Abstract
The deep learning (DL)-based prediction of accurate lymph node (LN) clinical target volumes (CTVs) for nasopharyngeal carcinoma (NPC) radiotherapy (RT) remains challenging. One of the main reasons is the variability of contours despite standardization processes by expert guidelines in combination with scarce data sharing in the community. Therefore, we retrospectively generated a 262-subjects dataset from four centers to develop the DL models for LN CTVs delineation. This dataset included 440 computed tomography images from different scanning phases, disease stages and treatment strategies. Three clinical expert boards, each comprising two experts (totalling six experts), manually delineated six basic LN CTVs on separate cohorts as the ground truth according to LN involvement and clinical requirements. Several state-of-the-art segmentation algorithms were evaluated on this benchmark, showing promising results for LN CTV segmentation. In conclusion, this work built a multicenter LN CTV segmentation dataset, which may be the first dataset for automatic LN CTV delineation development and evaluation, serving as a benchmark for future research.
Similar content being viewed by others
Background & Summary
Nasopharyngeal carcinoma (NPC) is the 23rd most common cancer, and radiotherapy (RT) is the primary treatment approach for it1,2. In the process of RT administration, delineating lymph node (LN) clinical target volumes (CTVs) on computed tomography (CT) is a crucial step. RT planning requires accurately delineating neck lymphatic drainage regions and calculating their radiation dose, as it maximizes the chance of a cure while minimizing radiation-related toxic effects. In the past, clinicians manually performed LN CTVs delineation with slice-by-slice on CT images. It is laborious and prone to intra- and interobserver variability3,4. Additionally, the online adaptive RT often involves repeated manual delineation on repeated planning CT images, thus increasing much burden on radiation oncologists and prolonging patient waiting time5,6.
Recently, deep learning (DL)-based segmentation has shown the potential to alleviate the manual delineation burden7,8,9,10,11. Furthermore, the application of DL for automated delineation has been extended to LN levels in head and neck cancer (HNC).Van der Veen et al.12 examined the performance of a DL model in segmenting LN levels in HNC patients and showed that it improved time efficiency and reduced interobserver variability. Additionally, Cardenas et al.13 developed a DL model to auto-delineate commonly used LN level combinations for HNC patients robustly and consistently. Those studies have investigated the accuracy of DL-based LN level segmentation in HNC. The training of those models relied on the delineation of LN levels following the 2013 updated consensus guideline14. However, it is important to note that those models and contours generated by them may not directly apply to NPC whose nodal metastatic characteristics differ from other HNC15,16. Lin et al. proposed a specific LN CTV delineation map to NPC based on 10,651 LNs distribution17. They observed that certain boundaries of levels Ib, II, IVa and V required adjustments. For instance, when delineating level Ib, the submandibular gland should be spared. Moreover, two works proposed additional modifications for levels IIb and V18,19. Hence, to facilitate the automated RT workflow in NPC, it is necessary to develop a DL model specifically tailored for LN CTVs delineation in this particular subtype following the most recent clinical guidelines.
In addition, specific combinations of LN levels selected according to LN metastatic risk are needed in clinical practice20,21,22. For instance, in NPC patients with N0, it is recommended to delineate a combination of bilateral levels II, III and Va. In patients with N1, the recommended delineation includes an ipsilateral combination of levels II-V and a contralateral combination of levels II, III and Va23. However, most previous DL models for NPC could not automatically output selective target volumes based on LN involvement24, which does not align with the clinical requirements.
Although existing studies reported promising segmentation performance for automatic HNC organ-at-risk (OAR) and tumor segmentation12,13,25, several potential challenges need to be noticed and handled before clinical application development. First, all of these studies developed the model and reported the performance on their collected private datasets12,13, it’s not clear what’s the encouraging solution for this task due to the lack of wide and reproductive research. It’s significantly different from many organ26,27 or tumor28 segmentation tasks which have many public datasets that can be used for model development and evaluation. However, few public HNC LN CTV datasets can be accessed for research until now29,30,31. Second, these public datasets mainly focused on survival and recurrence analysis using tumor-based radiomics. So, no study investigates CTV delineation in radiation therapy. Third, the robustness of the automatic segmentation models when dealing with patients with different stages or treatment strategies is not well evaluated. With the treatment ongoing, significant changes may occur in the location, size, and appearance of the tumor and LNs, such as reduction or disappearance23. It is desirable to develop a detailed LN CTV delineation dataset to investigate its potential influence in radiation therapy.
