Abstract
Chronic postsurgical pain (CPSP), with a high prevalence and rising epidemic of opioids crisis, is typically derived from acute postoperative pain. Our knowledge on the forming of chronic pain mostly derives from mechanistic studies of pain processing in the brain and spinal cord circuits, yet most pharmacological interventions targeting CNS came to be unhelpful in preventing CPSP. Revealing the peripheral mechanisms behind the transition from acute to chronic pain after surgery could shine a light on the novel analgesic regimens. Based on two recognized animal models in simulation of acute and chronic postsurgical pain, we provide a next-generation RNA sequencing (RNA-seq) data set to evaluate the time-course transcriptomic variation in the tissue of skin, muscle and dorsal root ganglion (DRG) in these two pain models. The aim of this study is to identify the potential origin and mechanism of the persistent postoperative pain, and further to explore effective and safer analgesic regimens for surgical patients.
Similar content being viewed by others
Background
Acute pain is ubiquitous in surgical patients, however, some of them may last for a long time and transitions to chronic pain. Chronic postsurgical pain (CPSP), which has been associated with a rising epidemic of opioid abuse and even overdose-related death, is now attaching global attention1. The 11th revision of the Internation Classification of Disease defines Chronic postsurgical pain (CPSP) as pain occurs or develops in the incision areas, persists beyond the tissue healing process, and cannot be explained by other causes such as infection or tumor recurrence2. The reported prevalence of CPSP varies from 6% to 40% in adults underwent all types of surgeries, while the overall number is prodigious3,4,5. Yet CPSP has been increasingly recognized as a world-wide health problem, as it not only induced discomfort and disability, but also contribute to mal-prescription of opioids. However, clinical understanding on reasons or interventions for CPSP is poor, and the molecular mechanism behind the transition from acute to chronic pain remains elusive6.
Animal studies in chronic pain has achieved substantial progress in recent years, both peripheral tissue damage and modification in the brain and spinal cord have been proved to play a role in the development of chronic pain7. In the central nerve system (CNS), the nociceptive signals are transmitted via ascending projection neurons from spinal cord to cortex, while the noxious signals are also modulated in each level, forming a complicate descending analgesic pathway8,9. However, although complete reviews of neurol pathways of pain are available, precise clinical interventions to them is difficult, and outcomes are somehow disappointing. Comparatively, peripheral interventions, such as local anesthetic technics, have achieved more promising outcomes in at least controlling CPSP, if not preventing it10,11. Meanwhile, the peripheral tissue damage and somatosensory nerve injury are definitely involved in the initiation and development of chronic pain12. Thus, studies focusing on unravel the process of peripheral-neural interaction in the shape of pain after tissue damage is necessary. Previous animal studies already provide us with available animal model for understanding iatrogenic chronic pain. The Lateral Paw Incision (LPI) model, used a 1 cm incision through the skin to muscle of the rat plantar hindpaw, which could result in a short-term incisional pain which last 48-hours or more in rats, while the Skin/Muscle Incision and Retraction (SMIR) model develops a persistent pain by skin-muscle incision and retraction in the thigh of the rat. These two models are ideal paradigms for understanding postoperative acute and chronic pain, as all of them involved skill-muscle injury and cause reliable nociceptive behaviors under hindpaw stimulation13,14. Previously, Dr. Huang reported and compared time-course expressions of several inflammatory-associated genes in the dorsal root ganglion (DRG) of LPI and SMIR rats, using q-PCR15. Besides, their study also applied next-generation RNA sequencing (RNA-seq) to evaluate the time-course transcriptomic variation in the tissue of skin, muscle and DRG, to explore the potential origin of the temporary and persistent postoperative pain, and further to expound pathogenic mechanisms behind the transition of acute to chronic pain. Inspired by the design of previous studies and the technology diffusion of gene sequencing, we implemented this RNA-seq to evaluate the time-course transcriptomic variation in the tissue of skin, muscle and DRG, and further to identify the potential origin and mechanism of CPSP.
Methods
All animal experiments were approved by Animal Care and Use Committee of Navel Medical University, and conformed to the guidelines of the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Male adult (250 to 300 g) Sprague-Dawley rats were housed in a mixed-sex rodent room, and were maintained on a 12-hr light-dark cycle (6am–6 pm) with free available to food and water. Rats were acclimatized to the rodent room for at least 1 week prior to the procedure. A total of 60 rats were used, after baseline mechanical sensitivity measurement, 51 of them were arranged for LPI and SMIR model, 15 were then excluded during the study process. Rats excluded mainly for failure of modeling or local infection. Ultimately 20 rats in SMIR and 16 in LPI group fulfilled the study schedules, with 4 rats were used for tissue collection and RNA-sequencing in each time-point. All rats were sacrificed by euthanasia. The study flow is presented in Fig. 1.
