Abstract
As one of the two most ancient groups of extant vertebrates, lamprey has become an important model organism in various fields of biology. In this paper, we present a comprehensive tissue-wide spatial metabolomics dataset for lampreys, where 14 distinct tissues were analyzed using liquid chromatography-mass spectrometry (LC-MS) in both positive and negative ion modes. The dataset has been fully validated using internal standard and pooled quality control samples and is readily accessible at the UCSD Metabolomics Workbench. This dataset serves as a valuable resource for researchers using lampreys as a model organism. Additionally, it acts as a benchmark metabolomics dataset for evaluating new algorithms and software tools and comparing them with previously published results. A lamprey spatial metabolomics database is also provided to support studies utilizing lampreys as an animal model, and to complement and validate other spatial metabolomics studies on lampreys conducted with mass spectrometry imaging or other techniques.
Similar content being viewed by others
Background & Summary
Lampreys are jawless fishes that diverged from the rest of the vertebrate lineage approximately 550 million years ago1,2. Together with hagfishes, they constitute the remaining members of a mostly extinct group of jawless fishes known as Agnatha3. Lampreys have long been recognized as key for understanding vertebrate evolution due to their basal position in vertebrate phylogeny4. With advancements in cell biology and biochemistry, the lamprey has emerged as an important model organism in various research fields, such as evolutionary biology, embryology, ecology, physiology, and biomedicine3,4,5,6.
Lampreys possess a powerful and unique immune system, as well as many valuable genes7,8,9,10. Studies on lampreys using genomics1,6,11,12, transcriptomics2,13, peptidomics14, and proteomics15,16 approaches have brought significant insights into the timing and mechanisms of evolution, the diversification of morphology, and the emergence of adaptive immunity17. These studies have also led to the discovery of many important human disease-related genes and associated proteins or peptide drugs, greatly contributing to human disease therapy. In contrast, metabolomics studies on lampreys are still limited, despite that metabolites have much more varied and far more critical roles as signaling molecules, immune modulators, endogenous toxins, and environmental sensors.
In a recent study, we applied a liquid chromatography-mass spectrometry (LC-MS) based tissue-wide spatial metabolomics approach to comprehensively profile 14 different lamprey tissues (Fig. 1a). In the study, we identified two groups of metabolites in the lamprey buccal gland - prostaglandins and the kynurenine pathway metabolites - that can be injected into the host fish to assist lamprey blood feeding18. These metabolites have been shown to function as vasodilators and anticoagulants, helping maintain vascular homeostasis and regulate inflammatory responses in the host fish. It is important to note that in that study, we focused exclusively on blood-feeding related metabolites in the buccal gland. However, there are many other metabolites in lampreys with various biologically and physiologically important functions, that are worth studying. For instance, 390 and 802 mass features were detected at levels over 100 times higher in the buccal gland than in the other 13 tissues in positive and negative ion modes, respectively. However, the identity and functions of most of these mass feature remain to be explored. In addition to the buccal gland, numerous tissue-specific metabolites have yet to be investigated. For example, we have identified a sulfated bile acid named petromyzonol sulfate, a unique sex pheromone to lampreys19,20, which is primarily concentrated in the supraneural body. This distribution, which has not been previously examined, may be closely related to its function as a pheromone, indicating that its anatomical localization could play a crucial role in its biological activity. Further study is needed to understand this potential connection.
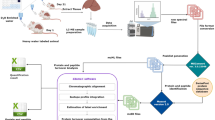
Schematic representation of LC-MS based lamprey spatial metabolomics workflow. (a) Fourteen different lamprey tissues were carefully dissected, and metabolites were extracted from each sample tissue. Twenty microliters of 2-chloro-L-phenylalanine (0.3 mg/mL) was added to each extract as an internal standard to monitor the data quality. (b) Block randomization was used in the experimental design to reduce the prevalence of unanticipated confounders. Pooled quality control (QC) samples were dispersed every 10 samples across the multiple batches to evaluate and correct inter-batch variations. The extracts were analyzed at both positive and negative ion modes. (c) The data quality was initially assessed using the R package RawHummus. Data preprocessing steps, including peak picking, retention time (RT) alignment, peak matching, and peak filling, were then conducted to generate a data matrix containing RT, mass-to-charge ratio (m/z) values, and peak intensity. The data matrix was used for univariate and multivariate statistical analyses. Next, mass features were identified using database searches, comparisons to authentic standards, and MS/MS. Identified mass features were used to construct the lamprey spatial metabolomics database. Abbreviation: E: eye; B: brain; M: muscle; S: supraneural body; N: notochord; X: blood; Bu: buccal gland; G: gill; H: heart; L: liver; O: ovary; I: intestine; K: kidney; and T: testis. These abbreviations are in accordance with the metadata.
