Abstract
The pink stem borer, Sesamia inferens Walker (Lepidoptera: Noctuidae), is one of the most notorious pest insects of rice and maize crops in the world. Here, we generated a high-quality chromosome-level genome assembly of S. inferens, using a combination of Illumina, PacBio HiFi and Hi-C technologies. The total assembly size was 973.18 Mb with a contig N50 of 33.39 Mb, anchored to 31 chromosomes, revealing a karyotype of 30 + Z. The BUSCO analysis indicated a high completeness of 98.90% (n = 5286), including 5172 (97.8%) single-copy BUSCOs and 58 (1.1%) duplicated BUSCOs. The genome contains 58.59% (564.58 Mb) repeat elements and 26628 predicted protein-coding genes. The chromosome-level genome assembly of S. inferens provides in-depth knowledge and will be a helpful resource for the Lepidoptera and pest control research communities.
Similar content being viewed by others
Background & Summary
Lepidoptera, encompassing butterflies and moths, is the second most diverse pest insect, with 180,000 described species. They commonly possess 31 chromosomes and constitute one-tenth of Earth’s described species1. Moreover, in both nature and agriculture settings, there is hardly any plant or crop that is not attacked by at least one lepidopteran pest2,3,4. Indeed, the larval stages (caterpillars) are major pests in forests, stored grains, and fiber and food crops. Besides, resistance to insecticide is an increasing problem and moths are among the most feared invasive species.
In the family of moths or Noctuidae, stem borers are notorious pest insects; the stem borer caterpillars damage crops by boring or tunneling inside their plant stems. The pink stem borer or Sesamia inferens Walker (Lepidoptera: Noctuidae) is very destructive for rice in the world5,6,7, but this polyphagous pest is also a major pest to a broad spectrum of crops, encompassing economically important graminaceous crops such as maize, sorghum, wheat, oats and sugarcane8,9,10. The adults can fly long distance, and the females release sex pheromone to attract the male for copulation, where the sex pheromones and the pheromone binding protein (PBP) gene family are relatively conserved in the Noctuidae and act according a lock-and-key principle11,12,13,14. After eclosion, adult moths engage in courtship and mating behavior in 0-day-old, with a mating rate reaching as high as 83.3%15; one female moth can produce 300–600 eggs. Hence, the females maximize their fitness by laying eggs preferentially on plants that maximize their offspring performance16,17,18,19,20,21. Our experimental results22, employing the age-stage, two-sex life table theory, and based on indoor experiments, along with statistical analyses of the offspring from oviposition and hatching of S. inferens, as well as other multiple parameters, have revealed its potential for widespread damage. Larvae tunnel through stalk internodes, weakening them and making them susceptible to breakage by strong winds, while also exposing plants to infection by the red rot fungus, leading to a significant decrease in sucrose content23. S. inferens successfully accomplished its entire developmental cycle on different gramineous crop hosts22 Symptoms known as “dead hearts” or “white heads”24, cause plant lodging and unfilled grains, leading to high yield losses25,26,27. Due to high levels of insecticide resistance and the hidden behavior of the insects into the plant stems, reducing the efficacy of chemical and biological control with parasitoids, the best options today for pest population suppression include field trapping using sex pheromones22,28 and the cultivation of trap plants29,30.
In this study, we present the first chromosome-level genome assembly and sex chromosome identification of S. inferens, providing valuable genomic resources for further research and development. The resulting assembly has a high quality, with a scaffold N50 size of 33.39 Mb. The completeness of the assembly was assessed using the BUSCO analysis, which revealed a high completeness of 98.90%. Repetitive elements were found to constitute a significant portion of the S. inferens genome, accounting for 58.59% of the total genome size. A total of 26628 protein-coding genes were identified in genome assembly. In conclusion, this chromosome-level assembly of the S. inferens genome does not only provide valuable genomic resources for understanding the biology and genetic basis for Lepidoptera, and supports the development of effective strategies for pest insect control based on sex pheromone traps and without use of chemical pesticides.
Methods
Sample collection and sequencing
Insect materials
Specimens of S. inferens were collected from the Shibanzhen, Bozhou District, Zunyi City, Guizhou Province, China. The larvae were collected from sorghum plants (Sorghum bicolor) (Fig. 1). The samples included 3rd instar larvae, pupae, and adult males and females. Among them, 3rd instar larvae were subjected to 24-hour starvation treatment. To ensure the thorough removal of microbial contaminants from the surface of the samples, both larvae and pupae were subjected to surface sterilization. This process involved immersion in 75% ethanol for 1 min followed by three subsequent rinses with sterile water. The detailed protocols are as follows.