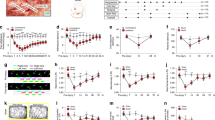
Experimental workflow. The Skin/Muscle Incision and Retraction (SMIR) and Lateral Paw Incision (LPI) animal models were established, and timepoints for behavior test and the followed tissue harvest as described (N = 4 in each timepoint). Following transcardial perfusion, tissues of the skin, muscle and dorsal root ganglion were extracted, stored and processed for RNA extraction and sequencing.
Lateral paw incision
The LPI surgery was performed as previously described. Rats were anesthetized with sevoflurane inhalation anesthetic for quick induction and recovery. A 1-cm longitudinal incision was made along the hairs bordering the lateral plantar surface of the hind paw using scalpel, the depth was through the skin, fascia to the underlying flexor muscle. Pressure applied for a minute to stop bleeding, and the skin was immediately sutured using 5–0 nylon sutures, with topical antibiotics administered. Animals were then separately placed in their own cages for recover. Sutures were removed 36 hours later by when the wound was mostly healed.
Skin/muscle incision and retraction
The SMIR surgery was performed as previously described. Rats were anesthetized with intraperitoneal sodium pentobarbital (CAS:57-33-0, Sigma–Aldrich, St. Louis, MO, USA), at doses of 50 mg/kg. Then rats were lied on their back on operating table. After hair removal and skin sterilization, a 15 mm skin incision was made 5 mm medial to the saphenous vein, to visualize the saphenous nerve and thigh muscle. Then, a short incision, 4 mm medial to the saphenous vein and nerve, was made on the muscle, to allow blunt dissection.
A blunt scissor was used at first to enlarge and deepen the cut, exposing the underlying fascia of the adductor muscles. A customed animal tissue dissecting retractor was then inserted into the full-thickness the of the skin-muscular incision, to retract it by 2 cm and persist for 1 hour. During the retraction, open wound was covered with wet sterile gauze to prevent fluid loss and infection. Animals were transferred to a temperature and closely monitored. After the 1 hour retraction, check for the validity of the skin and muscle. Then skin closure, resuscitation maintained as LPI rats.
Behavioral test
All animals were acclimatized to the behavioral test room for 1 hour before each behavioral test. Animals were placed on an elevated wire mesh surface, mechanical allodynia/ hyperalgesia was detected using Von-Frey monofilaments to the mid-plantar area of the hindpaw encircled by footpads. The filaments with increasing bending forces of 4 g, 6 g, 10 g and 15 g were applied in order, each filament was applied 10 times, and the number of hindpaw withdrawal was recorded, then the percentage of withdraw response was calculated.
One day prior to the procedures, baseline mechanical sensitivity of all rats was measured, and this time-point was denoted as 0 Day. Rats with abnormal baseline performance were removed. In LPI rats, the mechanical pain was measured at 6 hours, 2 days and 10 days after surgery. While in SMIR, the time points were 6 hours, 2 days, 10 days and 32 days after surgery. The time points were selected according to the previous researches. For rats met the following situations were determined as failure of modeling and removed from the study: significant restriction of movements or disability; local or general infection; delayed wound-healing; unreliable or unexpected allodynia response in each behavioral test. Notably, after the behavioral test and removal in each time-point (including 0 Day), 4 rats in each of the two groups were randomly picked up for tissue harvest.
Skin, muscle and DRG isolation
Rats were deeply anesthetized with isoflurane, transcardially perfused with nuclease-free saline (pH 7.4). The incised skin and muscle tissue were rapidly dissected, the C3-C6 DRGs of LPI and C2-C5 DRGs of SMIR were then extracted. All samples were immediately transferred to ice-cold RNAlater solution (AM7020, Thermo, Waltham, MA, USA) and refrigerated at 4 °C for 48 hours. Then samples were removed from RNAlater solution, placed into fresh 1.5 mL conical tubes and stored at −80°C.
