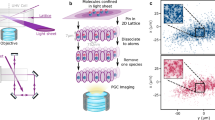
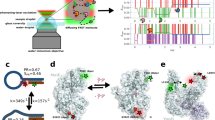
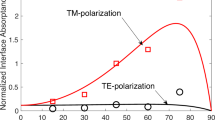
Abstract
Mass photometry (MP) has emerged as a powerful approach to study quaternary biomolecular structure, dynamics and interactions. The capabilities of the method ultimately hinge on the ability to accurately measure the tiny optical contrast generated by individual molecules at a glass–water interface, which enables mass-resolved quantification of biomolecular mixtures. Ideally, this capability is limited only by photon shot noise, but in practice depends on additional parameters and details of the assay. Here, we focus on the key factors affecting MP performance and present simple steps that can be taken to achieve optimal MP measurements in terms of mass resolution, quantitative detection limit, reproducibility and analyte concentration range without compromising the speed and simplicity of the technique. Each sample takes <10 min to analyse, with an additionial 2 h if amination of the glass surface is desired.
Key points
-
The protocol optimizes the interplay between surface and sample preparation, experimental approach and analysis parameters to maximize sensitivity and resolution when performing MP measurements.
-
All steps can be implemented into MP workflows with less than a day’s work, and many require only simple considerations at the time of the measurement.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout









Similar content being viewed by others
Data availability
The datasets for all figures are available at https://doi.org/10.5281/zenodo.15800728. All data are shared under the CC BBY 4.0 license.
References
Young, G. et al. Quantitative mass imaging of single biological macromolecules. Science 360, 423–427 (2018).
Asor, R., Loewenthal, D., van Wee, R., Benesch, J. L. P. & Kukura, P. Mass photometry. Annu. Rev. Biophys. 54, 379–399 (2025).
Asor, R. et al. Oligomerization-driven avidity correlates with SARS-CoV-2 cellular binding and inhibition. Proc. Natl Acad. Sci. USA 121, e2403260121 (2024).
Olerinyova, A. et al. Mass photometry of membrane proteins. Chem. 7, 224–236 (2021).
Sonn-Segev, A. et al. Quantifying the heterogeneity of macromolecular machines by mass photometry. Nat. Commun. 11, 1772 (2020).
Fineberg, A., Surrey, T. & Kukura, P. Quantifying the monomer–dimer equilibrium of tubulin with mass photometry. J. Mol. Biol. 432, 6168–6172 (2020).
Zinder, J. C. et al. Shelterin is a dimeric complex with extensive structural heterogeneity. Proc. Natl Acad. Sci. USA 119, e2201662119 (2022).
Kissling, V. M. et al. Mre11-Rad50 oligomerization promotes DNA double-strand break repair. Nat. Commun. 13, 2374 (2022).
Liebthal, M., Kushwah, M. S., Kukura, P. & Dietz, K. J. Single molecule mass photometry reveals the dynamic oligomerization of human and plant peroxiredoxins. iScience 24, 103528 (2021).
Hundt, N. et al. Direct observation of the molecular mechanism underlying protein polymerization. Sci. Adv. 8, 31 (2022).
Wagner, C., Fuchsberger, F. F., Innthaler, B., Lemmerer, M. & Birner-Gruenberger, R. Quantification of empty, partially filled and full adeno-associated virus vectors using mass photometry. Int. J. Mol. Sci. 24, 11033 (2023).
Ebberink, E. H. T. M., Ruisinger, A., Nuebel, M., Thomann, M. & Heck, A. J. R. Assessing production variability in empty and filled adeno-associated viruses by single molecule mass analyses. Mol. Ther. Methods Clin. Dev. 27, 491–501 (2022).
Häußermann, K., Young, G., Kukura, P. & Dietz, H. Dissecting FOXP2 oligomerization and DNA binding. Angew. Chem. Int. Ed. Engl. 58, 7662–7667 (2019).