We extracted genomic DNA from S. inferens 3rd instar larvae samples using the Genome DNA extraction Kit (TIANGEN) as per the product manual. After extraction, utilizing the NanoDrop One, we detected the purity, concentration, and nucleic acid absorption peaks of S. inferens genomic DNA, focusing on the OD260/280 and OD260/230 ratios. For precise concentration determination, we employed the Qubit 3.0 system. A comparative analysis between Qubit 3.0 fluorescence photometer and NanoDrop One was conducted to assess sample purity. Additionally, we performed agarose gel electrophoresis to ascertain the integrity of the genomic DNA. For sequencing preparations, we employed Qubit 3.0 for precise quantification and Agilent 2100 Bioanalyzer for size analysis to ensure the library’s compliance with anticipated dimensions. Upon successful library validation, we initiated sequencing on the PacBio Sequel II (Pacific Biosciences), aligning the sequencing output with the predefined target data volume. The processed genomic DNA was subsequently employed for the generation of a single-molecule real-time (SMRT) bell sequencing library, utilizing the SMRTbell Template Prep Kit 2.0 developed by Pacific Biosciences31. As a result, we obtained a total of 80.0 Gb Illumina short read sequencing and 504.2 Gb PacBio sequencing reads. In total, 64.70 million raw reads (approximately 97.05 Gb) were obtained for scaffolding in genome assembly.
Genome assembly
To achieve a high-quality assembly, we initiated the process with rigorous quality control of the initial raw reads. In the process of data quality control, several steps are implemented to ensure the integrity and reliability of the sequencing data. Initially, reads containing adapter sequences are eliminated. Following this, bases with consecutive quality scores below 20 at both ends of the sequencing read are subjected to trimming. Reads with a resulting length of less than 50 bp are subsequently excluded. Ultimately, only paired-end reads are retained for subsequent analysis. We used HiFiasm (v 0.15.1) to preliminarily assemble the S. inferens genome, which could resolve near-identical repeats and segmental duplications to generate better haplotype assemblies32. The HiFiasm outputs a primary assembly after performing all-versus-all read overlap alignment and correcting sequencing errors. Purge_Haplotigs software was used to complete genome deredundancy after initial assembly and error corrected, and the redundant heterozygous contigs were identified and removed according to reads depth distribution and sequence similarity33. The total length of HiFiasm, HiFiasm + purge haplotigs and HiFiasm + purge haplotigs + contamination removal was 99712 Mb, 97610 Mb and 97320 Mb, respectively (Table S1). The hybrid was used to obtain a de novo genome assembly for S. inferens with total length of 973.20 Mb and contig N50 length of 30.57 Mb (Table S2). The genome assembly quality was comprehensively evaluated through BUSCO alignment against the Lepidoptera_odb10 orthologue database, assessing the overall integrity of the assembled genome. After aligning the second-generation reads to the genome, mutations were identified using software samtools, picard and GATK. Homozygosity and heterozygosity rates for SNPs and InDels were then calculated separately. The homozygous SNP rate was found to be <0.001%, while the homozygous InDel rate was 0.001%. In contrast, the heterozygous SNP rate was 1.070%, and the heterozygous InDel rate was 0.247%.
Chromosomal-level genome scaffolding with Hi-C data
To obtain the genome at the chromosomal level, Hi-C technology (High-throughput/resolution chromosome conformation capture) was applied34,35. The Hi-C library was prepared using a modified method according to standard protocol36. The samples were 3rd instar larvae.
