Abstract
Bladder cancer (BCa), as the most common malignant tumor of the urinary system, has received significant attention in research on the clinical application of artificial intelligence algorithms. Nevertheless, it has been observed that certain investigations use data from various medical facilities to train models for BCa, which may pose a privacy risk. Given this concern, protecting patient privacy during machine learning algorithm training is a crucial aspect that requires substantial attention. One emerging machine learning paradigm that addresses this concern is federated learning (FL). FL enables multiple entities to collaboratively build machine learning models while preserving data privacy and security. In this study, we present a multicenter BCa magnetic resonance imaging (MRI) dataset. The dataset comprises 275 three-dimensional bladder T2-weighted MRI scans collected from four medical centers, and each scan includes diagnostic pathological labels for muscle invasion and pixel-level annotations of tumor contours. Four FL methods are used to assess the baseline of the dataset for both the task of diagnosing muscle-invasive bladder cancer and automatic bladder tumor lesion segmentation.
Similar content being viewed by others
Background & Summary
Bladder cancer (BCa) is the most common malignant tumor of the urinary system with an incidence ranking tenth in malignant tumors worldwide1. The diagnosis and treatment of BCa involves various facets, such as imaging-based diagnosis, pathological image analysis, prognostic prediction, treatment planning, and research on molecular markers and genomics2. In diagnostic imaging studies of BCa, the performance of artificial intelligence (AI) techniques, particularly deep learning (DL) methods, has demonstrated comparable efficacy to that of experienced radiologists3,4. In those investigations, researchers use multi-center data for the development of DL models, aiming to enhance their accuracy and adaptability. Furthermore, external datasets are also utilized for validation purposes. However, the rise in privacy concerns complicates the sharing of sensitive medical data between different centers. It becomes more and more difficult to collect extensive clinical data from numerous centers for the training of DL models5.
In response to data privacy concerns arising from multi-center data modeling processes, Google introduced Federated Learning (FL) in 20166. FL is a distributed machine learning paradigm that allows each center (act as “a client”) to train models locally and then combine the local models into a global model. FL achieves the goal of jointly training a global model without exchange of local data. Presently, the limited availability of multi-center, standardized datasets for medical imaging of BCa poses a significant challenge to the widespread application and advancement of FL in the field of BCa.
In this study, we present a standardized multi-center BCa magnetic resonance imaging (MRI) dataset7, derived from real clinical scenarios. The dataset gathers data from four different hospitals. These hospitals are located in three cities, and the data collection follows the same patient inclusion and exclusion criteria. This variability in data sources, combined with differing characteristics such as scanning equipment and data volume, makes the dataset particularly well-suited for FL applications, as it effectively captures and addresses the heterogeneity found in real-world clinical settings.
The dataset consists of 275 three-dimensional (3D) T2-weighted (T2W) MRI scans of 228 BCa patients, with each patient bearing one or more bladder tumors. Each tumor in the dataset includes labels for tumor muscle invasion and annotations of tumor lesion contouring. Each tumor is accompanied by pathological examination results for muscle invasion in bladder cancer. BCa is typically categorized into non-muscle invasive bladder cancer (NMIBC) and muscle invasive bladder cancer (MIBC), depending on how deeply the tumor has grown into the bladder’s muscle wall. The two different tumor types exhibit different treatment modalities, prognostic indicators, and survival8,9,10,11,12, making accurate preoperative predictions of muscle invasion crucial for the clinical management of BCa treatment and prognosis. Tumor segmentation plays a critical role in clinical treatment, especially in radiation therapy-based cancer and oncology treatments. With the release of the dataset, the development of automated bladder tumor segmentation based on T2-weighted imaging (T2WI) can be significantly advanced.
To the best of our knowledge, existing FL medical image datasets are released through the challenge competition and are not accessible after the competition. Consequently, the introduction of the open-access multi-center MRI medical imaging dataset, carries significant implications for advancing FL research in the domain of medical image analysis. The dataset includes labels for tumor muscle invasion and tumor lesion annotations, making it possible to efficiently train MIBC diagnostic models and automated tumor segmentation models. The diversity of dataset’s labels provides the ground for investigating multi-task learning13 and mixed supervised learning14. Sourced from four different centers, the BCa dataset enables researchers to delve into areas such as FL6, domain generalization15, and domain adaptation16.
