Abstract
Trait-based approaches are revolutionizing our understanding of high-diversity ecosystems by providing insights into the principles underlying key ecological processes, such as community assembly, species distribution, resilience, and the relationship between biodiversity and ecosystem functioning. In 2016, the Coral Trait Database advanced coral reef science by centralizing trait information for stony corals (i.e., Subphylum Anthozoa, Class Hexacorallia, Order Scleractinia). However, the absence of trait data for soft corals, gorgonians, and sea pens (i.e., Class Octocorallia) limits our understanding of ecosystems where these organisms are significant members and play pivotal roles. To address this gap, we introduce the Octocoral Trait Database, a global, open-source database of curated trait data for octocorals. This database houses species- and individual-level data, complemented by contextual information that provides a relevant framework for analyses. The inaugural dataset, OctocoralTraits v2.2, contains over 97,500 global trait observations across 98 traits and over 3,500 species. The database aims to evolve into a steadily growing, community-led resource that advances future marine science, with a particular emphasis on coral reef research.
Similar content being viewed by others
Background and Summary
Traits - the measurable properties of organisms - capture the biology and morphology of species and have been used for centuries to infer species boundaries based on shared characteristics1. Traits also dictate organismal performance and its interaction with the ecological niche2,3. Thus, exploring patterns of trait diversity across organisms and species also brings us closer to understanding the general principles that determine, among other processes, the patterns of abundance and distribution of species4,5, their evolution6, their resilience (e.g., resistance and recovery capacity)7,8, or their contribution to ecosystem functioning and services9,10. Consequently, the use of trait-based approaches in ecology has exponentially increased in recent decades, leading both to the development of new methods and an increase in the necessity of trait data among the scientific community11. The compilation of trait data across the Tree of Life remains nonetheless a major challenge12. In fact, trait databases are still restricted to relatively few taxonomic groups of mostly terrestrial organisms (e.g.13,14,15,16,17,18,19,20), and are even rarer in other systems traditionally less accessible to scientists, such as the marine realm. Nevertheless, considerable effort has recently been made to compile trait data for some marine groups (e.g., stony corals21, algae22, and fish23, among others). However, trait data gaps still exist for most taxa, and even when data are available for some species, it is often difficult and time-consuming to integrate them within a single study24. This is in part because trait information mostly comes from scattered sources that are often difficult to find or access, and often including different languages (e.g., taxonomic descriptions, local identification guides, grey literature, etc.). In addition, traits are often measured using differing methodologies and standards, and relevant contextual information is often lacking. This highlights the need for new trait databases that simplify the process for neglected yet ecologically important marine taxa.
Coral-dominated ecosystems are some of the most biodiverse ecosystems on Earth25. They provide millions of people with innumerable services including food provision, coastal protection, or recreation26. Yet, coral-dominated ecosystems are being severely transformed by global change, leading them into uncharted territories27,28,29. To advance the science of these systems more rapidly at this time of accelerating change, the Coral Trait Database was launched in 201621, which centralized diverse trait information for stony corals (i.e., Sub-Phylum Anthozoa, Class Hexacorallia, Order Scleractinia) into a single, open-access repository. The Coral Trait Database became the basis for research and education that has advanced coral reef science worldwide (e.g.8,27,30,31). However, these advances are limited because of the lack of information for the anthozoan Class Octocorallia, which significantly contributes to the biodiversity and functioning of many coral-dominated marine ecosystems.
Octocorals are a class of anthozoans that host more than 3,500 nominal species of mainly non-stony corals (e.g., soft corals, sea fans and sea pens) distributed across more than 75 families32. They can be found from the shallow waters to the deep sea and across all marine ecoregions33. Octocorals differ from hexacorals (i.e., scleractinians) in many traits; most notably they have polyps with eight pinnate tentacles rather than six or 6-fold non-pinnate tentacles, and they tend to have unconsolidated sclerites or semi-rigid skeletons rather than the hard skeletons of Scleractinia34,35. Like hexacorals, octocorals also provide critical functions to ecosystems and services to human societies. Octocorals can be foundation species that form three-dimensional habitats that are home, refuge, and spawning grounds for many associated taxa, which in turn increases biodiversity, stability, and food provision to humans36,37,38,39,40. Their complex morphologies can also locally modify water flows and light intensity, which may favor the settlement of sessile invertebrate larvae over opportunistic macroalgae, thus further promoting assemblage stability and resilience41,42. Octocorals can also directly influence nutrient cycling and the trophic network by acting as food source (e.g., for nudibranchs), and as suspension feeders that capture particulate organic matter and plankton from the water column43,44,45,46,47. Moreover, although octocorals are not typically considered reef-builders, they can contribute to the carbon cycle and reef calcification processes by photosynthesis and by fixing calcium carbonate (CaCO3) in their sclerites and skeletal structures48,49,50. Skeletons of some octocoral species such as Corallium rubrum have been used for jewelry by humans since ancient times51. Octocorals are also important sources of pharmaceutical products and have a high aesthetic value that enhances dive tourism and inspiration52,53.