Spatial metabolomics is an emerging field of omics research that enables detecting and mapping the spatial distribution of metabolites and lipids on the scales of cells, tissues, organs or entire organisms21. Mass spectrometry imaging (MSI) is the most popular technique for spatial metabolomics due to its excellent spatial resolution, while the number of detected metabolites is typically low and metabolite identification is challenging22,23. Conversely, despite its limitation in spatial resolution, the LC-MS-based spatial metabolomics approach delivers unmatched sensitivity and molecular coverage, thus allowing for a more comprehensive exploration of biological systems. In this study, we have performed a tissue-wide spatial metabolomics analysis of lampreys, where 14 unique lamprey tissues were dissected and analyzed using LC-MS (Fig. 1a). This approach allows us to explore and compare metabolites at tissue levels in an “untargeted manner”18. The dataset was collected from a well-designed experiment in which block randomization was applied to minimize the inter-batch variations24 (Fig. 1b). Internal standard and pooled quality control (QC) samples were used to monitor, evaluate, and correct system variations to ensure highly reproducible and robust metabolomics data (Fig. 2a,b).
Data quality evaluation and mass feature filtering. (a) The data quality was assessed by comparing the peak area of the internal standard (IS), 2-chloro-L-phenylalanine, which was added equally to each sample extract. Line plots displayed the log10-transformed peak area variations of the IS detected in positive and negative ion modes (n = 3 for each tissue), respectively. The red dashed lines indicated the average log10-transformed peak area of the IS. (b) Principal component analysis (PCA) of all samples revealed that the pooled QC samples were closely clustered in the PCA scores plot, indicating good reproducibility of the dataset. (c) A mass feature filtering step was conducted to remove unstable mass features with a coefficient of variation (CV) >30% in QC samples, features lacking an MS2 spectrum, and features with incorrect MS2 spectra (i.e., where the wrong precursor ion was selected for fragmentation). The bar plot displayed the number of unique mass features remaining after each filtering step in both positive and negative ion modes.
This dataset therefore has the following re-use potentials: (1) to serve as a resource dataset for researchers using lamprey as a model organism to explore potential biomarkers for their specific studies; (2) to provide a benchmark metabolomics dataset that is valid, comprehensive, fully transparent, and readily accessible, allowing for the evaluation of new algorithms and comparison with previously published results; and (3) to offer a spatial metabolomics database (https://www.lampreydb.com) that complements and validates other spatial metabolomics studies on lampreys using MSI-based technologies.
Methods
The sample preparation, data acquisition, preprocessing, statistical analysis, metabolite identification and database construction have been described previously. For more detailed description, please refer to the publication by Gou et al.18.
Materials
Adult Arctic lampreys (Lethenteron camtschaticum) in the spawning migration stage were collected from the Songhua River in Heilongjiang Province, China. Fourteen different tissues, heart (H), liver (L), kidney (K), brain (B), supraneural body (S), muscle (M), intestine (I), gill (G), eye (E), testis (T), ovary (O), buccal gland (Bu), blood (X), and notochord (N), were carefully dissected and rinsed in sterile phosphate-buffered saline (PBS: 10 mM phosphate buffer, 2.7 mM potassium chloride, 137 mM sodium chloride, pH 7.4). The secretion from the buccal gland was extracted using a syringe. All samples were snap-frozen in liquid nitrogen and stored at −80 °C until LC-MS analysis (Fig. 1a).