Bigelyte, G. et al. Miniature type V-F CRISPR-Cas nucleases enable targeted DNA modification in cells. Nat. Commun. 12, 6191 (2021).
Acharya, A. et al. Distinct RPA domains promote recruitment and the helicase-nuclease activities of Dna2. Nat. Commun. 12, 6521 (2021).
Hickman, A. B., Kailasan, S., Genzor, P., Haase, A. D. & Dyda, F. Casposase structure and the mechanistic link between DNA transposition and spacer acquisition by CRISPR-Cas. Elife 9, e50004 (2020).
Gizardin-Fredon, H. et al. Denaturing mass photometry for rapid optimization of chemical protein-protein cross-linking reactions. Nat. Commun. 15, 3516 (2024).
Wu, D. et al. Mass photometry: a powerful tool for carbohydrates-proteins conjugation monitoring and glycoconjugates molecular mass determination. Glycoconj. J. 40, 401–412 (2023).
Young, J. W. et al. Characterization of membrane protein interactions by peptidisc-mediated mass photometry. iScience 27, 108785 (2024).
Soltermann, F. et al. Quantifying protein–protein interactions by molecular counting with mass photometry. Angew. Chem. Int. Ed. Engl. 59, 10774–10779 (2020).
den Boer, M. A. et al. Comparative analysis of antibodies and heavily glycosylated macromolecular immune complexes by size-exclusion chromatography multi-angle light scattering, native charge detection mass spectrometry, and mass photometry. Anal. Chem. 94, 892–900 (2022).
Sendker, F. L. et al. Emergence of fractal geometries in the evolution of a metabolic enzyme. Nature 628, 894–900 (2024).
Melo, L. et al. Size distributions of gold nanoparticles in solution measured by single-particle mass photometry. J. Phys. Chem. B 125, 12466–12475 (2021).
Lai, S. H., Tamara, S. & Heck, A. J. R. Single-particle mass analysis of intact ribosomes by mass photometry and Orbitrap-based charge detection mass spectrometry. iScience 24, 103211 (2021).
Foley, E. D. B., Kushwah, M. S., Young, G. & Kukura, P. Mass photometry enables label-free tracking and mass measurement of single proteins on lipid bilayers. Nat. Methods 18, 1247–1252 (2021).
Heermann, T., Steiert, F., Ramm, B., Hundt, N. & Schwille, P. Mass-sensitive particle tracking to elucidate the membrane-associated MinDE reaction cycle. Nat. Methods 18, 1239–1246 (2021).
Kratochvíl, J., Asor, R., Helmi, S., Struwe, W. B. & Kukura, P. Lifting the concentration limit of mass photometry by PEG nanopatterning. Nano Lett. 24, 10032–10039 (2024).
Wu, D. & Piszczek, G. Standard protocol for mass photometry experiments. Eur. Biophys. J. 50, 403–409 (2021).
Kofinova, Z., Karunanithy, G., Ferreira, A. S. & Struwe, W. B. Measuring protein-protein interactions and quantifying their dissociation constants with mass photometry. Curr. Protoc. 4, e962 (2024).
Li, Y., Struwe, W. B. & Kukura, P. Single molecule mass photometry of nucleic acids. Nucleic Acids Res. 48, e97 (2020).
Sülzle, J., Elfeky, L. & Manley, S. Surface passivation and functionalisation for mass photometry. J. Microsc. 295, 14–20 (2024).
Ebberink, E. H. T. M. et al. Probing recombinant AAV capsid integrity and genome release after thermal stress by mass photometry. Mol. Ther. Methods Clin. Dev. 32, 101293 (2024).
Van Wee, R. et al. NIPBL and STAG1 enable loop extrusion by providing differential DNA-cohesin affinity. Proc. Natl Acad. Sci. USA 122, e2514190122 (2025).
Cole, D., Young, G., Weigel, A., Sebesta, A. & Kukura, P. Label-free single-molecule imaging with numerical-aperture-shaped interferometric scattering microscopy. ACS Photonics 4, 211–216 (2017).