Under intensifying global change, many octocoral species are playing important roles in terms of reef reconfiguration processes. For instance, in the Caribbean Sea, where stony corals have generally suffered marked declines in abundance following cumulative climatic impacts, most octocoral species have prevailed54,55,56. Similarly, some octocorals are becoming the new dominant groups in some impacted reefs of the Western and Central Indo-Pacific following a decline in hard coral cover57,58. Conversely, in other regions such as the Mediterranean Sea, habitat-forming octocorals are experiencing population collapses following recurrent marine heatwaves59.
Octocorals and their traits are thus an undeniably crucial piece of the puzzle to understand how coral-dominated ecosystems function, how they are being transformed by global change, and how we can improve their management in the Anthropocene. Here, we introduce the Octocoral Trait Database, a global, open-source database of curated trait data for octocorals. We release its first data descriptor, OctocoralTraits v2.2, which has been integrated with those data of the scleractinian corals in the Coral Trait Database (www.coraltraits.org), and hosts species- and individual-level data alongside contextual data that provide relevant framing for analyses. The primary goals of the new database are: (i) to aggregate diverse information on octocoral traits into a single open-access repository that ensures transparent and accurate archiving of coral trait data, (ii) to promote the appropriate crediting of original data sources, (iii) to continue engaging the reef coral research community in the gathering and quality control of trait data to facilitate future coral reef research, and iv) to expand the current stony coral version of the Coral Trait Database with octocoral data to promote the advancement of marine science, with a particular emphasis on coral reefs. Here, we publish a first global data release that contains a sample with more than 97,500 error-checked trait observations, including over 148,000 trait measurements across 128 traits (30 of which are contextual) and over 3500 valid octocoral species. Trait observations were compiled across all marine ecoregions and across all depth zones, from the deep sea to the shallow waters. Furthermore, while OctocoralTraits v2.2 serves as a static data descriptor, we envision it as an evolving data product that will continue to grow collaboratively to further facilitate the quantification of trait variation among coral species in the Anthropocene.
Methods
Ontology of the data descriptor
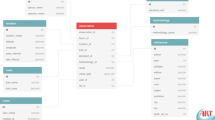
The structure of OctocoralTraits v2.2 matches that of the data descriptor published by Madin et al.21 for scleractinian corals, as well as those of other similar trait databases, such as AusTraits60. Specifically, it follows the principles of the Observation and Measurement Ontology61, where observations at the individual or species level bind associated measurements and may provide context for other observations (Fig. 1). For example, recording both the height and width of the same octocoral colony would be considered a single observation with two distinct measurements, each representing a different trait of the coral. If the water depth is also noted, it remains part of the same observation but is categorized as a contextual trait, as it does not directly pertain to the colony itself.
(a) General scheme on how data is structured in the database following the Observation-Measurement scheme61 and Madin et al.21. (b) Example of an observation. Each observation contains data for the data enterer, the species of interest, the scientific source, the type of access (i.e., optional variable that data enterers can control to keep data private before publication) and the location. In addition, each observation binds one or various measurements that apart from specifying the name of the measured trait (e.g., height of the colony), include information about: the standard used to measure it (e.g., m, cm…etc), the method used (e.g., ruler), the actual value measured, and the value type (e.g., raw data for a single measurement, expert/group opinion for single/consensus view of experts, mean, median, range, maximum, or minimum…). The number of replicates and estimates of precision (e.g., standard error, standard deviation…) can also be specified when applicable. Finally, one or multiple contextual traits (e.g., water depth, habitat type…), that might be relevant for explaining intra-specific variation in the trait/s of interest can also be associated to the observation.
Traits selection
OctocoralTraits v2.2 has been compiled to populate the Class Octocorallia within the Coral Trait Database with the first global data descriptor. To achieve this goal, we first identified a set of relevant octocoral traits based on: i) traits that were already present for scleractinian corals in the Coral Traits database and that also apply to octocorals, and ii) new traits identified by the team as relevant for octocorals in terms of their biomechanical, morphological, physiological, biogeographical, ecological, conservation and/or reproductive characteristics. The selected traits include both individual-level traits (i.e., heritable features of organisms) that are measured at the individual level and can potentially vary within species (e.g., growth rate, colony height) and species-level traits that are properties of species as entities and, therefore, invariant within species (e.g., species depth range, conservation status). The complete list of 98 selected traits can be found in Table S1, along with their definitions and detailed information on their types of associated variables (e.g., numeric vs categorical) and allowed values. Table S2 includes information on the 30 contextual traits (e.g., habitat characteristics such as depth), which provide additional information about the specific conditions under which individual-level traits were quantified. This information is crucial for understanding trait variation21.