To validate the usage of the dataset for FL studies, we conduct a comprehensive survey of classical FL methods. These methods include FedAvg6, a pioneering approach in the field, and SiloBN17, a FL method that effectively addresses the challenge of disparate data distributions through the incorporation of a batch normalization layer. Additionally, we examine FedProx18 for effective management of data heterogeneity and FedBN19 for enhanced privacy protection. By leveraging these four FL methods, we constructed corresponding baseline for the dataset, specifically in diagnosing MIBC and performing automatic segmentation of BCa.
Methods
Cohort
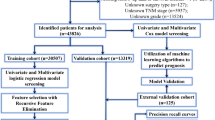
The dataset for this retrospective study was created under a waiver of informed consent, as the Ethics Committees determined the data to be non-sensitive and the study posed minimal risk to participants. The waiver was approved in accordance with the recommendations of the Ethics Committees of Dongguan Hospital affiliated with Southern Medical University (KYKT2019-027), the Ethics Committee of Sun Yat-sen University Cancer Prevention and Treatment Centre (B2023-552-01), the Ethics Committee of Zhuhai Hospital affiliated with Jinan University (2024-KT-34), and the Ethics Committee of Fifth Affiliated Hospital of Sun Yat-Sen University (L011-1). We conduct a retrospective collection of bladder T2WI data and clinical information from Dongguan Hospital, affiliated with the Southern Medical University (center 1), the Sun Yat-Sen University Cancer Centre (center 2), the Zhuhai Hospital affiliated with the Jinan University (center 3) and Fifth Affiliated Hospital of Sun Yat-Sen University (center 4) between November 2019 and July 2022, and included a total of 279 patients. All patients underwent either radical cystectomy, partial cystectomy, or transurethral resection of bladder tumor within 2 weeks after multiparametric MRI scanning.
The inclusion criteria for this study are as follows: (a) patients who are untreated or received only diagnostic transurethral resection of bladder tumor and (b) patients with bladder cancer confirmed by radical or partial cystectomy or transurethral resection of bladder tumor within 2 weeks of the multiparametric MRI. The following patients are excluded: (a) no surgical treatment, and pathological T stage could not be obtained (11 tumors); (b) histopathological type of non-urothelial carcinoma (inverted papilloma in two tumors, leiomyoma in two tumors, adenocarcinoma in three tumors, glandular cystitis in two tumors, and mesenchymal tumor in one case); (c) tumor recurrence after BCa surgery in 6 tumors; and (d) 11 patients with multiple tumors, but the corresponding pathological diagnosis results of the tumor are lost. The final dataset includes 228 patients. All patients are Asian due to the geographical location of the hospital. Table 1 presents data characteristics of tumors across each center.
MRIs
The images of T2WI are collected in four MRI scanners from four hospitals respectively. This result in a large data variability, due to the various imaging protocols used in different machines, scanners changes and updates. Shortly, the T2WIs are all performed in 3.0 T (100%). Summaries of the acquisition parameters for all the MRI modalities in the Table 2. The T2WIs have high resolution (1x1mm, or less) in horizontal planes, and typical slice thickness (3–5 mm) in clinical practice.
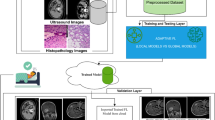
The images are fully de-identified by removing all direct and indirect identifiers protected under HIPAA (Health Insurance Portability and Accountability Act). The original DICOM (Digital Imaging and Communications in Medicine) files are converted to Neuroimaging Informatics Technology Initiative (Nifti) format (nii.gz) using dcm2niix (https://github.com/rordenlab/dcm2niix) with the anonymization option. Another round of visual quality control is preformed to secure complete anonymization, including 3D reconstruction of each image to guarantee that individuals could not be identified. The overall structure of the archive is represented in Fig. 1
Tumor annotations
The tumor annotations in the dataset are delineated on the T2WI images by an experienced radiologist (J.L., who has 14 years of work experience). These annotations are then reviewed and, if necessary, modified by another experienced radiologist (L.D. with 14 years of work experience). In instances of disagreement between the two radiologists, discussions are held until a consensus is reached, ensuring the quality of the annotations. Examples of T2WI and annotations are shown in Fig. 2.