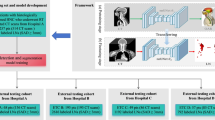
In this work, we aim to build a multicenter NPC LN CTVs segmentation benchmark for model development and evaluation. We retrospectively collected 262 patients with 440 CT images from four hospitals. To develop a specialized DL model for delineating LN CTVs in NPC, we modified the delineation of levels Ib, IIb, III, and the supraclavicular region based on previous studies17,18,19 and the 2013 consensus14. We invited three clinical expert boards to delineate a comprehensive set of six basic LN CTVs manually as the ground truth, an example of this dataset is shown in Fig. 1. To comprehensively investigate and evaluate the performance of recent works, the dataset consists of patients with different stages, treatment strategies, imaging modalities, and raters. Here, we split the dataset into 5 cohorts according to the sources, an internal cohort for training and validation and 4 external cohorts just for evaluation. After that, we trained and evaluated several state-of-the-art (SOTA) segmentation methods on the internal training and testing sets. Moreover, we investigated the robustness and generalization ability of these SOTA methods on four external testing sets. These results provided benchmark results and potential directions for future research.
Methods
Data ethics
This retrospective study was approved by the Ethics Committee on Biomedical Research of these hospitals (approve number, SCCHEC-02-2023-005), including Sichuan Cancer Hospital (SCH), Sichuan Provincial People’s Hospital (SPH), Anhui Provincial Hospital (APH), and Southern Medical University (SMU). Since this was a retrospective study with blurred face regions and all clinical and personal metadata removed, the informed consent was waived. The committee approved the open publication of this dataset.
Data collection
A total of 262 NPC patients were retrospectively collected between January 2019 and June 2022. The internal cohorts were collected from SCH and contained 300 CT images from 150 patients (Table 1). Each patient underwent an unenhanced CT (ueCT) scan followed by a contrast-enhanced CT (ceCT) scan in the same body position within several minutes. The detailed imaging protocols are that all NPC patients underwent RT and were immobilized supine using a thermoplastic head, neck, and shoulder mask. CT examinations were then performed, covering a scanning range from 3 cm above the suprasellar cistern to 2 cm below the sternal end of the clavicle. The CT images from SCH were acquired using a Brilliance CT Big Bore system from Philips Healthcare (Philips Healthcare, Best, the Netherlands), with the following scanning conditions: bulb voltage at 120 kV, current ranging from 275 to 375 mA, scan thickness of 3.0 mm, and an image matrix of 512 × 512. An injected contrast agent, iohexol, was used during the ceCT examination. Similarly, CT images from SPH, APH and SMU were acquired using a Somatom Definition AS 40 system from Siemens Healthcare (Siemens Healthcare, Forcheim, Germany), with the following conditions: bulb voltage at 120 -140 kV, 280-380 mAs current, 3.0 mm slice thickness, and an image matrix of 512 × 512. Iohexol was used as a contrast agent. In addition, to make possible dosimetric studies in the future, the electron density (ED) to Hounsfield unit (HU) of the CT conversion table for the four hospitals is present in Fig. 2.
LN CTVs delineation
During the RT treatment process, different patients need to delineate different LN CTVs according to the clinical stages for individual RT. To obtain individual LN CTVs for each NPC patient, we designed and manually delineated six basic LN CTVs based on the clinical anatomical structures, including the left (L)_Ib, L_II+III+Va, L_IV+Vb+Vc, right (R)_Ib, R_II+III+Va and R_IV+Vb+Vc. The residual seven LN CTVs can be generated by an anatomical-guided adaptive combination. For instance, combining L_II+III+Va with L_IV+Vb+Vc gives L_II-V. Hence, in addition to the six CTVs, the DL model can produce L_Ib-V, L_II-V, R_Ib-V, R_II-V, bilateral (B)_II+III+Va, B_Ib-V, and B_II-V. In total, 13 LN CTVs can automatically be produced by the DL model. The clinical and anatomical significance of 13 LN CTVs are shown in Table 2 and it provides the adaptive combination anatomical guidelines. Based on the above definitions, all radiation oncologists used ITK-SNAP32 to delineate CTV manually.