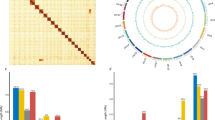
Cells were treated with paraformaldehyde to fixed DNA conformation for 10 mins and stopped crosslinking by 2.5 M glycine for 20 mins. Subsequent to cell lysis, Crosslink DNAs were cut with a restriction enzyme and produced fill ends with biotin, DNA fragments were ligated using DNA ligase. To reverse the cross-linked state of DNA, proteinase digestion was applied, followed by purification of DNA, which was subsequently randomly sheared into fragments ranging from 300–500 bp. Biotin-labeled DNA was selectively captured using streptavidin magnetic beads, which was used to build the library and subsequent sequencing via the Illumina platform. We used bowtie 2 (v 2.2.3)37 to map the paired-end reads to the preliminary assembly. Then, HiC-Pro (v 2.7.8)38 was used to detect the ligation site of unmapped reads, which were mainly composed of the chimeric regions spanning across the ligation junction. High-quality clean data 94.998 Gb (read length: 150 bp) were generated after sequencing and filtering, then used for preliminary assembly by applying a 3D-DNA pipeline35 and LACHESIS39 using default parameters. We employed Juicer to construct the chromosome interaction map and then utilized Juicebox for visual correction. This allowed us to identify potential errors in contig sequence, direction, or assembly within the contig, ensuring the accuracy and reliability of our genome assembly. After the assisted assembly of the genome, a comprehensive genome-wide interaction map was constructed using Juicer40. Analysis of Hi-C data revealed assembly errors in the 3D-DNA assembly process, encompassing contig order, orientation, and internal arrangement. Performed manual visual error correction using JuiceBox (v 2.13.07)40,41. The corrected genome-wide interaction map exhibits enhanced intra-chromosomal interactions, with stronger interactions occurring between contigs in closer linear proximity. A chromatin contact matrix was manually curated in JuiceBox and the 31 scaffolds are clearly distinguishable in the heatmap in Fig. 2a, the interaction signal around the diagonal is strongly apparent. Contig distribution on genome chromosomes in Fig. 2b. The 88 contigs were divided, anchored, sorted, oriented, and merged into 31 chromosomes using LACHESIS and corrected by JuiceBox. The chromosomal heatmap showed good collinearity on the diagonal, which confirms the high quality of scaffolding. The final genome assembly was 973.18 Mb with a scaffold N50 of 33.39 Mb (Table 1, Fig. 1).
Genome assembly of Sesamia inferens. (a) Hi-C assembly of chromosome interactive heat map. Abscissa and ordinate represent order of each bin on corresponding chromosome group. Color block illuminates intensity of interaction from white (low) to red (high). (b) Contig distribution on genome chromosomes. The grey color represents the length of the corresponding chromosome, while other colors represent contigs of different length ranges. (c) Association analysis of GC content and coverage depth of second-generation reads. (d) Association analysis of GC content and coverage depth of third-generation reads.
Sex chromosomes identification
In this study, we performed whole-genome resequencing of 10 male and 10 female adult of S. inferens using the Illumina platform and producing a total of 294.63 Gb clean data. Quality-controlled sequencing reads were aligned to the reference genome scaffolds using BWA software (v 0.7.17)42. The resulting BAM files were utilized for further coverage analysis. Coverage rates for males and females were calculated separately using Samtools (v 1.10)43. The inherent copy number differences between the sexes for sex chromosomes, where the Z chromosome exhibits a higher copy number in males, while the W chromosome is present only in females, were analyzed44,45,46. The log ratio of male to female coverage (log2(M:F)) was computed, and changepoint analysis was performed using the R package “changepoint” (https://CRAN.R-project.org/package=changepoint) to detect points of variation. Chromosomes were categorized based on their log2(M:F) values: chromosomes with values ranging from 0 to ±0.1 were considered autosomes; those with values less than −0.25 were designated as W chromosomes; and those with values greater than or equal to 0.25 were identified as Z chromosomes (ZZ: ♂; ZW: ♀). Based on the log2(M:F) ratio, chromosome 1 was identified as the Z chromosome, and chromosome 31 as the W chromosome (Fig. 3).
Identification of sex chromosomes in Sesamia inferens genome assembly. 10 male and 10 female adults were resequenced, and the obtained reads were analyzed for coverage comparison. Chromosomes with a log2 (M:F read counts) value of 0 were regarded as autosomes (black dots), that with a value less than or equal to the −0.25 were considered W chromosome (red dot), and that with a value greater than or equal to 0.25 were considered Z chromosome (blue dot).
Transcriptome sequencing
To assist in the annotation of genome structure, transcriptomic libraries were prepared from the 3rd instar larvae, pupae, adult males and adult females of S. inferens. Each sample designated for sequencing had an individual library constructed for the procedure. Total RNA was isolated from individual S. inferens sample utilizing the TRIzol (Invitrogen, Carlsbad, CA, USA) reagent method. Following homogenization, samples were allowed to stand at ambient conditions before chloroform was introduced. The mixture underwent centrifugation at 12,000 g at 4 °C, allowing for phase separation. The aqueous phase was subsequently subjected to isopropanol precipitation and centrifugation. The RNA pellet obtained was rinsed in 75% ethanol (prepared in RNase-free water) and centrifuged twice to ensure purity. The air-dried pellet was reconstituted in DEPC-treated water, and its integrity and concentration were quantified using a NanoDrop-2000 spectrophotometer at 260 nm. The RNA samples that had good quality were then utilized for cDNA library construction. Sequencing was carried out on the Illumina NovaSeq 6000 platform47. The obtained spliced transcript was used for genome structure annotation to provide evidence of transcription level.