All these patients are pathologically confirmed with BCa. For patients who underwent transurethral resection of a bladder tumor, a piece of detrusor muscle tissue at the tumor base is also removed for histopathologic examination to evaluate for detrusor muscle invasion. Pathologic specimens are obtained by TURBT in 222 tumours or by surgical resection in the other 57 tumors. Since each patient may have multiple tumors, the BCa dataset includes data for 275 tumors, with 160 tumors from Center 1, 48 from Center 2, 32 from Center 3, and 35 from Center 4. A total of 27 patients exhibit multiple tumors in the study cohort.
Typical tumors from four centers are shown in Fig. 2. Each center utilizes unique MRI equipment and scanning parameters, as detailed in Table 2.
In the BCa dataset, to protect the privacy of patients, basic clinical information (e.g., gender and age) is not disclosed. Figure 3 presents the distribution of tumor characteristics among the four centers.
Data Records
The dataset7 is deposited in Zenodo (https://zenodo.org/records/10409145). Because the data are originally assembled under a waiver of patient consent, the dataset is released under a CC-BY license, allowing for open access and use with proper attribution. The data structure, format, and naming are shown as follows (Fig. 4):
The process of our data collection and processing is illustrated in Fig. 1.
-
1.
Within the “FedBCa” directory, image data in the Nifti format (nii.gz) is sorted into four subdirectories, each corresponding to a different data collection center:
“Center 1” stores T2W images collected from Center 1.
“Center 2” stores T2W images collected from Center 2.
“Center 3” stores T2W images collected from Center 3.
“Center 4” stores T2W images collected from Center 4.
-
2.
Each “Center X” folder is meticulously organized to encompass two subfolders and an Excel spreadsheet.
The “Image” subfolder is dedicated to storing T2W image data specific to the center, containing a collection of subject images in the Nifti format (nii.gz).
The “Annotation” subfolder within each center’s directory contains the manual annotation data for the images, offering a precise delineation of tumors, also in the Nifti format (nii.gz).
“Center_X_label.xlsx” records the filenames of the image data, their corresponding annotation filenames, and includes the pathological labels of MIBC.
Technical Validation
Quality control for images and annotations
In this study, rigorous quality control is applied to MRI images and annotations. Firstly, to ensure that the population of research subjects is sufficiently consistent on key characteristics, all MRI images are selected based on uniform inclusion and exclusion criteria. Secondly, each image underwent quality assessment to ensure the absence of motion blur or artifacts, and to maintain sufficient clarity for accurately depicting details of the regions of interest. For image annotations, experienced radiologists are tasked with precise tumor localization and annotation. To guarantee the accuracy and consistency of annotations, a double-review process is employed, where one radiologist performs the annotation and another experienced radiologist reassess each annotation to ensure reliability. We calculate intra-rater reliability using the Dice similarity coefficient, which indicates if the same voxels are being selected as part of the lesion mask or not. For Dice calculation, we compare the annotations of two radiologists for all 275 cases, and the intra-rater Dice coefficient is 0.870. We also calculated the intraclass correlation coefficient (ICC) for the lesion volumes. The ICC ranges from 0–1; 1 is total agreement. The intra-rater ICC is 0.988. These quality control measures aim to enhance the validity and credibility of the dataset in bladder cancer diagnosis research.
Experimental verification in federated learning tasks
To assess the enhancement of accuracy and generalization provided by FL, we utilize FL methods, centralized training (mixed data from four centers), and single-center training to develop a MIBC prediction model or automated tumor segmentation model, respectively. To build the baseline of FL in the dataset, we conduct a survey on classical FL methods.
These methods include FedAvg6, SiloBN17, FedProx18, and FedBN19, each with distinct algorithm designs and implementation details. FedAvg6 is a foundational algorithm that trains a global model across multiple clients while keeping data localized. FedAvg involves initializing a global model, performing local training on each client, sending model updates to the server, and averaging these updates to form a new global model, iterating until convergence. SiloBN17 addresses data heterogeneity in multi-center medical investigations by combining a local batch normalization (BN) layer with center-specific statistics. This approach results in a model that is jointly trained and tailored to each center. SiloBN enhances robustness under varying data conditions while minimizing the risk of information leakage by avoiding the sharing of center-specific activation statistics. FedProx18 improves the handling of non-IID data through a re-parameterization module and targeted parameter modifications for individual clients. FedProx also allows for varying quantities of local tasks across devices and stabilizes the method with an approximation term. FedBN19 facilitates feature transfer among heterogeneous clients by enabling the exchange of extracted model attributes instead of raw data. Local BN is employed to align feature distributions across clients, ensuring consistency and supporting local model training.