RNA isolation and sequencing
Total RNA was isolated from each thymic sample using the RNAmini kit (52906, Qiagen, Germany). Gel electrophoresis and Qubit (Q33226, Thermo, Waltham, MA, USA) were used to exam the RNA quality and RNA integrity number (RIN). All samples had RIN values ≥ 7.0 (Table 2) and at least 10 ng total RNA. For RNA sequencing, RNA samples from four biological replicates at each time point were separated into independent pools, at equal amounts. Strand-specific libraries were constructed using the TruSeq RNA sample preparation kit (RS-122-2001, Illumina, San Diego, CA, USA), and sequencing was carried out by 2 × 150 bp paired-end using the lllumina Novaseq 6000 instrument. The raw data was trimmed by Skewer and the data quality was checked by FastQC (Supplementary Figs. 1–36). Clean reads were aligned to the Rattus norvegicus, genome assembly Rnor_6.0, using STAR. Mapping results were provided in Table 2, with the base ratios of Q20 and Q30 were counted as well. length was.. The expression of the transcript was calculated by FPKM (Fragments Per Kilobase of exon model per Million mapped reads) using Perl. Differentially expression transcripts (DETs) were determined using the MA-plot-based method with Random Sampling (MARS) model in the DEGseq package between different time points (as described in Table 3). Generally, in MARS model, M = log2C1 - log2C2, and A = (log2C1 + log2C2)/2 (C1 and C2 denote the counts of reads mapped to a specific gene obtained from two samples). The thresholds for determining DEGs are P < 0.05 and absolute fold change ≥ 2. Numbers of up and down-regulated DEGs in all comparisons presented in Table 3.
Data Records
Technical Validation
All animals had general check-up and behavioral test to verify the pain model, to make sure all samples send for sequencing represented reliable LPI/SMIR animal. The results of mechanical allodynia/hyperalgesia of all rats are exhibited in Fig. 2. Besides, as our samples represent heterogeneous populations of cells in the skin, muscle and DRG, the location of incision and extracted tissues were all defined in advance, and all tissues were extracted in certain volume and weight. The name scheme and NCBI data accession of all 108 samples were summarized in Table 1 and more specifically in Supplementary Table S1. The quality of RNA was examined using gel electrophoresis and Qubit. All samples showed clear band in the gel. The measured RNA concentration and RNA integrity numbers (RIN) of all samples were high (Table 2, Supplementary Table S2), which were appropriate for deep sequencing. RNA-seq data quality was determined using FastQC. A representative FastQC report is depicted in Fig. 3. As indicated, the reads had universally high-quality values (Fig. 3A). Meanwhile, the representative distribution of read length showed peak at 150 bp (Fig. 3B), while the distribution of GC content was similar to the theoretical distribution, indicates very low contamination (Fig. 3C). Then, the coverage uniformity along with transcripts was measured, with no significant 5′ or 3′ end bias identified (Fig. 3D). The regional distribution of all reads mapped to the reference genome were also displayed (Fig. 3E). We summarized the general RNA quantification and quality control results of animal models and tissues in Table 2, while the specific sequencing data of each sample were also presented in Supplementary Table S2. The results suggested that the average input fragment size per sample was 150 bp with a mean read depth of 44.36 million, and a high percentage of reads were mapped to the reference rat genome (97.69% alignment; Table 2). Besides, the lowest Q20/Q30 reaches 97.45%/93.50%, which indicates more confident base call (Supplementary Table S2). Table 3 provides summary data of differential expressed genes during the comparisons. PCA plots for the 6 groups are provided in Fig. 4.
Mechanical pain assessment of the LPI and SMIR animals sending for sequencing. A total of 108 36 rats were finally used for sequencing, with 16 in LPI while another 20 in SMIR. By each time-point of tissue harvest, the withdrawal threshold of the ipsilateral (red line) and contralateral (blue line) hindpaw of all rats were measured using Von-Frey monofilaments, following an increasing bending forces of 4 g, 6 g, 10 g and 15 g in order.
Representative quality check for RNA-seq. (A) Representative quality score distribution for read1 150 bp bases. (B) Representative distribution of read1 length. (C) Representative distribution of GC content for each read1. (D) Regional distribution of all reads mapped to the reference genome. (E) Coverage uniformity along with transcripts.
Principle component analysis (PCA) illustrates the clustering of (A) LPI_SKI; (B) LPI_MUS; (C) LPI_DRG; (D) SMIR_SKI; (E) SMIR_MUS; (F) SMIR_DRG. In each PCA plot, the first two principal components are shown, and the percent of total variation explained by each component is shown in the axis titles. Samples with similar characteristics appear close to each other, and samples with dissimilar characteristics are farther apart.
Code availability
The following open access software was used in this study, and we used default parameters with no custom code was used beyond the tools listed.
1. skewer (version 0.2.2) was used to trim adapters and flter quality read: https://sourceforge.net/projects/skewer/files/?source=navbar.
2. FastQC (version 0.11.5) was used to check the quality of clean reads: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
3. STAR (version 2.5.2b) was used to map sequence reads to the Rattus Rnor6 genome: https://github.com/alexdobin/STAR.
4. StringTie (version 2.2.1) was used to count the original sequence reads of known genes: http://ccb.jhu.edu/software/stringtie.
5. DESeq2 (version 1.16.1) was used to identify diferentially expressed genes: https://bioconductor.org/packages/release/bioc/html/DESeq2.html.