Genome quality assessment
The best five hits of BLASTN again NCBI NT database were from Atethmia, Cosmia, Mythimna, Amphipyra and Xestia (Table 2). Moreover, we compared the Lepidoptera_odb10 database using BUSCO. The assessment showed 98.9% of BUSCO genes were successfully detected, of which 98.9% were single copy and 1.1% duplicated (Table 3). The results of these evaluations indicate that the genome assembly has a high level of completeness and accuracy.
The assembled S. inferens genome size is 973.18 Mb with a scaffold N50 of 33.39 Mb (Fig. 1, Table S2), close to the estimated size in other Lepidoptera48. Using blobtools (v. 1.1.160), we created a blobplot to evaluate possible contamination of the contigs used for genome assembly (Fig. 2c,d). Taken together, these confidently confirm the accuracy of the chromosome scaffolding.
Repeat sequence annotation
We identified repeat sequences and transposable elements (TEs) using the methods of de novo assembly35 and homologous prediction. First, we used RepeatModeler (v 2.0.2) (https://github.com/Dfam-consortium/RepeatModeler) to predict the repeat sequence with default parameters. Then, RepBase database49 and RepeatMasker (v 4.1.2) (https://github.com/rmhubley/RepeatMasker) were used to annotate the sequence homologs. The results showed that 564.58 Mb are repeat sequences, accounting for 58.59% of the S. inferens genome. Among these repeat sequences, most (24.51%) are long interspersed nuclear elements (LINEs), followed by 12.92% of unclassified elements, 10.75% of long terminal repeats (LTRs)50,51,52, 5.55% of short interspersed nuclear elements (SINEs), 5.38% of rolling-circles and only 4.82% of DNA elements (Table 4).
Gene prediction and function assignment
We annotated protein coding genes in the S. inferens genome using a pipeline that combines de novo prediction, homology searching and transcriptome evidence48. The repeat-masked genome was then subjected to further analysis according to the MAKER (v 3.01.03) genome annotation pipeline53,54. First, we utilized BRAKER (v 2) to construct the parametric species model for the S. inferens genome55,56,57. Next, we employed Trinity (v 2.14.0) to perform transcript splicing with the default parameters for genome structure annotation58,59. The obtained spliced transcript was used for genome structure annotation to provide evidence of transcription level. Finally, we executed MAKER incorporating the transcriptome, genome, parametric model of species, and the protein sequences of 10 other Lepidoptera (Abrostola tripartita, Bombyx mori, Cnaphalocrocis medinalis, Habrosyne pyritoides, Helicoverpa armigera, Hyphantria cunea, Plutella xylostella, Spodoptera exigua, Spodoptera frugiperda and Spodoptera litura) with good annotations down from InsectBase 2.0 (http://v2.insect-genome.com) as input data to predict genes48. A total of 26628 protein coding genes were annotated following the pipeline combined with above-mentioned three methods. Our comparative analysis between our genome assembly and the previously published chromosome-level assembly of S. inferens60 highlighted several key differences. Specifically, our genome assembly exhibited a larger genome size of 973.18 Mb compared to the previously published size of 865.04 Mb. Additionally, while the previous assembly consisted of 1135 contigs and 69 scaffolds, our assembly comprised 88 scaffolds. Notably, our assembly featured a higher Contig N50 value of 30.17 Mb and a slightly lower Scaffold N50 value of 33.39 Mb. Furthermore, our analysis included the identification of the sex chromosomes of S. inferens, providing further elucidation of its karyotype, as detailed in Table S3.
Phylogeny
OrthoFinder61,62,63 (v 2.5.1) was used to analyze the orthologous and paralogous genes of 10 insect genomes, including Drosophila melanogaster (assembly accession: GCF_000001215.4), P. xylostella (assembly accession: GCA_019096205.1), A. tripartita (assembly accession: GCA_905340225.1), B. mori (assembly accession: GCF_014905235.1), C. medinalis (IBG_00192), H. pyritoides (assembly accession: GCA_907165245.1), H. cunea (assembly accession: GCA_003709505.1), S. frugiperda (assembly accession: GCF_011064685.1), S. exigua (assembly accession: GCA_011316535.1), S. litura (assembly accession: GCA_002706865.1), H. armigera (assembly accession: GCF_002156985.1), and D. melanogaster was selected as an outgroup (Fig. 4).