He, Y. et al. Multiscale modeling and analysis for high-fidelity interferometric scattering microscopy. J. Phys. D. Appl. Phys. 54, 274002 (2021).
Becker, J. et al. A quantitative description for optical mass measurement of single biomolecules. ACS Photonics 10, 2699–2710 (2023).
Tokuriki, N. et al. Protein folding by the effects of macromolecular crowding. Protein Sci. 13, 125–133 (2004).
Jarmoskaite, I., Alsadhan, I., Vaidyanathan, P. P. & Herschlag, D. How to measure and evaluate binding affinities. Elife 9, 1–34 (2020).
Claasen, M., Kofinova, Z., Contino, M. & Struwe, W. B. Analysis of protein complex formation at micromolar concentrations by coupling microfluidics with mass photometry. J. Vis. Exp. 2024 https://doi.org/10.3791/65772 (2024).
Acknowledgements
This work was funded by the European Research Council (ERC) Consolidator Grant PHOTOMASS 819593 (to P.K. and K.I.); the Engineering and Physical Research Council (EPSRC) Leadership Fellowship EP/T03419X/1 (to P.K. and J.C.T.); the Biotechnology and Biological Sciences Research Council BB/W00349X/1 (to P.K. and S.T.); the Wellcome Trust grant number 218514/Z/19/Z (to R.v.W.); UK Research and Innovation (UKRI) under the UK government’s Horizon Europe funding guarantee through project Marie Skłodowska-Curie Actions (MSCA) Postdoctoral Fellowship NanoMassCreator (101062868) EP/X025713/1 (to J.K.); a Clarendon scholarship, a Menasseh Ben Israel scholarship and a Kingsgate scholarship (to D.L.); and an EPRSC Doctoral Training Partnership (to J.B.). For the purpose of Open Access, the author has applied a CC BY public copyright license to any Author Accepted Manuscript (AAM) version arising from this submission. The authors thank K. Zouboulis, F. Naughton-Allen and A. O’Shea for feedback on the manuscript and discussion, and M. Kushwah for providing the Dyn1 ΔPRD protein.
Author information
Authors and Affiliations
Contributions
All authors agree that their authorship orders can be exchanged on their CVs. All authors contributed to the conceptualization. Data acquisition, analysis, interpretation and writing of corresponding protocol sections were as follows: Fig. 1, J.C.T.; Figs. 2, 3d, 4g–i, D.L.; Fig. 3a–c,e–g, J.B.; Fig. 4a–f, R.v.W.; Fig. 5, K.I.; Figs. 6, 7 and 9, J.K.; Fig. 8, S.T.; writing of the introduction, S.T.; review and editing of the final manuscript, S.T. and P.K.; supervision, S.T. and P.K.
Corresponding authors
Ethics declarations
Competing interests
P.K. is a nonexecutive director, shareholder of and consultant to Refeyn Ltd. J.L.P.B is an academic founder, shareholder, and advisor to Refeyn Ltd. The other authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Francesca Vallese, Grzegorz Piszczek, Alexander Buell and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key references
Asor, R. et al. Proc. Natl. Acad. Sci. USA 121, e2403260121 (2024): https://doi.org/10.1073/pnas.2403260121
Sonn-Segev, A. et al. Nat. Commun. 11, 1772 (2020): https://doi.org/10.1038/s41467-020-15642-w
Olerinyova, A. et al. Chem. 7, 224–236 (2021): https://doi.org/10.1016/j.chempr.2020.11.011
Young, G. et al. Science 360, 423–427 (2018): https://doi.org/10.1126/science.aar5839
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kratochvíl, J., van Wee, R., Thiele, J.C. et al. Best practice mass photometry: a guide to optimal single-molecule mass measurement. Nat Protoc (2025). https://doi.org/10.1038/s41596-025-01255-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41596-025-01255-4