Data acquisition
OctocoralTraits v2.2 was assembled using only scientific sources, ensuring peer-reviewed level of quality control. Specifically, in April 2022, we searched the Web of Science for scientific works using the general query: “octocoral*” OR “soft coral” OR “sea pen” OR “gorgonian” OR “sea fan”. We conducted a single, general search to balance the creation of an initial global dataset of octocoral trait data for the Coral Trait Database with maintaining a manageable workload for the team. This search yielded over 11,000 scientific sources, which were then manually screened at the abstract level using the abstrackr software62 to exclude sources unrelated to the traits or organisms of interest. This process led to an initial selection of 2,360 potentially relevant publications. However, many of them were subsequently excluded upon further review for not being related to the organisms or traits of interest. For example, many sources mentioned ‘soft corals’ in the abstract but referred to organisms outside of the Class Octocorallia, such as zoanthids.
From the selected sources, the data extraction process primarily involved capturing values from text and result tables. In the few cases where information was only available in figures, data were extracted by establishing a scale based on axis values using ImageJ2 software63. Finally, as data compilation progressed, some additional articles of interest—those not captured by the initial search but referenced within reviewed sources—were also included, along with some newly published articles unavailable at the time of the initial search. Similarly, although the initial search focused primarily on publications in English, some relevant publications in other languages (e.g., Spanish, German) were also included where team members were sufficiently confident with those languages. The complete list of 796 used data sources can be found at Supp. Table S332,36,49,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163,164,165,166,167,168,169,170,171,172,173,174,175,176,177,178,179,180,181,182,183,184,185,186,187,188,189,190,191,192,193,194,195,196,197,198,199,200,201,202,203,204,205,206,207,208,209,210,211,212,213,214,215,216,217,218,219,220,221,222,223,224,225,226,227,228,229,230,231,232,233,234,235,236,237,238,239,240,241,242,243,244,245,246,247,248,249,250,251,252,253,254,255,256,257,258,259,260,261,262,263,264,265,266,267,268,269,270,271,272,273,274,275,276,277,278,279,280,281,282,283,284,285,286,287,288,289,290,291,292,293,294,295,296,297,298,299,300,301,302,303,304,305,306,307,308,309,310,311,312,313,314,315,316,317,318,319,320,321,322,323,324,325,326,327,328,329,330,331,332,333,334,335,336,337,338,339,340,341,342,343,344,345,346,347,348,349,350,351,352,353,354,355,356,357,358,359,360,361,362,363,364,365,366,367,368,369,370,371,372,373,374,375,376,377,378,379,380,381,382,383,384,385,386,387,388,389,390,391,392,393,394,395,396,397,398,399,400,401,402,403,404,405,406,407,408,409,410,411,412,413,414,415,416,417,418,419,420,421,422,423,424,425,426,427,428,429,430,431,432,433,434,435,436,437,438,439,440,441,442,443,444,445,446,447,448,449,450,451,452,453,454,455,456,457,458,459,460,461,462,463,464,465,466,467,468,469,470,471,472,473,474,475,476,477,478,479,480,481,482,483,484,485,486,487,488,489,490,491,492,493,494,495,496,497,498,499,500,501,502,503,504,505,506,507,508,509,510,511,512,513,514,515,516,517,518,519,520,521,522,523,524,525,526,527,528,529,530,531,532,533,534,535,536,537,538,539,540,541,542,543,544,545,546,547,548,549,550,551,552,553,554,555,556,557,558,559,560,561,562,563,564,565,566,567,568,569,570,571,572,573,574,575,576,577,578,579,580,581,582,583,584,585,586,587,588,589,590,591,592,593,594,595,596,597,598,599,600,601,602,603,604,605,606,607,608,609,610,611,612,613,614,615,616,617,618,619,620,621,622,623,624,625,626,627,628,629,630,631,632,633,634,635,636,637,638,639,640,641,642,643,644,645,646,647,648,649,650,651,652,653,654,655,656,657,658,659,660,661,662,663,664,665,666,667,668,669,670,671,672,673,674,675,676,677,678,679,680,681,682,683,684,685,686,687,688,689,690,691,692,693,694,695,696,697,698,699,700,701,702,703,704,705,706,707,708,709,710,711,712,713,714,715,716,717,718,719,720,721,722,723,724,725,726,727,728,729,730,731,732,733,734,735,736,737,738,739,740,741,742,743,744,745,746,747,748,749,750,751,752,753,754,755,756,757,758,759,760,761,762,763,764,765,766,767,768,769,770,771,772,773,774,775,776,777,778,779,780,781,782,783,784,785,786,787,788,789,790,791,792,793,794,795,796,797,798,799,800,801,802,803,804,805,806,807,808,809,810,811,812,813,814,815,816,817,818,819,820,821,822,823,824,825,826,827,828,829,830,831,832,833,834,835,836,837,838,839,840,841,842,843,844,845,846,847,848,849,850,851,852,853,854,855,856,857,858,859.
This final list of data sources included published research articles, taxonomic monographs, scientific books, theses, identification guides, published conference abstracts and reports. Moreover, for the geographical traits’ marine province and marine realm, trait data compilation from the aforementioned sources was supplemented by information from the online, public, scientific platform OBIS (https://obis.org846). Specifically, we first retrieved occurrence data from the platform using the robis R package860 in RStudio software v2023.06.0 + 421861, utilizing the Aphia_ID code of each species as the taxon ID. Spatial outliers were then flagged with the CoordinateCleaner R package862 and manually examined for accuracy by consulting original sources. Verified data were subsequently classified into realms and/or provinces using the meowR package863 and incorporated into the data descriptor.