Clinically delineation guidelines
In terms of level IIb, we adapted the ranges according to the previous work19. Specifically, the cranial boundary of level IIb is set at the cranial edge of C1 (the first cervical spine), the medial boundary is defined by the lateral edges of the rectus capitis lateralis, obliquus capitis superior, obliquus capitis inferior, and elevator scapulae muscles and the posterior range extends to the C1 and C2 levels, while the gap between sternocleidomastoid and splenius capitis muscles could be removed (Fig. 3A, red arrow). The caudal, lateral, and anterior boundaries remain consistent with the 2013 consensus guidelines. Regarding level Ib, we made modifications based on the previous studies17,18. First, we spared the submandibular gland (SMG) during delineation (Fig. 3D, yellow arrow). Second, based on the distance between all positive LNs and anatomical landmarks in level Ib, the Medial Mandibular sub-level was omitted (Fig. 3D, green arrow). For level III, we primarily modified the anterior boundary following the previous work17. The anterior edge of level III is defined as the region close to the anterior edge of the carotid sheath (Fig. 3E, purple arrow), and we spared the space between the anterior of the sternocleidomastoid muscle and the anterior neck ribbon muscles (Fig. 3E, dark blue arrow). Regarding level V, we used the cervical fascia anatomy-oriented nodal delineation. The lateral border of level V is defined by the scapular hyoid muscle (Fig. 3G, light blue arrow), and the medial border is close to the anterior border of the carotid sheath. To ensure the precise contours, S.C. Zhang (MD, with more than twenty years of experience in head and neck radiation therapy), Y. Zhao (MD, with more than ten years of experience in head and neck radiation therapy) and W.J. Liao (MD, with more than ten years of experience in head and neck radiation therapy), who were the main authors of the three clinical guideline papers17,18,19 involved in LN levels delineation in NPC, participated in the ground truth delineation for LN CTVs in this study. These experts first manually delineated several clinical classical patients to provide annotation references for multiple expert boards to minimize the variations in annotation style. These modifications and references were applied to enhance the accuracy and clinical relevance of the DL model.
Examples of experts’ annotation at some key anatomical levels. The red arrow in A represents the tight muscle gap between the sternocleidomastoid and splenius capitis muscles, which can be excluded when delineating IIb. The yellow arrow and green arrow in D represent the submandibular gland (SMG) and Medial Mandibular sub-level, respectively. These two areas can be removed when delineating Ib. The purple arrow in E represents the carotid sheath. The dark blue arrow represents the space between the anterior of the sternocleidomastoid muscle and the anterior neck ribbon muscles, and this area can be spared when delineating III. The light blue in G represents the scapular hyoid muscle, which is the lateral border of level V.
Data split
We randomly split into 120 patients for training and 30 patients for internal testing (validation cohort). External cohorts with different modalities encompassed two external testing cohorts: 1) Validation cohort 1, collected from SPH and comprised 60 ceCT images; 2) Validation cohort 2 was collected from APH and included 32 ueCT images. External cohorts with different treatment strategies consisted of two additional external testing cohorts (Validation cohorts 3 and 4), including CT images obtained before and post treatments: 1) Validation cohort 3, collected from SMU, comprised 12 patients, each with induction chemotherapy (IC)-naïve and post-IC ceCT images (referred to as CTpreIC and CTpostIC, respectively); 2) Validation cohort 4, collected from SCH, included eight patients who underwent three repeated ceCT scans during adaptative RT. These scans were performed after 0, 15-20, and 25-30 fractions and labeled as CTRA, CTRB, and CTRC, respectively (the detailed scan intervals are presented in Table 3). Table 1 presents the detailed clinical and imaging characteristics of each cohort over the LNCTVSeg dataset. The inclusion of CT images from various centers and manufacturers enhances the heterogeneity and representativeness of our data. Meanwhile, it can be observed that the clinical and image characteristics of these cohorts are distributed in a similar space. To protect the patients’ private information, we blurred the face region and removed all metadata about clinical and personal33.
Data Records
The dataset without private information is available on Figshare34. Then, all files are stored with the format NIfTI-1 (32-bit floating point). Each cohort is stored as a folder and named “Internal_Cohort”, and “Testing_Cohort_X” (“X” is from 1 to 4), Raw CT images in the training or testing sets are stored in the “imagesTr” or “imagesTs” folders of each cohort with the name “lnctvseg_cx_xxxx_yyyy.nii.gz”, respectively, where “cx” denotes the center number x is the digit number from 1 to 4, “xxxx” is the index number of each patient the range is 1 to the total number of patients in “cx”, “yyyy” is set to “0000” (non-contrast CT) or “0001” (contrast-enhanced CT). The corresponding labels are stored in the “labelsTr” and “labelsTs” folders with the name “lnctvseg_cx_xxxx.nii.gz”, respectively. The clinical baseline and CTV labelling index are presented in Tables 1 and 2.