Phylogenetic trees were constructed based on single-copy orthologous gene families. The phylogenetic tree was constructed by maximum likelihood (ML) using IQ-TREE (v 2.1.2) with the best model (JTT + F + R5) and 1000 rapid bootstrap replicates to assess the robustness of the tree64. Additionally, we used Astral-III65 to merge all gene trees obtained through OrthoFinder into a unified species tree. It is essential to emphasize that the two trees generated from these methods must be congruent, validating the consistency and accuracy of our phylogenetic analysis. Divergence time was estimated by MCMCtree66 program in the PAML package (v 4.8) based on the multiple sequence alignment protein sequences. The calibration time points of D. melanogaster (99.15 MYA), P. xylostella (81.66 MYA), S. inferens (13.26 MYA), B. mori (49.25 MYA) and H. pyritoides (40.98 MYA) were obtained from TimeTree67 (http://timetree.org/) (Fig. 4). Gene family contraction and expansion were analyzed using CAFE (v 4.2), incorporating the results from OrthoFinder and the phylogenetic tree with divergence time information68. Finally, iTOL (https://itol.embl.de/#) was used to visualize and enhance the appearance of the phylogenetic tree. S. inferens exhibits an explanation of 825, which is equivalent to half of that observed in S. frugiperda, lower than in P. xylostella, and on par with that in D. melanogaster. The expansion of gene families is considered a pivotal factor contributing to biodiversity and evolution. Data regarding gene family expansion in S. inferens reveals a relatively rapid rate of renewal and iteration. This accelerated gene family evolution enables the organism to adapt to the diverse and continually changing challenges presented by its environment. This result aligns with the phenomenon observed in the field, where the infection of S. inferens has transitioned gradually from localized edge infections to widespread field infestations69.
Data Records
The raw sequencing data and genome assembly of S. inferens have been deposited at the National Center for Biotechnology Information (NCBI). Illumina, PacBio and Hi-C data for S. inferens genome sequencing have been deposited in the NCBI Sequence Read Archive with accession number SRR26501366, SRR27137600 and SRR27032946 under BioProject accession number PRJNA101423470.
Illumina transcriptome data 3rd instar larva (SRR26056362), pupa (SRR26056882), female adult (SRR26050603), male adult (SRR26056479) are available under Bioproject PRJNA101423470.
Genome resequencing data for female adults (SRR28744322, SRR28744323, SRR28744324, SRR28744325, SRR28744326, SRR28744327, SRR28744328, SRR28744328, SRR28744330, SRR28744331) and male adults (SRR28778051, SRR28778052, SRR28778053, SRR28778054, SRR28778055, SRR28778056, SRR28778057, SRR28778058, SRR28778059, SRR28778060) are available under Bioproject PRJNA101423470.
This Whole Genome Shotgun project has been deposited at GenBank under the accession JAYKGN00000000071. The version described in this paper is version JAYKGN010000000.
The annotation file is available in figshare72.
Technical Validation
After extraction, the DNA purity, concentration and integrity were detected using NanoDrop One, Qubit 3.0 fluorescence photometer and Agilent 2100 Bioanalyzer (Agilent Technologies, CA, USA), respectively. RNA integrity and concentration were quantified using a NanoDrop One spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States). High-quality DNA and RNA were used for sequencing.
We used three methods to assess the completeness and quality of the assembly. First, a data accuracy assessment was conducted to confirm the belonging of the assembly results to the target species. The genome sequence was fragmented at 10 kb intervals, and the resulting sequences were aligned to the NCBI nucleotide database (NT library) using Blast software73. Second, a sequence consistency evaluation was performed by aligning second and third-generation data to the assembled genome using BWA (v 0.7.17)42 and Minimap2 (v 2.24)74. As depicted in Table 5, The alignment statistics for the second-generation reads show a mapping rate of 99.67%, a paired mapping rate of 92.40%, an average sequencing depth of 69.38 X, and 99.98% coverage. For third-generation reads, the mapping rate was 99.98%, the average sequencing depth was 26.77 X, and the coverage was 100.00%. Higher mapping and coverage rates indicate a higher consistency between the assembly results and the reads, reflecting better assembly performance. Third, the quality of the genome sequence was evaluated by BUSCO (v 4)75,76,77,78 with Lepidoptera_odb10 and default parameters. In addition, after aligning second-generation reads to the genome, mutations were identified using samtools, picard, and GATK (v 4.4.0.0)79. The rates of homozygous and heterozygous SNPs and InDels were calculated. The homozygous SNP rate was <0.01%, the homozygous InDel rate was 0.001%, the heterozygous SNP rate was 1.070%, and the heterozygous InDel rate was 0.247%.