Species taxonomy
For the static OctocoralTraits v2.2 used in this descriptor, all trait observations are associated with accepted species of octocorals. The accepted name and taxonomy for each species were determined based on the most authoritative and up-to-date lists of octocoral species, namely the 2024 World List of Octocorallia864 and the World Register of Marine Species (WORMS, as of May 2024865) (http://www.marinespecies.org). In cases where source publications contained trait data for combinations that are no longer valid (e.g., Pseudopterogorgia americana), species names were updated in the database to reflect the currently accepted name (e.g., Antillogorgia americana). To ensure the traceability of taxonomic updates, all trait observations also include information on the Aphia ID code for the original species that appeared in the scientific source from which trait data was obtained. These Aphia ID codes are linked to the WORMS platform.
Data integration
To progressively integrate trait data from various scientific sources into a unified, structured dataset, we implemented a data processing workflow following Madin et al.21. Specifically, we: (i) manually assigned observation IDs to link related measurements, (ii) reformatted location coordinates to the decimal degree system when necessary, (iii) standardized terminology for traits, locations, methodologies, and standards across sources, (iv) updated taxonomy as needed (see previous section for details), and (v) harmonized values for categorical traits, as terminology can vary significantly among sources (refer to Table S1 for accepted values). A notable challenge was the extensive variety of nomenclature used in the scientific literature to describe growth forms across the Octocorallia tree of life, with different terms often describing the same concepts. To address this, we introduced a hierarchical classification system that aims to facilitate the use of octocoral morphologies in ecological studies by consolidating hundreds of terms into a manageable set of ecologically meaningful morphologies. It includes seven high-level Types of Growth (Trait 1) based on spatial occupation patterns, further subdivided into 17 basic (Trait 2) and 32 detailed (Trait 3) growth forms for finer resolution (see Tables S4-S11 for detailed descriptions). For example, a coral described as “fan-shaped” by an author in the original source would be hierarchically classified as “erect branched” (Trait 1; high level), “arborescent” (Trait 2; medium level), and “branched planar” (Trait 3; fine level) under the Type of Growth, Growth Form Basic, and Growth Form Detailed traits, respectively. Additionally, a fourth trait, labeled simply as Growth Form (Trait 4), retains the original terminology from the data source to maintain traceability.
A similar challenge was encountered with the vast existing terminology for octocoral skeletons. Here, we built upon the work of McFadden et al.32 to propose a summarized classification comprising 20 basic categories that encompass the observed variation in this trait across octocorals (see accepted values for the “Type of Skeleton” trait in Table S1).
Lastly, for quantitative traits (e.g., colony height), where standardization to a preferred unit is straightforward, we did not enforce a specific unit, leaving the choice of which standard to use up to end users.
Data Records
Access
The OctocoralTraits v2.2 data descriptor is publicly accessible as a ZIP file deposited in Zenodo866. This ZIP file, named OctocoralTraits, contains a primary core table in .csv format (OctocoralTraits_v2_2.csv) with trait observations linked to measurements, along with multiple interrelated tables (also in .csv format) that provide additional details on both the observations (e.g., location, species, resource) and the measurements (e.g., trait, methodology, standard, value type, precision type). Specific information about each of these core and interrelated tables is provided in Tables 1–8.
Data coverage
The OctocoralTraits v2.2 data release integrates more than 97,500 error-checked trait observations and over 148,000 trait measurements across 128 traits. These data encompass over 3,500 valid octocoral species and 75 families, with observations spanning all marine ecoregions and depth zones, from deep-sea habitats to shallow waters. OctocoralTraits v2.2 therefore offers a broad phylogenetic (Figs. 2 and 3), geographic (Fig. 4), and bathymetric (Fig. 5) coverage. However, considerable data gaps persist, since many species lack comprehensive trait data (Fig. 2), some traits are underrepresented across octocoral families (e.g., growth rate) (Fig. 3), and certain realms (e.g., polar) and depth zones (e.g., mesophotic) remain poorly sampled across the scientific community (Figs. 4 and 5).
Species-complete tree with phylogenetic distribution of trait data coverage (i.e., number of trait observations per species). The 15 largest families of the Class Octocorallia are labeled by colour. The two orders within the class, Scleralcyonacea and Malacalcyonacea, are also indicated. Arrowheads indicate that, for this first data release, Keratoisis grayi (4708 records), Muricea californica (437 records) and Corallium rubrum (280 records) have a disproportionately larger number of trait observations that do not fit in the plot. The phylogenetic tree was added to this figure solely for visualization purposes, organizing families by evolutionary relationships to enhance the interpretability of trait distributions across lineages, without implying further phylogenetic analysis. It corresponds to an adaptation of the family-resolved Maximum likelihood tree of Octocorallia inferred from 1059 bp alignment of mitochondrial gene mtMutS32, with species being incorporated as polytomies. (*) To facilitate visualization, all sea pens have been grouped into a single Pennatuloidea superfamily. Data for species belonging to genera that are currently incertae sedis have not been included in the figure. Finally, a complementary figure showing the phylogenetic distribution of trait data coverage as the number of different traits with data per species can be found in Fig. S1.