Technical Validation
Experiment settings
To benchmark the LN CTV segmentation task, we use the nnUNet as a baseline and also investigate the results of several recent transformer-based architectures, including 1) 3D UXNET35, a hypermodel combines large kernel size convolutions and hierarchical transformer for medical image segmentation, which can enlarge global receptive fields and reduce the model parameters and perform very well on several medical datasets; 2) REPUXNET36, a pure 3D CNN model that performs element-wise scaling in large kernel weights to enhance the learning convergence and effectively adapt large receptive field for volumetric segmentation, which achieved the SOTA performance on abdominal organ and lesion segmentation; 3) SwinUNETR37, a new pure transformer model that merges the merits of Swin Transformers and UNet for medical image segmentation and shows encouraging performance on several medical segmentation tasks. For a fair comparison, we used their official implementations with provided pre-trained models and trained them with the same settings. Considering the huge variations in CT imaging protocols across different centers, encompassing different vendors, parameters, and the presence or absence of contrast enhancement, we extended the baseline nnUNet with mixup-based data augmentation38,39 to improve the generalization ability on heterogeneous images.
Evaluation metrics
Three metrics were used to evaluate segmentations, including DSC, the 95% Hausdorff distance (HD95), and normalized surface Dice (NSD) with a tolerance distance 2 mm40. The DSC measures the volumetric overlap between two segmentations, and the HD95 (mm) measures the boundary distance between two segmentations. Generally, a superior model should generate a higher value for DSC (maximum of 1) and a lower value for HD95 (minimum of 0). The NSD quantifies the overlap of the surfaces between two segmentations, with a perfect match scoring 1. If a target is missed, the DSC, HD95 and NSD will be set to 0, 100 and 0, respectively. We employed the DSC, HD95 and NSD implementations of the MedPy.
Intraobserver variability
In this study, twelve CT images were randomly selected from SCH and re-contoured by the same expert who had taken part in ground truth generation after two months. The DSC was calculated to assess intraobserver variability between the original and recontoured delineations for various LN CTVs. Meanwhile, we also measured the DSC between the original and the DL-generated contours (nnUNet with mixup). The results are listed in Table 4. The DSC between the original and the model-predicted contours was higher than between the original and recontoured LN CTVs, although a statistical difference was not obtained. Moreover, the variance in observed DSC for LN CTVs was lower for DL auto-segmentation than for recontouring by the expert, except for bilateral levels Ib.
Interobserver variability
we randomly selected 30 patients from SCH and invited two expert groups (SPH and APH) to independently segment the LN CTVs for these patients. Together with the initial segmentations by the SCH group, we obtained LN CTVs delineated by three expert groups (SCH, SPH, and APH). We calculated the DSC to assess interobserver variability, as shown in Table 5. For example, the DSC among the three groups around from 80.0% to 89% for four basic LN CTVs except for levels_Ib. It is indicated that although multiple expert groups delineated the target areas, the consistency of the target areas delineated by different groups was relatively good when all experts strictly followed our delineation principles and requirements.
Experimental results
Table 6 shows the quantitative segmentation results of these methods both in the internal and external cohorts. It can be found that the baseline nnUNet and the modified nnUNet perform better than the other three transformer-based models in terms of mean DSC (mDSC) on all testing cohorts. Meanwhile, the modification of the baseline also improved the baseline performance by a slight margin. Besides, the results in the term of NSD also show a similar trend compared with DSC, where the CNN-based segmentation models consistently outperform transformer-based models. (Fig. 4 shows some segmentation results.)
Visual examples of the nnUNet(mixup) predictions in internal testing (A), external testing 1(B), and external testing 2 cohorts (C). We each ranked all patients in the internal testing cohort, external testing cohort 1, and external testing cohort 2 and selected the median scores of patients in the term of DSC. Yellow lines denoted the experts’ delineated contours, and green lines denoted the model-predicted ones.
Limitation
Unlike previous studies including level Ia segmentation13,25, our study did not involve it, which might limit the dataset’s and model’s broader applicability. That’s because the patients in this dataset consist of NPC, one of the main subtypes of HNC. In NPC, LN metastases are rarely observed in this area, and thus this area is not commonly included in NPC RT.
Usage Notes
The Dataset is publicly available on Figshare under the Creative Commons 4.0 Attribution (CC-BY-4.0). All NIfTI files can be visualized via ITK-SNAP32 and other packages.