Metabolite extraction
To extract the samples, 30 mg of each sample was placed into a 2 mL Eppendorf tube along with 20 μL of internal standard (2-chloro-L-phenylalanine, 0.3 mg/mL) and 400 μL of an extraction solution composed of 80% methanol in water (V/V) (Fig. 1a). Two small steel balls were then added to each tube. The tubes were pre-cooled at −20 °C for 2 minutes before grinding the samples at 60 Hz for 2 minutes using a Tissuelyser-48 grinding miller (Jingxing Limited Company, Shanghai, China). The mixture was briefly vortexed and then sonicated at room temperature (25–28 °C) for 10 minutes. Subsequently, the samples were centrifuged at 13,000 rpm and 4 °C for 10 minutes. Following centrifugation, 300 μL of the supernatant was transferred to a brown glass vial and dried using a freeze concentration centrifugal dryer. The dried residue was reconstituted in 300 μL of a methanol and water mixture (1/4, V/V), vortexed for 30 seconds, and then placed at −20 °C for 2 hours. The samples were centrifuged again at 13,000 rpm and 4 °C for 5 minutes. The supernatants (150 μL) were carefully extracted using crystal syringes, filtered through a 0.22 μm PTFE filter (Acrodisc® CR 13 mm; PALL), and transferred into LC vials for LC-MS analysis. Pooled QC samples were created by combining 20 μL aliquots from each extracted sample.
LC-MS analysis
A Dionex Ultimate 3000 UHPLC system coupled with a Q-Exactive quadrupole-Orbitrap mass spectrometer, equipped with a heated electrospray ionization (ESI) source (Thermo Fisher Scientific, Waltham, MA, USA), was utilized for spatial metabolomics analysis in both positive and negative ion modes (Fig. 1b). The separation was carried out on an ACQUITY UPLC HSS T3 column (1.8 μm, 2.1 × 100 mm). The binary gradient elution system consisted of (A) water with 0.1% formic acid (V/V) and (B) acetonitrile with 0.1% formic acid (V/V). The following gradient program was used: 5% B from 0–2 min, 5–25% B from 2–4 min, 25–50% B from 4–8 min, 50–80% B from 8–10 min, 80–100% B from 10–14 min, held at 100% B for 1 min, followed by 100% to 5% B from 15–15.1 min, and holding at 5% B from 15.1–18 min. The flow rate was maintained at 0.35 mL/min, with the column temperature set to 45 °C. All samples were kept at 4 °C during analysis, and a 2 μL injection volume was used. The mass spectrometer scanned a mass range from m/z 66.7 to 1000.5, with a resolution of 70,000 for full MS scans and 35,000 for HCD MS/MS scans. The collision energy was adjusted to 10, 20, and 40 eV. The mass spectrometer operated under the following conditions: spray voltage of 3800 V (+) and −3000 V (−); sheath gas flow rate of 35 arbitrary units; auxiliary gas flow rate of 8 arbitrary units; and a capillary temperature of 320 °C. The quality control (QC) samples were injected at regular intervals (every 10 samples) throughout the analytical run to ensure the repeatability of the data (Fig. 1b).
Data preprocessing and metabolite annotation
Three distinct software tools were employed for data preprocessing and metabolite identification: MS-DIAL (v.4.9)25,26, Progenesis QI (v.2.4, Waters), and Compound Discoverer (v.3.3.3, Thermo Scientific). Among them, Progenesis QI and MS-DIAL were primarily used for metabolite identification. For MS-DIAL, the raw data were converted into.abf files using the Reifycs ABF Converter (Figure S1). Then, the .abf files were imported to the MS-DIAL platform. For Progenesis QI and Compound Discoverer, the raw data were directly used for data analysis (Fig. 1c). The specific parameters used for lamprey metabolomics data processing using the three software are detailed in Supplementary Table S1.
Lamprey spatial metabolomics database construction
The LampreyDB database was constructed using MySQL (v.8.0) and Django (v.3.0.6) as the backend framework. The frontend was developed with HTML and JavaScript. Custom scripts, along with the Python library Beautiful Soup (https://pypi.org/project/beautifulsoup4/), were used to create the interactive anatomical heatmap. Additional visual elements, such as MS spectrum plots, were generated using the Plotly Python library (https://plotly.com/python/). The database is hosted on Microsoft Azure cloud service.