Family-complete heatmap with phylogenetic distribution of trait data coverage for each trait of interest. Parentheses following family names indicate the number of species within a given family. Purple cells correspond with the traits of interest released in this data descriptor, with color gradient indicating the % of species with data for a given trait, within a given family. Dark yellow cells indicate traits that are only possible in sea pens (e.g., the number of polyps per polyp leaf) and therefore which are not possible in other octocorals. (*) refers to Conservation trait category, while (**) refers to Stoichiometric trait category. To facilitate visualization, all sea pens have also been grouped into a single Pennatuloidea superfamily (as in Fig. 2). See Table S1 for the trait definitions and Fig. S2 for trait data coverage across genera whose family assignment is incertae sedis. The phylogenetic tree used to order families across the x axis corresponds to the family-resolved Maximum likelihood tree of Octocorallia inferred from 1059 bp alignment of mitochondrial gene mtMutS32. As for Fig. 2, it was added solely for visualization purposes, organizing families by evolutionary relationships to enhance the interpretability of trait distributions across lineages, without implying further phylogenetic analysis.
(a) Relative percentage of compiled trait observations among global and georeferenced estimates. (b) Geographic distribution of georeferenced trait data across marine realms. (c) Total number of trait observations per marine realm. (*) in panel c denotes that the number of trait observations for that region is higher than shown in the plot (i.e., Temperate Northern Atlantic; 5,633 records). Additional notes: As mentioned in Table 3, Global estimates are associated with traits that do not (or very rarely) vary within species regardless of the geographical context. Thus, a global estimate for a given trait can be assumed to be fairly or totally constant across the species. For instance, the presence of a skeletal axis is an inherent characteristic of gorgonians regardless of where the trait is quantified. Apart from the global estimates, OctocoralTraits v2.2 contains over 10,000 data points of geo-referenced records. These records are associated with traits that may vary within species depending on local/regional environmental parameters. Therefore, the geographic context of the trait observation is needed to provide relevant framing for analysis.
(a) Bathymetric (i.e., across light zones) distribution of compiled trait data across the 54 marine provinces where data has been collected. Numbers have been added to each marine province with data, corresponding exactly to the list provided by Spalding et al.868. Specifically: 1. Arctic, 2. Northern European Seas, 3. Lusitanian, 4. Mediterranean Sea, 5. Cold Temperate Northwest Atlantic, 6. Warm Temperate Northwest Atlantic, 8. Cold Temperate Northwest Pacific, 9. Warm Temperate Northwest Pacific, 10. Cold Temperate Northeast Pacific, 11. Warm Temperate Northeast Pacific, 12. Tropical Northwestern Atlantic, 13. North Brazil Shelf, 14. Tropical Southwestern Atlantic, 16. West African Transition, 17. Gulf of Guinea, 18. Red Sea and Gulf of Aden, 19. Somali/Arabian, 20. Western Indian Ocean, 22. Central Indian Ocean Islands, 24. Andaman, 25. South China Sea, 26. Sunda Shelf, 28. South Kuroshio, 29. Tropical Northwestern Pacific, 30. Western Coral Triangle, 31. Eastern Coral Triangle, 32. Sahul Shelf, 33. Northeast Australian Shelf, 34. Northwest Australian Shelf, 35. Tropical Southwestern Pacific, 36. Lord Howe and Norfolk Islands, 37. Hawaii, 38. Marshall, Gilbert, and Ellis Islands, 39. Central Polynesia, 40. Southeast Polynesia, 43. Tropical East Pacific, 44. Galapagos, 45. Warm Temperate Southeastern Pacific, 47. Warm Temperate Southwestern Atlantic, 48. Magellanic, 50. Benguela, 51. Agulhas, 53. Northern New Zealand, 54. Southern New Zealand, 55. East Central Australian Shelf, 56. Southeast Australian Shelf, 57. Southwest Australian Shelf, 58. West Central Australian Shelf, 59. Subantarctic Islands, 60. Scotia Sea, 61. Continental High Antarctic, 62. Subantarctic New Zealand. (b) Mean percentage (±SD) of trait observations per light zone across the 54 provinces with data.
Technical Validation
OctocoralTraits v2.2 is a curated data descriptor developed to contribute to the Coral Traits Database. Therefore, it has undergone the same editorial control and quality assurance processes established for that initiative (see https://www.coraltraits.org/procedures and Madin et al.21). Specifically, all compiled data comes from scientific sources that have been published (e.g., articles) or undergone rigorous peer review (e.g., PhD theses), ensuring that data accuracy was validated before integration. Moreover, to address potential unnoticed errors in prior validation and those that may arise during the harmonization of the data into a unified, comprehensive dataset, additional measures were taken to ensure a fully curated version for publication. First, whenever data from a new source was compiled, the .csv files were manually reviewed to confirm that all variables had the correct and intended data types. Following the finalization of data compilation, custom R scripts (available with the data descriptor on the Zenodo repository866) were applied to: i) detect duplicate measurements and/or observations, ii) identify inconsistencies in data structure (e.g., an observation linked to multiple species, locations, or resources), and iii) detect and flag outliers in quantitative traits and invalid values in categorical traits. Flagged values were then manually checked against the original sources by trait editors, and when necessary, original authors were consulted. Any confirmed errors identified through the technical validation process were corrected or removed, resulting in the curated OctocoralTraits v2.2 version released here.