Code availability
No specific script was used in this work. All commands and pipelines used in data processing were executed according to the manual and protocols of the corresponding bioinformatic software, and default parameters were applied if not mentioned in the Methods described above.
References
Wright, C. J. et al. Comparative genomics reveals the dynamics of chromosome evolution in Lepidoptera. Nat Ecol Evol. 8(4), 777–790 (2024).
Bilal, M. et al. Indoxacarb-loaded fluorescent mesoporous silica nanoparticles for effective control of Plutella xylostella L. with decreased detoxification enzymes activities. Pest. Manag. Sci. 76, 3749–3758 (2020).
Oerke, E. Crop losses to pests. J. Agr. Sci. 144, 31–43 (2005).
Mulhair, P. O. et al. Diversity, duplication, and genomic organization of homeobox genes in Lepidoptera. Genome Res. 33, 32–44 (2023).
Han, L. et al. Lethal and Sub - Lethal Effects of Transgenic Rice Containing cry1Ac and CpTI Genes on the Pink Stem Borer, Sesamia inferens (Walker). Agr. Sci. China 10, 384–393 (2011).
Yang, L. et al. Floating chitosan-alginate microspheres loaded with chlorantraniliprole effectively control Chilo suppressalis (Walker) and Sesamia inferens (Walker) in rice fields. Sci. Total. Environ. 783, 147088 (2021).
Soujanya, P. L. et al. Role of morphological traits and cell wall components in imparting resistance to pink stem borer, Sesamia inferens Walker in maize. Front. Plant. Sci. 14, 1167248 (2023).
Chai, H. N. & Du, Y. Z. The complete mitochondrial genome of the pink stem borer, Sesamia inferens, in comparison with four other Noctuid moths. Int. J. Mol. Sci. 13, 10236–10256 (2012).
Mahesh, P. et al. Natural Incidence of Sesamia inferens Walker, in Sugarcane Germplasm. Sugar Tech. 15, 384–389 (2013).
Cheraghali, Z. et al. Genetic diversity of populations of the stem borer Sesamia nonagrioides (Lepidoptera: Noctuidae) in southern and southwestern Iran, using RAPD-PCR. North-West J. Zool. 11(1), 70–75 (2015).
Wang, H. et al. Structural basis for action by diverse antidepressants on biogenic amine transporters. Nature 503, 141–145 (2013).
Diéguez, M., Pàmies, O. & Moberg, C. Self-Adaptable Tropos Catalysts. Accounts. Chem. Res. 54, 3252–3263 (2021).
Jiang, N. et al. Revisiting the sex pheromone of the fall armyworm Spodoptera frugiperda, a new invasive pest in South China. Insect Sci. 29, 865–878 (2021).
Ando, T., Inomata, S. & Yamamoto, M. Lepidopteran sex pheromones. Topics Curr. Chem. 239, 51–96 (2004).
Nagayama, A. et al. Emergence and mating behavior of the pink borer, Sesamia inferens (Walker) (Lepidoptera: Noctuidae). Appl. Entomol. Zool. 39, 625–629 (2004).
Mayhew, P. J. Adaptive patterns of host-plant selection by phytophagous insects. Oikos 79, 417–428 (1997).
Gripenberg, S., Mayhew, P. J., Parnell, M. & Roslin, T. A meta-analysis of preference-performance relationships in phytophagous insects. Ecol. Lett. 13, 383–393 (2010).
Valladares, G. Host-Plant Selection in the Holly Leaf-Miner: Does Mother Know Best? J. Anim. Ecol. 60, 227–240 (1991).
Jaenike, J. On optimal oviposition behavior in phytophagous insects. Theor. Popul. Biol. 14, 350–356 (1978).
Sekhar, J. C. et al. Differential Preference for Oviposition by Sesamia inferens Walker on Maize Genotypes. Annals of Plant Protection. Sciences 17, 46–49 (2009).
Zang, L., Wang, S., Zhang, F. & Desneux, N. Biological Control with Trichogramma in China: History, Present Status and Perspectives. Annu. Rev. Entomol. 66, 463–484 (2020).
Chen, L. et al. Demography and fitness of Sesamia inferens Walker (Lepidoptera: Noctuidae) on three important gramineous crops. CABI Agric Biosci 4, 49 (2023).