Data Records
Both RAW data files and open-source text mzML files have been archived in at the UCSD Metabolomics Workbench, an international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools27, under the Study ID ST003413. The data can be accessed directly via its Project https://doi.org/10.21228/M8MN9F28. The zipped RAW (8.45 GB) dataset contains 94 s, with 47 acquired in positive ion mode and 47 in negative ion mode. Similarly, the zipped mzML dataset (5.67 GB) includes the same number of files, with 47 acquired in positive ion mode and 47 in negative ion mode. The metadata is the same as shown in Fig. 1a and as described in the original publication.
Technical Validation
Five pooled quality control (QC) samples were prepared and analyzed at the start and end of the LC-MS analytical run, as well as at intermittent points throughout, in both positive and negative ion modes. They were then analyzed with R package RawHummus29 to systematically monitor the data quality and inter-batch variation. RawHummus automatically selects six mass features evenly across the entire RT range for a fast and more unbiased quality check. The six features were then used to evaluate 12 quality metrics which are closely related to LC peak shape, retention time (RT), mass accuracy, detector sensitivity and fragmentation to evaluate the chromatogram, MS1 and MS2 of the QC data. The resulting data quality evaluation reports showed that the maximum retention time shift was 0.03 and 0.05 min, maximum mass shifts were 0.96 and 0.92 ppm, and maximum ion intensity variations were 15.06% and 13.22% for QC data acquired positive and negative ion modes, suggesting a high level of consistency and reliability in the analytical data (Supplementary File S1, S2). In addition, an internal standard, 2-chloro-L-phenylalanine, was used for rapid inter-batch variation evaluation. The result showed that the log10 transformed peak intensity variation was within ±5% in positive ion mode, and within ± 10%, further confirming the stability of the dataset (Fig. 2a). Principal component analysis (PCA) of all the samples showed that the pooled QC samples were closely clustered in the score plot, suggesting an excellent data reproducibility (Fig. 2b).
A total of 6,568 and 3,143 unique mass features were detected in positive and negative ion modes, respectively (Fig. 2c). A data cleaning step was then performed to eliminate the unstable mass features (i.e., those with a CV > 30% in QC samples), features lacking MS2 spectra (i.e., those were not subjected to fragmentation), and features with wrong MS2 spectra (where wrong precursor ion was selected for fragmentation). The resulting 2,621 (positive ion mode) and 1,835 (negative ion mode) mass features were retained for subsequent metabolite identification and statistical analysis (Fig. 2c).
Usage Notes
The lamprey metabolomics data files have been archived at the UCSD Metabolomics Workbench27, and is readily accessible at https://doi.org/10.21228/M8MN9F28. The instrument.RAW files are accessible using Thermo specific software (i.e., FreeStyle and Xcalibur). The RAW data can be preprocessed and analyzed using proprietary software such as Compound Discoverer (Thermo) and Progenesis QI (Waters), as well as open-source tools like XCMS30, MS-DIAL25,26, MZmine (Schmid et al. 2023), and MetaboAnalyst31. It is worth noting that data conversion may be necessary in order to use certain software, such as ABF format (https://www.reifycs.com/abfconverter/) for MS-DIAL, and mzXML or mzML format32 for XCMS (Figure S2). The preprocessed data in Excel format can be found in Supplementary Tables S2, S3.
In addition, we have provided a lamprey spatial metabolomics webserver (LampreyDB, https://www.lampreydb.com), which consists of detailed chemical, qualitative, quantitative, and spatial distribution information of each identified metabolite in lamprey (Fig. 3). The current version of LampreyDB includes information on over 1,000 metabolites, encompassing 2,031 records from both positive and negative ion modes. Users can query and check their metabolites of interest or identify unknown peaks using this database.