Usage Notes
By hosting the dataset on the public repository Zenodo, we adhere to FAIR principles867 and enable reuse under a CC-BY license with appropriate attribution, specifically citing this data descriptor. Additionally, the descriptor includes direct references to all original scientific sources used to compile the global harmonized dataset, found in the resource_id.csv table (also referenced in this manuscript64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163,164,165,166,167,168,169,170,171,172,173,174,175,176,177,178,179,180,181,182,183,184,185,186,187,188,189,190,191,192,193,194,195,196,197,198,199,200,201,202,203,204,205,206,207,208,209,210,211,212,213,214,215,216,217,218,219,220,221,222,223,224,225,226,227,228,229,230,231,232,233,234,235,236,237,238,239,240,241,242,243,244,245,246,247,248,249,250,251,252,253,254,255,256,257,258,259,260,261,262,263,264,265,266,267,268,269,270,271,272,273,274,275,276,277,278,279,280,281,282,283,284,285,286,287,288,289,290,291,292,293,294,295,296,297,298,299,300,301,302,303,304,305,306,307,308,309,310,311,312,313,314,315,316,317,318,319,320,321,322,323,324,325,326,327,328,329,330,331,332,333,334,335,336,337,338,339,340,341,342,343,344,345,346,347,348,349,350,351,352,353,354,355,356,357,358,359,360,361,362,363,364,365,366,367,368,369,370,371,372,373,374,375,376,377,378,379,380,381,382,383,384,385,386,387,388,389,390,391,392,393,394,395,396,397,398,399,400,401,402,403,404,405,406,407,408,409,410,411,412,413,414,415,416,417,418,419,420,421,422,423,424,425,426,427,428,429,430,431,432,433,434,435,436,437,438,439,440,441,442,443,444,445,446,447,448,449,450,451,452,453,454,455,456,457,458,459,460,461,462,463,464,465,466,467,468,469,470,471,472,473,474,475,476,477,478,479,480,481,482,483,484,485,486,487,488,489,490,491,492,493,494,495,496,497,498,499,500,501,502,503,504,505,506,507,508,509,510,511,512,513,514,515,516,517,518,519,520,521,522,523,524,525,526,527,528,529,530,531,532,533,534,535,536,537,538,539,540,541,542,543,544,545,546,547,548,549,550,551,552,553,554,555,556,557,558,559,560,561,562,563,564,565,566,567,568,569,570,571,572,573,574,575,576,577,578,579,580,581,582,583,584,585,586,587,588,589,590,591,592,593,594,595,596,597,598,599,600,601,602,603,604,605,606,607,608,609,610,611,612,613,614,615,616,617,618,619,620,621,622,623,624,625,626,627,628,629,630,631,632,633,634,635,636,637,638,639,640,641,642,643,644,645,646,647,648,649,650,651,652,653,654,655,656,657,658,659,660,661,662,663,664,665,666,667,668,669,670,671,672,673,674,675,676,677,678,679,680,681,682,683,684,685,686,687,688,689,690,691,692,693,694,695,696,697,698,699,700,701,702,703,704,705,706,707,708,709,710,711,712,713,714,715,716,717,718,719,720,721,722,723,724,725,726,727,728,729,730,731,732,733,734,735,736,737,738,739,740,741,742,743,744,745,746,747,748,749,750,751,752,753,754,755,756,757,758,759,760,761,762,763,764,765,766,767,768,769,770,771,772,773,774,775,776,777,778,779,780,781,782,783,784,785,786,787,788,789,790,791,792,793,794,795,796,797,798,799,800,801,802,803,804,805,806,807,808,809,810,811,812,813,814,815,816,817,818,819,820,821,822,823,824,825,826,827,828,829,830,831,832,833,834,835,836,837,838,839,840,841,842,843,844,845,846,847,848,849,850,851,852,853,854,855,856,857,858,859;). We therefore encourage users to reference the original sources whenever feasible, along with this data descriptor. Finally, the OctocoralTraits data descriptor is an evolving data product, continuously linked to ongoing data entry and validation. We encourage users to access the latest versions of OctocoralTraits files, which will be regularly updated in the same Zenodo repository866. Furthermore, since OctocoralTraits v2.2 is part of the Coral Trait Database collaborative effort, subsequent new versions of the dataset will also be accessible and downloadable at (www.coraltraits.org).