6. Pheatmap was used to plot the heatmap: https://cran.r-project.org/web/packages/pheatmap/index.html.
7. TopGO was used for GO functional enrichment analysis: http://www.bioconductor.org/packages/release/bioc/html/topGO.html.
References
Glare, P., Aubrey, K. R. & Myles, P. S. Transition from acute to chronic pain after surgery. Lancet 393, 1537–1546, https://doi.org/10.1016/S0140-6736(19)30352-6 (2019).
Schug, S. A. et al. The IASP classification of chronic pain for ICD-11: chronic postsurgical or posttraumatic pain. Pain 160, 45–52, https://doi.org/10.1097/j.pain.0000000000001413 (2018).
Richebé, P., Capdevila, X. & Rivat, C. Persistent Postsurgical Pain: Pathophysiology and Preventative Pharmacologic Considerations. Anesthesiology 129, 590–607, https://doi.org/10.1097/ALN.0000000000002238 (2018).
van Driel, M. E. C. et al. Development and validation of a multivariable prediction model for early prediction of chronic postsurgical pain in adults: a prospective co hort study. British journal of anaesthesia 129, 407–415, https://doi.org/10.1016/j.bja.2022.04.030 (2022).
van Helden, E. V. et al. Chronic postsurgical pain after minimally invasive adrenalectomy: prevalence and impact on quality of life. BMC Anesthesiol 22, 153, https://doi.org/10.1186/s12871-022-01696-4 (2022).
Buvanendran, A. Chronic postsurgical pain: are we closer to understanding the puzzle? Anesth Analg 115, 231–232, https://doi.org/10.1213/ANE.0b013e318258b9f7 (2012).
Kang, Y., Trewern, L., Jackman, J., McCartney, D. & Soni, A. Chronic pain: definitions and diagnosis. BMJ 381, e076036, https://doi.org/10.1136/bmj-2023-076036 (2023).
Barroso, J., Branco, P. & Apkarian, A. V. Brain mechanisms of chronic pain: critical role of translational approach. Transl Res 238, 76–89, https://doi.org/10.1016/j.trsl.2021.06.004 (2021).
Descalzi, G. et al. Epigenetic mechanisms of chronic pain. Trends Neurosci 38, 237–246, https://doi.org/10.1016/j.tins.2015.02.001 (2015).
Carley, M. E. et al. Pharmacotherapy for the Prevention of Chronic Pain after Surgery in Adults: An Updated Systematic Review and Meta-analysis. Anesthesiology 135, 304–325, https://doi.org/10.1097/ALN.0000000000003837 (2021).
Steyaert, A. & Lavand’homme, P. Prevention and Treatment of Chronic Postsurgical Pain: A Narrative Review. Drugs 78, 339–354, https://doi.org/10.1007/s40265-018-0866-x (2018).
Antony, A. B. et al. Neuromodulation of the Dorsal Root Ganglion for Chronic Postsurgical Pain. Pain Med 20, S41–S46, https://doi.org/10.1093/pm/pnz072 (2019).
Flatters, S. J. L. Characterization of a model of persistent postoperative pain evoked by skin/muscle incision and retraction (SMIR). Pain 135, 119–130, https://doi.org/10.1016/j.pain.2007.05.013 (2008).
Wang, C.-F., Pancaro, C., Gerner, P. & Strichartz, G. Prolonged suppression of postincisional pain by a slow-release formulation of lidocaine. Anesthesiology 114, 135–149, https://doi.org/10.1097/ALN.0b013e3182001996 (2011).
Huang, L., Wang, C.-F., Serhan, C. N. & Strichartz, G. Enduring prevention and transient reduction of postoperative pain by intrathecal resolvin D1. Pain 152, 557–565, https://doi.org/10.1016/j.pain.2010.11.021 (2010).
Meng, X., Bu, L., Shen, L. & Tao, K. A transcriptome data set for comparing skin, muscle and DRG between acute and chronic postsurgical pain rats. Gene Expression Omnibus https://identifiers.org/geo:GSE267799 (2024).
Acknowledgements
This study was supported by National Natural Science Foundation of China (NO. 82301449), and basic medical research project of Naval Medical University (2022QN095).
Author information
Authors and Affiliations
Contributions
Study concept and design: X.Y. Meng, K.M. Tao; drafting of the manuscript: X.Y. Meng; implement the trial: X.Y. Meng; analysis and interpretation of data: X.Y. Meng, L. Bu and L. Shen; revision of the manuscript: K.M. Tao. X.Y. Meng, L. Bu and L. Shen contributed equally to this work, all authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Meng, Xy., Bu, L., Shen, L. et al. A transcriptome data set for comparing skin, muscle and dorsal root ganglion between acute and chronic postsurgical pain rats. Sci Data 11, 1229 (2024). https://doi.org/10.1038/s41597-024-04078-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41597-024-04078-2