Overview of the LampreyDB spatial metabolomics database. (a) LampreyDB consists of 2,032 records of chemical, spectral, and distribution information for 1,000 metabolites detected from different lamprey tissues, with 1,706 derived from positive ion mode and 326 from negative ion mode. (b) Users can query their metabolites of interest by searching chemical formulas or exact m/z values in the database. (c) LampreyDB provides extensive chemical information for each metabolite in the database, including metabolite name, class, chemical formula, retention time, accurate m/z value, SMILES, InChIKey, and chemical structure. (d) The interactive MS/MS spectrum for each metabolite allows users to visualize and analyze fragmentation patterns, facilitating the identification and comparison of metabolites within the database. (e) The interactive anatomical heatmap enables users to visually inspect and compare the spatial distribution of metabolites.
Code availability
The code for statistical analysis is available at https://github.com/YonghuiDong/LampreyStat. The code for lamprey spatial metabolomics database is available at https://github.com/YonghuiDong/LampreyDB.
References
Smith, J. J. et al. The sea lamprey germline genome provides insights into programmed genome rearrangement and vertebrate evolution. Nat Genet 50, 270–277 (2018).
Ajmani, N., Yasmin, T., Docker, M. F. & Good, S. V. Transcriptomic analysis of gonadal development in parasitic and non-parasitic lampreys (Ichthyomyzon spp.), with a comparison of genomic resources in these non-model species. G3 Genes|Genomes|Genetics 11, jkab030 (2021).
McCauley, D. W., Docker, M. F., Whyard, S. & Li, W. Lampreys as Diverse Model Organisms in the Genomics Era. BioScience 65, 1046–1056 (2015).
York, J. R., Lee, E. M.-J. & McCauley, D. W. The Lamprey as a Model Vertebrate in Evolutionary Developmental Biology. in Lampreys: Biology, Conservation and Control (ed. Docker, M. F.) 481–526, https://doi.org/10.1007/978-94-024-1684-8_6 (Springer Netherlands, Dordrecht, 2019).
Xu, Y., Zhu, S.-W. & Li, Q.-W. Lamprey: a model for vertebrate evolutionary research. Dongwuxue Yanjiu 37, 263–269 (2016).
York, J. R., Thresher, R. E. & McCauley, D. W. Applying functional genomics to the study of lamprey development and sea lamprey population control. Journal of Great Lakes Research 47, S639–S649 (2021).
Guo, P. et al. Dual nature of the adaptive immune system in lampreys. Nature 459, 796–801 (2009).
Rast, J. P. & Buckley, K. M. Lamprey immunity is far from primitive. Proc. Natl. Acad. Sci. USA 110, 5746–5747 (2013).
Guan, X., Lu, J., Sun, F., Li, Q. & Pang, Y. The Molecular Evolution and Functional Divergence of Lamprey Programmed Cell Death Genes. Front. Immunol. 10, 1382 (2019).
Ravi, V. et al. Lampreys, the jawless vertebrates, contain three Pax6 genes with distinct expression in eye, brain and pancreas. Sci Rep 9, 19559 (2019).
Smith, J. J. et al. Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat Genet 45, 415–421 (2013).
Timoshevskaya, N. et al. An improved germline genome assembly for the sea lamprey Petromyzon marinus illuminates the evolution of germline-specific chromosomes. Cell Reports 42, 112263 (2023).
Lawrence, M. J. et al. Differences in the transcriptome response in the gills of sea lamprey acutely exposed to 3-trifluoromethyl-4-nitrophenol (TFM), niclosamide or a TFM:niclosamide mixture. Comparative Biochemistry and Physiology Part D: Genomics and Proteomics 48, 101122 (2023).
Wang, Y. et al. Peptidomics Analysis Reveals the Buccal Gland of Jawless Vertebrate Lamprey as a Source of Multiple Bioactive Peptides. Marine Drugs 21, 389 (2023).
Xiao, R., Pang, Y. & Li, Q. W. The buccal gland of Lampetra japonica is a source of diverse bioactive proteins. Biochimie 94, 1075–1079 (2012).
Li, B. et al. Proteomic analysis of buccal gland secretion from fasting and feeding lampreys (Lampetra morii). Proteome Sci 16, 9 (2018).
Osório, J. & Rétaux, S. The lamprey in evolutionary studies. Dev Genes Evol 218, 221–235 (2008).