Despite the meticulous curation process aimed at identifying and rectifying potential errors, the inherent complexity of data acquisition and compilation may warrant an additional layer of scrutiny from end-users. It is recommended, therefore, that users apply standard validation procedures and cross-referencing methodologies to ensure the accuracy and reliability of the data for their specific analyses. This precautionary measure, which may include consulting referenced original sources or the corresponding author of this publication in case of doubt, aligns with the best practices in data utilization, reinforcing the robustness of the database for scientific inquiry and research endeavors. Finally, special caution is advised when using data from geographical traits (e.g., marine realms and provinces), particularly those derived from the OBIS platform, as not all observations on that platform have been verified for accurate species identification or up-to-date taxonomy.
Code availability
All code used for curating the OctocoralTraits v2.2 data descriptor, as well as for generating the figures included in this manuscript is publicly available alongside the compiled data descriptor at the following Zenodo repository866: (https://doi.org/10.5281/zenodo.14228404). Additionally, the code used to build the online dynamic product, which also contains data for scleractinian corals (www.coraltraits.org), can be found at (https://github.com/jmadinlab/coraltraits2).
References
Linnaeus, C. Systema naturae, sive Regna tria naturae. Systematice proposita per Classes, Ordines, Genera, & Species. Lugduni Batavorum, Apud Theodorum Haak (1735).
Violle, C. & Jiang, L. Towards a trait-based quantification of species niche. J. Plant Ecol. 2(2), 87–93 (2009).
Lailvaux, S. P. & Husak, J. F. The life history of whole-organism performance. Q. Rev. Biol. 89(4), 285–318 (2104).
Sporbert, M. et al. Different sets of traits explain abundance and distribution patterns of European plants at different spatial scales. J. Veg. Sci. 32, e13016 (2021).
Koffel, T., Umemura, K., Litchman, E. & Klausmeier, C. A. A general framework for species-abundance distributions: linking traits and dispersal to explain commonness and rarity. Ecol. Lett. 25, 2359–2371 (2022).
Darwin, C. On the Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life. 1st edn (John Murray, 1859).
Pacifici, M. et al. Species’ traits influenced their response to recent climate change. Nat. Clim. Chang. 7, 205–208 (2017).
McWilliam, M., Pratchett, M. S., Hoogenboom, M. O. & Hughes, T. P. Deficits in functional trait diversity following recovery on coral reefs. Proc. R. Soc. B 287, 20192628 (2020).
Tilman, D. et al. Diversity and productivity in a long-term grassland experiment. Science 294, 843–845 (2001).
Díaz, S. et al. Incorporating plant functional diversity effects in ecosystem service assessments. Proc. Natl. Acad. Sci. USA 104, 20684–20689 (2007).
Green, S. J., Brookson, C. B., Hardy, N. A. & Crowder, L. B. Trait-based approaches to global change ecology: moving from description to prediction. Proc. R. Soc., B 289, 20220071 (2022).
Keller, A. et al. Ten (mostly) simple rules to future-proof trait data in ecological and evolutionary sciences. Methods Ecol. Evol. 14(2), 444–458 (2023).
Kattge, J. et al. TRY–a global database of plant traits. Glob. Change Biol. 17, 2905–2935 (2011).
Oliveira, B., São-Pedro, V., Santos-Barrera, G., Penone, C. & Costa, G. C. AmphiBIO, a global database for amphibian ecological traits. Sci. Data 4, 170123 (2017).
Meiri, S. Traits of lizards of the world: Variation around a successful evolutionary design. Glob. Ecol. Biogeogr. 27(10), 1168–1172 (2018).
Gallagher, R. V. et al. Open Science principles for accelerating trait-based science across the Tree of Life. Nat. Ecol. Evol. 4, 294–303 (2020).
Polme, S. et al. FungalTraits: a user-friendly traits database of fungi and fungus-like stramenopiles. Fungal Divers 105(1), 1–16 (2020).
Pekár, S. et al. The World Spider Trait database: a centralized global open repository for curated data on spider traits. Database 2021, baab064 (2021).
Herberstein, M. E. et al. AnimalTraits - a curated animal trait database for body mass, metabolic rate and brain size. Sci. Data 9(1), 265 (2022).
Tobias, J. A. et al. AVONET: morphological, ecological and geographical data for all birds. Ecol. Lett. 25, 581–597 (2022).
Madin, J. et al. The Coral Trait Database, a curated database of trait information for coral species from the global oceans. Sci. Data 3, 160017 (2016).
Vranken, S. et al. AlgaeTraits: a trait database for (European) seaweeds. Earth Syst. Sci. Data 15, 2711–2754 (2023).
Frimpong, E. A. & Angermeier, P. L. Fish Traits: A database of ecological and life-history traits of freshwater fishes of the United States. Fisheries 34(10), 487–495 (2009).
Madin, J. et al. A trait-based approach to advance coral reef science. TREE 31(6), 419–428 (2016).
Reaka-Kudla, M.L. The global biodiversity of coral reefs: a comparison with rain forests, in Biodiversity II: Understanding and Protecting Our Biological Resources, eds. M.L. Reaka-Kudla, D.E. Wilson, and E.O. Wilson, (Washington, DC: Joseph Henry Press), pp. 83–108 (1997).