References
Sun, X.-S. et al. The association between the development of radiation therapy, image technology, and chemotherapy, and the survival of patients with nasopharyngeal carcinoma: a cohort study from 1990 to 2012. International Journal of Radiation Oncology* Biology* Physics 105, 581–590 (2019).
Chen, Y.-P. et al. Nasopharyngeal carcinoma. The Lancet 394, 64–80 (2019).
van der Veen, J., Gulyban, A. & Nuyts, S. Interobserver variability in delineation of target volumes in head and neck cancer. Radiotherapy and Oncology 137, 9–15 (2019).
Peng, Y.-l. et al. Interobserver variations in the delineation of target volumes and organs at risk and their impact on dose distribution in intensity-modulated radiation therapy for nasopharyngeal carcinoma. Oral oncology 82, 1–7 (2018).
Ding, G. X. et al. A study on adaptive imrt treatment planning using kv cone-beam ct. Radiotherapy and Oncology 85, 116–125 (2007).
Huang, H. et al. Determining appropriate timing of adaptive radiation therapy for nasopharyngeal carcinoma during intensity-modulated radiation therapy. Radiation Oncology 10, 1–9 (2015).
Lin, L. et al. Deep learning for automated contouring of primary tumor volumes by mri for nasopharyngeal carcinoma. Radiology 291, 677–686 (2019).
Liao, W. et al. Automatic delineation of gross tumor volume based on magnetic resonance imaging by performing a novel semisupervised learning framework in nasopharyngeal carcinoma. International Journal of Radiation Oncology* Biology* Physics 113, 893–902 (2022).
Litjens, G. et al. A survey on deep learning in medical image analysis. Medical image analysis 42, 60–88 (2017).
Chen, X. et al. A deep learning-based auto-segmentation system for organs-at-risk on whole-body computed tomography images for radiation therapy. Radiotherapy and Oncology 160, 175–184 (2021).
Luo, X. et al. Segrap2023: A benchmark of organs-at-risk and gross tumor volume segmentation for radiotherapy planning of nasopharyngeal carcinoma. arXiv preprint arXiv:2312.09576 (2023).
Van der Veen, J., Willems, S., Bollen, H., Maes, F. & Nuyts, S. Deep learning for elective neck delineation: More consistent and time efficient. Radiotherapy and Oncology 153, 180–188 (2020).
Cardenas, C. E. et al. Generating high-quality lymph node clinical target volumes for head and neck cancer radiation therapy using a fully automated deep learning-based approach. International Journal of Radiation Oncology* Biology* Physics 109, 801–812 (2021).
Grégoire, V. et al. Delineation of the neck node levels for head and neck tumors: a 2013 update. dahanca, eortc, hknpcsg, ncic ctg, ncri, rtog, trog consensus guidelines. Radiotherapy and Oncology 110, 172–181 (2014).
Tang, L. et al. The volume to be irradiated during selective neck irradiation in nasopharyngeal carcinoma: analysis of the spread patterns in lymph nodes by magnetic resonance imaging. Cancer 115, 680–688 (2009).
King, A. D. et al. Neck node metastases from nasopharyngeal carcinoma: Mr imaging of patterns of disease. Head & Neck: Journal for the Sciences and Specialties of the Head and Neck 22, 275–281 (2000).
Lin, L. et al. Delineation of neck clinical target volume specific to nasopharyngeal carcinoma based on lymph node distribution and the international consensus guidelines. International Journal of Radiation Oncology* Biology* Physics 100, 891–902 (2018).
Zhao, Y. et al. Level ib ctv delineation in nasopharyngeal carcinoma based on lymph node distribution and topographic anatomy. Radiotherapy and Oncology 172, 10–17 (2022).
Zhang, J. et al. Level iib ctv delineation based on cervical fascia anatomy in nasopharyngeal cancer. Radiotherapy and Oncology 115, 46–49 (2015).
Chen, J.-z. et al. Results of a phase 2 study examining the effects of omitting elective neck irradiation to nodal levels iv and vb in patients with n0-1 nasopharyngeal carcinoma. International Journal of Radiation Oncology* Biology* Physics 85, 929–934 (2013).
Li, J.-G. et al. A randomized clinical trial comparing prophylactic upper versus whole-neck irradiation in the treatment of patients with node-negative nasopharyngeal carcinoma. Cancer 119, 3170–3176 (2013).
Huang, C.-L. et al. Upper-neck versus whole-neck irradiation at the contralateral uninvolved neck in patients with unilateral n3 nasopharyngeal carcinoma. International Journal of Radiation Oncology* Biology* Physics 116, 788–796 (2023).
