein structure alignment algorithm that can handle multiple-chains, inverse alignments, Cα only models, alternative alignments, and non-sequential alignments. BMC Bioinformatics 14, 24 (2013).
Dong, R., Peng, Z., Zhang, Y. & Yang, J. mTM-align: an algorithm for fast and accurate multiple protein structure alignment. Bioinformatics 34, 1719–1725 (2018).
Konagurthu, A. S., Whisstock, J. C., Stuckey, P. J. & Lesk, A. M. MUSTANG: a multiple structural alignment algorithm. Proteins 64, 559–574 (2006).
Menke, M., Berger, B. & Cowen, L. Matt: local flexibility aids protein multiple structure alignment. PLoS Comput. Biol. 4, e10 (2008).
Zhang, C., Shine, M., Pyle, A. M. & Zhang, Y. US-align: universal structure alignments of proteins, nucleic acids, and macromolecular complexes. Nat. Methods 19, 1109–1115 (2022).
Zhang, C. & Pyle, A. M. A unified approach to sequential and non-sequential structure alignment of proteins, RNAs, and DNAs. iScience https://doi.org/10.1016/j.isci.2022.105218 (2022).
Das, R. et al. Assessment of three-dimensional RNA structure prediction in CASP15. Proteins 91, 1747–1770 (2023).
Studer, G., Tauriello, G. & Schwede, T. Assessment of the assessment—all about complexes. Proteins 91, 1850–1860 (2023).
Zhang, C., Freddolino, P. L. & Zhang, Y. COFACTOR: improved protein function prediction by combining structure, sequence and protein–protein interaction information. Nucleic Acids Res. 45, W291–W299 (2017).
Zhang, C. X., Zheng, W., Freddolino, P. L. & Zhang, Y. MetaGO: predicting gene ontology of non-homologous proteins through low-resolution protein structure prediction and protein protein network mapping. J. Mol. Biol. 430, 2256–2265 (2018).
Laskowski, R. A., Watson, J. D. & Thornton, J. M. ProFunc: a server for predicting protein function from 3D structure. Nucleic Acids Res. 33, W89–W93 (2005).
Yang, J., Roy, A. & Zhang, Y. Protein–ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics 29, 2588–2595 (2013).
Roy, A. & Zhang, Y. Recognizing protein-ligand binding sites by global structural alignment and local geometry refinement. Structure 20, 987–997 (2012).
Brylinski, M. & Skolnick, J. A threading-based method (FINDSITE) for ligand-binding site prediction and functional annotation. Proc. Natl Acad. Sci. USA 105, 129–134 (2008).
Zhang, W., Bell, E. W., Yin, M. & Zhang, Y. EDock: blind protein-ligand docking by replica-exchange monte carlo simulation. J. Cheminform. 12, 37 (2020).
Baspinar, A., Cukuroglu, E., Nussinov, R., Keskin, O. & Gursoy, A. PRISM: a web server and repository for prediction of protein–protein interactions and modeling their 3D complexes. Nucleic Acids Res. 42, W285–W289 (2014).
Guerler, A., Govindarajoo, B. & Zhang, Y. Mapping monomeric threading to protein-protein structure prediction. J. Chem. Inf. Model. 53, 717–725 (2013).
Zhou, X. G., Hu, J., Zhang, C. X., Zhang, G. J. & Zhang, Y. Assembling multidomain protein structures through analogous global structural alignments. Proc. Natl Acad. Sci. USA 116, 15930–15938 (2019).
Zhou, X. et al. I-TASSER-MTD: a deep-learning-based platform for multi-domain protein structure and function prediction. Nat. Protoc. 17, 2326–2353 (2022).
Pearce, R., Huang, X. Q., Setiawan, D. & Zhang, Y. EvoDesign: designing protein–protein binding interactions using evolutionary interface profiles in conjunction with an optimized physical energy function. J. Mol. Biol. 431, 2467–2476 (2019).
Zhang, J., Liang, Y. & Zhang, Y. Atomic-level protein structure refinement using fragment-guided molecular dynamics conformation sampling. Structure 19, 1784–1795 (2011).