Nagayama, A. et al. Reinvestigation of sex pheromone components and attractiveness of synthetic sex pheromone of the pink borer, Sesamia inferens Walker (Lepidoptera: Noctuidae) in Okinawa. Appl. Entomol. Zool. 41, 399–404 (2006).
Dey, A. et al. Molecular diversity of Sesamia inferens (Walker, 1856) (Lepidoptera: Noctuidae) from India. 3 Biotech. 11, 134 (2021).
Jiang, M. X. & Cheng, J. A. Interactions between the striped stem borer Chilo suppressalis (Walk.) (Lep., Pyralidae) larvae and rice plants in response to nitrogen fertilization. Anz. Schadl-j. Pest. Sc 76, 124–128 (2003).
Rao, A. B. Technique of scoring for resistance in maize stalk borer (S. inferens.). In: Techniques for scoring for resistance to the major insect pests of maize. (AICMIP, IARI, New Delhi, 1983).
Siddiqui, K. H. & Marwaha, K. K. The Vistas of Maize Entomology in India. (Kalyani Publishers, 1993).
Wang, C. et al. Characterization of the pheromone receptors in Mythimna loreyi reveals the differentiation of sex pheromone recognition in Mythimna species. Insect Sci. 31(1), 173–185 (2024).
Gurr, G. M. et al. Multi-country evidence that crop diversification promotes ecological intensification of agriculture. Nat. Plants 2, 16014 (2016).
Tang, L. D. et al. Dead-end trap plants as an environment-friendly IPM tool: A case study of the successful use of vetiver grass in China. Entomologia Generalis 44(1), 81–93 (2024).
Eid, J. S. et al. Real-Time DNA Sequencing from Single Polymerase Molecules. Science 323, 133–138 (2009).
Cheng, H. Y. et al. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods 18, 170–175 (2021).
Roach, M. J., Schmidt, S. A. & Borneman, A. R. Purge Haplotigs: allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinformatics 19, 460 (2018).
van Berkum, N. L. et al. Hi-C: A Method to Study the Three-dimensional Architecture of Genomes. J. Vis. Exp. 39, 1869 (2010).
Dudchenko, O. et al. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science 356, 92–95 (2017).
Wingett, S. et al. HiCUP: pipeline for mapping and processing Hi-C data. F1000Res. 4, 1310 (2015).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Kelly, S. T. & Yuhara, S. HiCUP-Plus: a fast open-source pipeline for accurately processing large scale Hi-C sequence data. bioRxiv. (2022).
Burton, J. N. et al. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat. Biotechnol. 31, 1119–1125 (2013).
Robinson, J. T. et al. Juicebox.js Provides a Cloud-Based Visualization System for Hi-C Data. Cell Syst. 6, 256–258.e1 (2017).
Durand, N. C. et al. Juicebox Provides a Visualization System for Hi-C Contact Maps with Unlimited Zoom. Cell Syst. 3(1), 99–101 (2016).
Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv: Genomics (2013).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25(16), 2078–2079 (2009).
Xu, H. et al. Chromosome-level genome assembly of an agricultural pest, the rice leaffolder Cnaphalocrocis exigua (Crambidae, Lepidoptera). Mol. Ecol. Resour. 22(1), 307–318 (2022).
Zhao, X. et al. A chromosome-level genome assembly of rice leaffolder, Cnaphalocrocis medinalis. Mol. Ecol. Resour. 21(2), 561–572 (2021).
Mongue, A. J. et al. Neosex chromosomes in the monarch butterfly, Danaus plexippus. G3 (Bethesda). 7(10), 3281–3294 (2017).
Rao, S. S. P. et al. A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell 159, 1665–1680 (2014).
Mei, Y. et al. InsectBase 2.0: a comprehensive gene resource for insects. Nucleic. Acids. Res. 50, D1040–D1045 (2021).
Bao, W., Kojima, K. K. & Kohany, O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 6, 11 (2015).
Xu, Z. & Wang, H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic. Acids. Res. 35, W265–268 (2007).
Ou, S. & Jiang, N. LTR_retriever: A Highly Accurate and Sensitive Program for Identification of Long Terminal Repeat Retrotransposons. Plant Physiol. 176, 1410–1422 (2017).
Ou, S., Chen, J. & Jiang, N. Assessing genome assembly quality using the LTR Assembly Index (LAI). Nucleic. Acids. Res. 46, e126 (2018).
Cantarel, B. L. et al. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18, 188–196 (2007).