Tang, L.-L. et al. Elective upper-neck versus whole-neck irradiation of the uninvolved neck in patients with nasopharyngeal carcinoma: an open-label, non-inferiority, multicentre, randomised phase 3 trial. The Lancet Oncology 23, 479–490 (2022).
Men, K. et al. Deep deconvolutional neural network for target segmentation of nasopharyngeal cancer in planning computed tomography images. Frontiers in oncology 7, 315 (2017).
Weissmann, T. et al. Deep learning for automatic head and neck lymph node level delineation provides expert-level accuracy. Frontiers in Oncology 13, 1115258 (2023).
Luo, X. et al. Word: A large scale dataset, benchmark and clinical applicable study for abdominal organ segmentation from ct image. arXiv preprint arXiv:2111.02403 (2021).
Liao, W. et al. Comprehensive evaluation of a deep learning model for automatic organs at risk segmentation on heterogeneous computed tomography images for abdominal radiotherapy. International Journal of Radiation Oncology* Biology* Physics (2023).
Menze, B. H. et al. The multimodal brain tumor image segmentation benchmark (brats). IEEE transactions on medical imaging 34, 1993–2024 (2014).
Grossberg, A. J. et al. Imaging and clinical data archive for head and neck squamous cell carcinoma patients treated with radiotherapy. Scientific data 5, 1–10 (2018).
Bejarano, T., De Ornelas-Couto, M. & Mihaylov, I. B. Longitudinal fan-beam computed tomography dataset for head-and-neck squamous cell carcinoma patients. Medical physics 46, 2526–2537 (2019).
Elhalawani, H. et al. Matched computed tomography segmentation and demographic data for oropharyngeal cancer radiomics challenges. Sci. data 4, 170077 (2017).
Yushkevich, P. A. et al. User-guided 3d active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 31, 1116–1128 (2006).
Collins, S. A., Wu, J. & Bai, H. X. Facial de-identification of head ct scans. Radiology 296, 22–22 (2020).
Luo, X. et al. A multicenter dataset for lymph node clinical target volume delineation of nasopharyngeal carcinoma. figshare https://doi.org/10.6084/m9.figshare.26793622.v3 (2019).
Lee, H. H., Bao, S., Huo, Y. & Landman, B. A. 3d ux-net: A large kernel volumetric convnet modernizing hierarchical transformer for medical image segmentation. arXiv preprint arXiv:2209.15076 (2022).
Lee, H. H. et al. Scaling up 3d kernels with bayesian frequency re-parameterization for medical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention, 632–641 (Springer, 2023).
Tang, Y. et al. Self-supervised pre-training of swin transformers for 3d medical image analysis. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 20730–20740 (2022).
Zhang, H., Cisse, M., Dauphin, Y. N. & Lopez-Paz, D. mixup: Beyond empirical risk minimization. arXiv preprint arXiv:1710.09412 (2017).
Zhang, L. et al. Generalizing deep learning for medical image segmentation to unseen domains via deep stacked transformation. IEEE transactions on medical imaging 39, 2531–2540 (2020).
Antonelli, M. et al. The medical segmentation decathlon. Nat. communications 13, 4128 (2022).
Acknowledgements
This work was supported by the National Natural Science Foundation of China under Grant 82203197 and 62271115, the Sichuan Science and Technology Program, China (Grant 2023NSFSC1852 and 2023NSFSC0720), and the Sichuan Provincial Cadre Health Research Project (Grant/award number: 2023-803).
Author information
Authors and Affiliations
Contributions
L.W.J., L.X.D., and Z.S.T. conceived of the presented idea; L.W.J., Z.Y., Q.Y.J., H.Y., H.H., L.L., and Z.S.C. performed and confirmed the segmentations; L.X.D., and F.J. performed full data anonymization; L.W.J., X.J.F., and H.Y. collected data; L.W.J., and L.X.D. wrote the initial draft. W.G.T., Z.S.C., and Z.S.T. completed the critical review and revision of the manuscript and datasets, as well as proofreading.
Corresponding author
Ethics declarations
Competing interests
Te authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Luo, X., Liao, W., Zhao, Y. et al. A multicenter dataset for lymph node clinical target volume delineation of nasopharyngeal carcinoma. Sci Data 11, 1085 (2024). https://doi.org/10.1038/s41597-024-03890-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-024-03890-0
This article is cited by
-
Multicentre evaluation of deep learning CT autosegmentation of the head and neck region for radiotherapy
npj Digital Medicine (2025)