We use these four FL methods to build corresponding baseline of the dataset in diagnosing MIBC and performing automatic segmentation of BCa. Subsequently, we compare the performance of these methods on the test set (Tables 3 & 4).
In this study, we conduct all experiments using PyTorch for training on NVIDIA A100 GPUs. The models are trained in a Python environment (version 3.8; https://www.python.org/), utilizing PyTorch (version 1.13.1; https://pytorch.org/). Our computing system is equipped with Intel Xeon Gold 6326 processors.
We refine the preprocessing of bladder MR images, adapting to different tasks in this study. Each slice of the 3D T2WI is cropped to uniform dimensions. For classification task, original T2WI slices are cropped to create 128 × 128 patches centered around the tumor annotations. For segmentation tasks, the cropping frame size of T2WI slices is set at 160 × 160. The cropped frame, centered around annotations, is randomly offset by 10 to 15 pixels in the x-y axes. Figure 5 shows an overview of the experimental process.
An overview of the experimental procedure. Each center acts as a client. For each round of communication, a certain percentage of clients are randomly selected to the train local model and send the local model to the server. The server aggregates the new global model and updates the model of client.
We use image augmentation techniques, including horizontal and vertical flipping, image cropping, and affine transformations, to optimize the utilization of our data representation. For model optimization, we utilize the Adam optimizer with a fixed learning rate of 1e-05. In model training, the Cross-entropy loss20 function is adopted for classification tasks, while Dice loss21 is utilized for segmentation tasks. The batch size is set to 24, and the training is conducted over 500 epochs.Considering the limited sample size from center 2, 3, and 4, we select U-Net22 network, which is effective with small datasets, as the backbone for our segmentation tasks. We select ResNet-5023, a well-regarded classification network, as the backbone for the classification tasks. We randomly select 40% of data from each center for testing in classification tasks. For segmentation tasks, a randomly selected subset of 30% patients from each center is used to assess the performance of the model.
To balance computational efficiency and model accuracy, we set the proportion of clients participating in federated aggregation per round is set to 0.5, meaning approximately half of the clients participate in each global model aggregation. The number of local training epochs before each aggregation is set to 1, indicating that the local model trains for one epoch before aggregation. The batch size for local model training is set to 24. In this study, we utilize the Area Under the ROC Curve (AUC) to evaluate the performance of the classification models, and Dice similarity coefficient (DSC) to evaluate the segmentation performance.
The classification task results for the dataset are presented in Table 3. The Centralized training, which combines the training data of four centers, exhibits the highest AUC, with a mean value of 0.866. Among FL methods, SiloBN achieves the highest average AUC (0.849), followed by FedBN (AUC = 0.842). FedAvg and FedProx show competitive performance with AUCs of 0.839 and 0.824, respectively.
The prediction model trained on a single center demonstrates average AUCs ranging from 0.783 to 0.811. Among these, the model trained by Center 1 achieves the highest diagnostic accuracy. Notably, the diagnostic accuracy of all models trained on a single center is lower than the FL method.
Centralized training achieves the highest automatic segmentation accuracy (DSC = 0.841), as detailed in Table 4. The model trained by the data from Center 1 achieves the highest single-center training results (DSC = 0.770), which may be due to its larger data volume. All the four FL methods outperform single-center training. Among them, the FedProx method achieves a segmentation accuracy (DSC = 0.840) second only to centralized training. FedBN and SiloBN show competitive performance with DSCs of 0.837 and 0.831, respectively. It is noteworthy that the FL methods not only achieve superior segmentation accuracy over single-center training on average DSC, but this trend is consistently observed across each center. Figure 6 presents the segmentation results of four typical cases of the dataset with different methods, indicating that the models trained by centralized training and FL are more accurate in segmentation.
It is worth noting that models trained at a single center do not always perform well on test data from their own center, both in Classification and Segmentation tasks. The analysis of the data reveals several reasons for this issue. Firstly, each center’s dataset may not capture the full range of variability in the overall data distribution, leading to models that are overly specialized and fail to generalize well even within the same center. For example, the model trained at Center 1 has an AUC of 0.720 on its own test data but performs better on data from other centers, achieving an AUC of 0.900 on Center 3’s test data. Secondly, small sample sizes and data noise within each center can affect the model’s ability to learn robust features, leading to suboptimal performance. This is evident in the model trained at Center 4, which has an AUC of 0.750 on its own test data. These performance discrepancies highlight the challenges of single-center training and emphasize the advantages of centralized and federated learning approaches in developing more robust and generalizable models.