Gou, M. et al. Spatial Metabolomics Reveals the Multifaceted Nature of Lamprey Buccal Gland and Its Diverse Mechanisms for Blood-Feeding. Commun Biol 6, 881 (2023).
Li, K., Wang, H., Brant, C. O., Ahn, S. & Li, W. Multiplex quantification of lamprey specific bile acid derivatives in environmental water using UHPLC–MS/MS. Journal of Chromatography B 879, 3879–3886 (2011).
Johnson, N. S., Yun, S.-S., Buchinger, T. J. & Li, W. Multiple functions of a multi-component mating pheromone in sea lamprey Petromyzon marinus. Journal of Fish Biology 80, 538–554 (2012).
Alexandrov, T. Spatial Metabolomics and Imaging Mass Spectrometry in the Age of Artificial Intelligence. Annu. Rev. Biomed. Data Sci. 3, 61–87 (2020).
Taylor, M. J., Lukowski, J. K. & Anderton, C. R. Spatially Resolved Mass Spectrometry at the Single Cell: Recent Innovations in Proteomics and Metabolomics. J. Am. Soc. Mass Spectrom. 32, 872–894 (2021).
Dong, Y. & Aharoni, A. Image to insight: exploring natural products through mass spectrometry imaging. Natural Product Reports 39, 1510–1530 (2022).
Burger, B., Vaudel, M. & Barsnes, H. Importance of Block Randomization When Designing Proteomics Experiments. J. Proteome Res. 20, 122–128 (2021).
Tsugawa, H. et al. A cheminformatics approach to characterize metabolomes in stable-isotope-labeled organisms. Nat Methods 16, 295–298 (2019).
Tsugawa, H. et al. A lipidome atlas in MS-DIAL 4. Nat Biotechnol 38, 1159–1163 (2020).
Sud, M. et al. Metabolomics Workbench: An international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res 44, D463–D470 (2016).
Meng, G. et al. A Comprehensive Spatially Resolved Metabolomics Dataset for Lampreys. https://doi.org/10.21228/M8MN9F (2024).
Dong, Y. et al. RawHummus: an R Shiny app for automated raw data quality control in metabolomics. Bioinformatics 38, 2072–2074 (2022).
Smith, C. A., Want, E. J., O’Maille, G., Abagyan, R. & Siuzdak, G. XCMS: Processing Mass Spectrometry Data for Metabolite Profiling Using Nonlinear Peak Alignment, Matching, and Identification. Anal. Chem. 78, 779–787 (2006).
Pang, Z. et al. MetaboAnalyst 6.0: towards a unified platform for metabolomics data processing, analysis and interpretation. Nucleic Acids Research gkae253, https://doi.org/10.1093/nar/gkae253 (2024).
Adusumilli, R. & Mallick, P. Data Conversion with ProteoWizard msConvert. in Proteomics: Methods and Protocols (eds. Comai, L., Katz, J. E. & Mallick, P.) 339–368, https://doi.org/10.1007/978-1-4939-6747-6_23 (Springer, New York, NY, 2017).
Acknowledgements
This study was funded by the Chinese National Natural Science Foundation (Grant Nos. 31772884, 32070518), the Liaoning Climbing Scholar initiative, the Distinguished Professor of Liaoning Award (No. XLYC2002093), the Educational Department of Liaoning Province (Project No. LJKZ0962), the Open Fund of the Key Laboratory of Biotechnology and Bioresources Utilization at Dalian Minzu University, and the Ministry of Education (No. KF2022003).
Author information
Authors and Affiliations
Contributions
Y.D. and M.G. conceived the project. X.D., M.G. and Y.P. designed the methods and experiments. X.D. and M.G. performed the experiments. Y.D., M.G., Y.W. and X.W. analyzed the data. Y.D. and X.W. wrote the initial draft, and all authors participated in editing the manuscript. Y.P. supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Gou, M., Wang, X., Duan, X. et al. A Comprehensive Spatially Resolved Metabolomics Dataset for Lampreys. Sci Data 11, 1075 (2024). https://doi.org/10.1038/s41597-024-03925-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-024-03925-6