Liu, Z., Zhang, C., Zhang, Q., Zhang, Y. & Yu, D.-J. TM-search: an efficient and effective tool for Protein Structure Database search. J. Chem. Inf. Model. 64, 1043–1049 (2024).
Zhu, Y., Tong, C., Zhao, Z. & Lu, Z. MineProt: a stand-alone server for structural proteome curation. Database https://doi.org/10.1093/database/baad059 (2023).
Greene, L. H. et al. The CATH domain structure database: new protocols and classification levels give a more comprehensive resource for exploring evolution. Nucleic Acids Res 35, D291–D297 (2007).
Zhang, C., Zhang, X., Freddolino, P. L. & Zhang, Y. BioLiP2: an updated structure database for biologically relevant ligand–protein interactions. Nucleic Acids Res. https://doi.org/10.1093/nar/gkad630 (2023).
van Kempen, M. et al. Fast and accurate protein structure search with Foldseek. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-01773-0 (2023).
Yang, J. M. & Tung, C. H. Protein structure database search and evolutionary classification. Nucleic Acids Res. 34, 3646–3659 (2006).
Li, Z., Jaroszewski, L., Iyer, M., Sedova, M. & Godzik, A. FATCAT 2.0: towards a better understanding of the structural diversity of proteins. Nucleic Acids Res. 48, W60–W64 (2020).
Meng, E. C. et al. UCSF ChimeraX: tools for structure building and analysis. Protein Sci. 32, e4792 (2023).
Selmer, M., Al-Karadaghi, S., Hirokawa, G., Kaji, A. & Liljas, A. Crystal structure of Thermotoga maritima ribosome recycling factor: a tRNA mimic. Science 286, 2349–2352 (1999).
Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. & Sussman, J. L. JSmol and the next-generation web-based representation of 3D molecular structure as applied to Proteopedia. Isr. J. Chem. 53, 207–216 (2013).
DeLano, W. L. Pymol: an open-source molecular graphics tool. CCP4 Newsletter Pro. Crystallogr. 40, 82–92 (2002).
Sayle, R. A. & Milnerwhite, E. J. Rasmol—biomolecular graphics for all. Trends Biochem. Sci. 20, 374–376 (1995).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996).
Hanson, R. M. Jmol—a paradigm shift in crystallographic visualization. J. Appl. Crystallogr. 43, 1250–1260 (2010).
Acknowledgements
The authors thank X. Wei and Z. Perry for technical assistances to compile US-align for Mac OS. This work used the Advanced Cyberinfrastructure Coordination Ecosystem: Services and Support (ACCESS) program, which is supported by National Science Foundation (2138259, 2138286, 2138307, 2137603 and 2138296). This work is supported in part by the National Institute of Allergy and Infectious Diseases (AI134678 to L.F. and Y.Z.), Ministry of Education (T1 251RES2309 to Y.Z.), and the National University of Singapore startup grants (WBS #A-8001129-00-00, #A-0010130-15-00, #A-8000974-00-00 to Y.Z.). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the paper.
Author information
Authors and Affiliations
Contributions
Y.Z. conceived the project. C.Z. developed the program and prepared the server. C.Z., L.F. and Y.Z. drafted the manuscript and approved the final version. All collaborators of this study who fulfilled the criteria for authorship inclusion required by Nature Portfolio journals have been included as authors. Roles and responsibilities were agreed among collaborators ahead of the research. Local and regional research relevant to this study is referenced.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks John Dzimianski and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key references
Zhang, C. et al. Nat. Methods 19, 1109–1115 (2022): https://doi.org/10.1038/s41592-022-01585-1
Zhang, Y. et al. Nucleic Acids Res. 33, 2302–2309 (2005): https://doi.org/10.1093/nar/gki524
Zhang, Y. et al. Proteins 57, 702–710 (2004): https://doi.org/10.1002/prot.20264
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, C., Freddolino, L. & Zhang, Y. A graphic and command line protocol for quick and accurate comparisons of protein and nucleic acid structures with US-align. Nat Protoc (2025). https://doi.org/10.1038/s41596-025-01189-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41596-025-01189-x