Usage Notes
The FAIR (Findable, Accessible, Interoperable, and Reusable) Principles24 have gained widespread adoption in the realm of open data management. Existing FL datasets such as those utilized in FeTS challenge (https://fets-ai.github.io/Challenge/) and FL Breast Density Challenge (https://zenodo.org/records/6362204) from the MICCAI challenge do not fulfill the principle of “Accessible” after the competition.
We share a multi-center bladder T2WI dataset with labels for tumor muscle invasion and tumor lesion annotations, in alignment with the broad aim of the biomedical community to share FAIR data. Despite the inherent challenges in image processing, the image heterogeneity is an important feature of the dataset as it guarantees that tools developed using these images can be applied broadly. As shown in Fig. 3, BCa from different centers differed in grey value distribution, tumor size, tumor number and NMIBC/MIBC on T2WI. Sourced from four centers, the dataset proposed in this study facilitates research into FL6, domain generalization15, and domain adaptation16.
We have organized the data in accordance with the structure used in the Medical Segmentation Decathlon25, a popular abdominal organ segmentation competition. To facilitate the sharing and replication of findings, we have segregated the data into training and testing sets. Additionally, we provide the code for FL model training, which can be accessed at https://github.com/MedcAILab/FedBCa. Our data are deposited in Zenodo (https://zenodo.org/), which can be easily used by the AI community and is user-friendly organized to improve access to non-expert data analysts.
The dataset introduced in this study, being the first open-source multi-center bladder T2WI dataset, exhibits substantial research potential. In this study, we mine the usage of this dataset for FL studies. The strength of FL lies in its ability to train a global model, which outperforms the diagnostic accuracy and generalization performance of a model trained solely at a single center, while ensuring data privacy. Our findings serve as validation for the aforementioned advantages. To this end, we build a FL model training framework FedBCa (https://github.com/MedcAILab/FedBCa) based on PyTorch (https://pytorch.org/). Given the provision of preprocessed image data, users are only required to adjust the data paths within the code. The FedBCa framework is a publicly available, user-friendly FL training tool, with detailed user instructions provided, as well as code for four classical FL methods.
Code availability
We have released a code repository for automated classification and segmentation of MIBC with federated learning (https://github.com/MedcAILab/FedBCa). The applied neural network is based on the U-Net described in has been adapted to yield segmentation results. The ResNet described in has been adapted to yield classification results. Our implementation uses PyTorch based framework for deep learning in healthcare imaging. This new implementation is devised to provide a starting point for researchers interested in federated learning using state-of-the art federated learning frameworks for medical image processing.
References
Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians 68, 394–424 (2018).
Sherif A., Jonsson M. N. & NP, W. Treatment of muscleinvasive bladder cancer. Expert Review of Anticancer Therapy (2007).
Li, J. et al. Predicting muscle invasion in bladder cancer by deep learning analysis of MRI: comparison with vesical imaging–reporting and data system. European Radiology 33, 2699–2709 (2023).
Li, J. et al. Predicting muscle invasion in bladder cancer based on MRI: A comparison of radiomics, and single-task and multi-task deep learning. Computer Methods and Programs in Biomedicine 233, 107466 (2023).
Dehmer, G. J. et al. The National Cardiovascular Data Registry Voluntary Public Reporting Program. Journal of the American College of Cardiology 67, 205–215 (2016).
McMahan, Brendan et al. Communication-efficient learning of deep networks from decentralized data. Artificial intelligence and statistics. PMLR, 2017.
Cao, K. et al. (2023). A multi-center MRI dataset for bladder cancer and baseline evaluations of federated learning in its clinical application: Zenodo. https://doi.org/10.5281/zenodo.13622759 (2024)
Witjes, J. A. et al. European Association of Urology Guidelines on Muscle-invasive and Metastatic Bladder Cancer: Summary of the 2020 Guidelines. European Urology 79, 82–104 (2021).
Cookson, M. S. et al. The treated natural history of high risk superficial bladder cancer: 15-year outcome. Journal of Urology 158, 62–67 (1997).