Holt, C. & Yandell, M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12, 491 (2011).
Brůna, T. et al. BRAKER2: automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database. Nar. Genom. Bioinform. 3, lqaa108 (2020).
Hoff, K. J. et al. Whole-Genome Annotation with BRAKER. Methods Mol. Biol. 1962, 65–95 (2019).
Hoff, K. J. et al. BRAKER1: Unsupervised RNA-Seq-Based Genome Annotation with GeneMark-ET and AUGUSTUS. Bioinformatics 32, 767–769 (2015).
Grabherr, M. G. et al. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 29, 644–652 (2011).
Haas, B. J. et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512 (2013).
Li, H. R. et al. A chromosome-level genome assembly of Sesamia inferens. Sci Data 11, 134, https://doi.org/10.1038/s41597-024-02937-6 (2024).
Emms, D. M. & Kelly, S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 238 (2019).
Emms, D. M. & Kelly, S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16, 157 (2015).
Mier, P. & Pérez-Pulido, A. J. orthoFinder: a new automated tool for searching orthologous proteins useful for functional annotation. F1000Res. 5, 1743 (2014).
Nguyen, L. T. et al. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 32, 268–274 (2014).
Zhang, C. et al. ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics 19(Suppl 6), 153 (2018).
Puttick, M. N. MCMCtreeR: functions to prepare MCMCtree analyses and visualize posterior ages on trees. Bioinformatics 35, 5321–5322 (2019).
Kumar, S. et al. TimeTree: A Resource for Timelines, Timetrees, and Divergence Times. Mol. Biol. Evol. 34, 1812–1819 (2017).
Han, M. V. et al. Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Mol. Biol. Evol. 30, 1987–1997 (2013).
Sun, M. et al. Characterization and Expression of Genes Encoding Three Small Heat Shock Proteins in Sesamia inferens (Lepidoptera: Noctuidae). Int. J. Mol. Sci. 15, 23196–23211 (2014).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRP460199 (2023).
Yu, X. R. Sesamia inferens isolate XY-2023, whole genome shotgun sequencing project. GenBank https://identifiers.org/ncbi/insdc:JAYKGN000000000 (2023).
Yu, X. R. Chromosome-level genome assembly of Pink stem borer, Sesamia inferens Walker, 1856 (Lepidoptera: Noctuidae). figshare https://doi.org/10.6084/m9.figshare.24418837.v1 (2023).
Johnson, M. et al. NCBI BLAST: a better web interface. Nucleic. Acids. Res. 36, W5–9 (2008).
Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2017).
Simão, F. A. et al. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015).
Manni, M. et al. BUSCO Update: Novel and Streamlined Workflows along with Broader and Deeper Phylogenetic Coverage for Scoring of Eukaryotic, Prokaryotic, and Viral Genomes. Mol. Biol. Evol. 38, 4647–4654 (2021).
Seppey, M., Manni, M. & Zdobnov, E. M. BUSCO: Assessing Genome Assembly and Annotation Completeness. Methods Mol. Biol. 1962, 227–245 (2019).
Manni, M. et al. BUSCO: Assessing Genomic Data Quality and Beyond. Curr. Protoc. 1, e323 (2021).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Acknowledgements
This research was supported by the National Key R&D Program of China (2023YFE0104800), Guizhou Provincial Basic Research Program (Natural Science) (No. QianKeHeJiChu-ZK[2024]YiBan047), Program of Introducing Talents to Guizhou University (Nos. GuiDaRenJiHeZi(2021)14, GuiDaRenJiHeZi(2023)39) and Guizhou Highland Specialty Vegetable Green Production Science and Technology Innovation Talent Team (Qiankehe Platform Talent-CXTD [2022]003).
Author information
Authors and Affiliations
Contributions
Lian-Sheng Zang and Qing-Rong Bai contributed to the research design. Xiao-Rui Yu, Liang-De Tang and Qing-Rong Bai collected the samples. Xiao-Rui Yu and Xu Chen analyzed the data. Xiao-Rui Yu, Xu Chen and Qing-Rong Bai wrote the draft manuscript. Guy Smagghe and Lian-Sheng Zang revised the manuscript. All co-authors contributed to this manuscript and approved it.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yu, XR., Chen, X., Bai, QR. et al. Chromosome-level genome assembly and sex chromosome identification of the pink stem borer, Sesamia inferens (Lepidoptera: Noctuidae). Sci Data 11, 810 (2024). https://doi.org/10.1038/s41597-024-03625-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-024-03625-1