Chang, S. S. et al. Diagnosis and Treatment of Non-Muscle Invasive Bladder Cancer: AUA/SUO Guideline. The Journal of Urology 196(4), 1021–1029 (2016).
Babjuk, M. et al. EAU Guidelines on Non–Muscle-invasive Urothelial Carcinoma of the Bladder: Update 2016. European Urology 71, 447–461 (2017).
Chou, R. et al. Treatment of muscle‐invasive bladder cancer: A systematic review. Cancer 122, 842–851 (2016).
Xu, Q. et al. Multi-Task Joint Learning Model for Segmenting and Classifying Tongue Images Using a Deep Neural Network. Ieee Journal of Biomedical and Health Informatics 24, 2481–2489 (2020).
Wicaksana, J. et al. FedMix: Mixed Supervised Federated Learning for Medical Image Segmentation. Ieee Transactions On Medical Imaging:1 (2022).
Blanchard, G., Lee, G. & Scott, C. Generalizing from Several Related Classification Tasks to a New Unlabeled Sample. Advances in Neural Information Processing Systems 2178–2186 (2011).
Saenko, K., Kulis, B., Fritz, M. & Darrell, T. Adapting visual category models to new domains. Computer Vision–ECCV 2010: 11th European Conference on Computer Vision, Heraklion, Crete, Greece, September 5–11, 2010, Proceedings, Part IV 11 (2010).
Andreux, M., Jean, O. D. T., Beguier, C. & Tramel, E. W. Siloed Federated Learning for Multi-Centric Histopathology Datasets. Ithaca: Cornell University Library, arXiv.org. Reprinted. https://doi.org/10.1007/978-3-030-60548-3_13 (2020).
Li, T. et al. Federated optimization in heterogeneous networks. MLSys (2020).
Li, X., Jiang, M., Zhang, X., Kamp, M. & Dou, Q. FedBN: Federated Learning on Non-IID Features via Local Batch Normalization. ICLR (2021).
Mao, A., Mohri, M. & Zhong, Y. Cross-Entropy Loss Functions: Theoretical Analysis and Applications. International conference on Machine learning. PMLR, 2023.
Milletari, F. Navab, N. & Ahmadi, S.-A. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation2016 Fourth International Conference on 3D Vision (3DV), 565–571, https://doi.org/10.1109/3DV.2016.79 (2016).
Ronneberger, O., Fischer, P. & Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. Paper presented at the Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015 (2015).
He, K., Zhang, X., Ren, S. & Sun J. Deep Residual Learning for Image Recognition. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 770–778 (2016).
Wilkinson, M. D. et al. The FAIR Guiding Principles for scientific data management and stewardship. Scientific Data 3, 160018 (2016).
Antonelli, M. et al. The Medical Segmentation Decathlon. Nature Communications 13, 4128 (2022).
Acknowledgements
This work is supported in part by the National Natural Science Foundation of China (Grant Number 62371303), the Dongguan Science and Technology of Social Development Program (Grant Number 20211800905212), the Shenzhen-Hong Kong Institute of Brain Science-Shenzhen Fundamental Research Institutions of China (Grant Number 2023SHIBS0003) and the Macao Polytechnic University Grant (Grant Number RP/FCA-10/2023).
Author information
Authors and Affiliations
Contributions
In this study, K.C., J.L., C.Z. design the method and draft the manuscript. W.Z., J.Z., G.W., C.Z., J.L., L.D., and S.Y. collect and process the dataset. B. H, B.H., H.Z. and Y.S., review and edit the manuscript. B.H., and Y.Z. coordinate and supervise the whole work. All authors are involved in critical revisions of the manuscript and have read and approved the final version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Cao, K., Zou, Y., Zhang, C. et al. A multicenter bladder cancer MRI dataset and baseline evaluation of federated learning in clinical application. Sci Data 11, 1147 (2024). https://doi.org/10.1038/s41597-024-03971-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-024-03971-0
This article is cited by
-
Bladder lesion detection using EfficientNet and hybrid attention transformer through attention transformation
Scientific Reports (2025)
-
FAME: A Federated Adversarial Learning Framework for Privacy-Preserving MRI Reconstruction
Applied Magnetic Resonance (2025)
-
Precise vesical wall staging of bladder cancer in the era of precision medicine: has it been fulfilled?
Abdominal Radiology (2024)