Moberg, F. & Folke, C. Ecological goods and services of coral reef ecosystems. Ecol. Econ. 29, 215–233 (1999).
Hughes, T. P. et al. Global warming transforms coral reef assemblages. Nature 556, 492–496 (2018).
Cramer, K. L. et al. The transformation of Caribbean coral communities since humans. Ecol. Evol. 11, 10098–10118 (2021).
Gómez-Gras, D. et al. Climate change transforms the functional identity of Mediterranean coralligenous assemblages. Ecol. Lett. 24(5), 1038–1051 (2021).
Darling, E. S. et al. Relationships between structural complexity, coral traits, and reef fish assemblages. Coral Reefs 36, 561–575 (2017).
Madin, J. S. et al. Selecting coral species for reef restoration. J. Appl. Ecol. 60(8), 1537–1544 (2023).
McFadden, C. S., van Ofwegen, L. P. & Quattrini, A. M. Revisionary systematics of Octocorallia (Cnidaria: Anthozoa) guided by phylogenomics. Bull. Soc. Syst. Biol. 1(3) (2022).
Pérez, C. D., de Moura Neves, B., Cordeiro, R. T., Williams, G. C. & Cairns, S. D. Diversity and distribution of Octocorallia. in The Cnidaria, Past, Present and Future: The World of Medusa and Her Sisters, eds. S. Goffredo and Z. Dubinsky (Cham: Springer International Publishing), 109–123 (2016).
Conci, N., Vargas, S. & Wörheide, G. The biology and evolution of calcite and aragonite mineralization in Octocorallia. Front. Ecol. Evol. 9, 623774 (2021).
Quattrini, A. M. et al. Palaeoclimate ocean conditions shaped the evolution of corals and their skeletons through deep time. Nat. Ecol. Evol. 4, 1531–1538 (2020).
Baillon, S., Hamel, J. F., Wareham, V. E. & Mercier, A. Deep cold-water corals as nurseries for fish larvae. Front. Ecol. Environ. 10(7), 351–356 (2012).
Schweitzer, C. C. & Stevens, B. G. The relationship between fish abundance and benthic community structure on artificial reefs in the Mid-Atlantic Bight, and the importance of sea whip corals Leptogorgia virgulata. PeerJ. 7, e7277 (2019).
Neves, B. M. et al. Cold-water soft corals (Cnidaria: Nephtheidae) as habitat for juvenile basket stars (Echinodermata: Gorgonocephalidae). Front. Mar. Sci. 7, 547896 (2020).
Slattery, M. & Lesser, M. P. Gorgonians are foundation species on sponge-dominated mesophotic coral reefs in the Caribbean. Front. Mar. Sci. 8, 654268 (2021).
de Pádua, S. M. F. et al. The alien octocoral Carijoa riisei is a biogenic substrate multiplier in artificial Brazilian shipwrecks. Aquat. Ecol. 56, 183–200 (2022).
Ponti, M. et al. Ecological shifts in Mediterranean coralligenous assemblages related to gorgonian forest loss. PLoS ONE 9, e102782 (2014).
Ponti, M., Turicchia, E., Ferro, F., Cerrano, C. & Abbiati, M. The understorey of gorgonian forests in mesophotic temperate reefs. Aquatic Conserv. Mar. Freshw. Ecosyst. 28(5), 1153–1166 (2018).
Fabricius, K., Yahel, G. & Genin, A. In situ depletion of phytoplankton by an azooxanthellate soft coral. Limnol. Oceanogr. 43(2), 354–356 (1998).
Gili, J. M. & Coma, R. Benthic suspension feeders: their paramount role in littoral marine food webs. Trends Ecol Evol 13(8), 316–321 (1998).
Fabricius, K. E. & Dommisse, M. Depletion of suspended particulate matter over coastal reef communities dominated by zooxanthellate soft corals. Mar. Ecol. Prog. Ser. 196, 157–167 (2000).
Corry, M. et al. Functional role of the soft coral Dendronephthya australis in the benthic food web of temperate estuaries. Mar. Ecol. Prog. Ser. 593, 61–72 (2018).
Finlay-Jones, H., Raoult, V., Harasti, D. & Gaston, T. F. What eats a cauliflower coral? An assessment of predation on the endangered temperate soft coral, Dendronephthya australis. Mar. Freshw. Res. 73(3), 307–318 (2021).
Matsumoto, A. K. Estimation of in situ distribution of carbonate produced from cold-water octocorals on a Japanese seamount in the NW Pacific. Mar. Ecol. Prog. Ser. 399, 81–102 (2010).
Shoham, E., Prohaska, T., Barkay, Z., Zitek, A. & Benayahu, Y. Soft corals form aragonite-precipitated columnar spiculite in mesophotic reefs. Sci. Rep. 9(1), 1241 (2019).
Atrigenio, M. P., Conaco, C., Guzman, C., Yap, H. T. & Aliño, P. M. Distribution and abundance of Heliopora coerulea (Cnidaria: Coenothecalia) and notes on its aggressive behavior against scleractinian corals: Temperature mediated? Reg. Stud. Mar. Sci. 40, 101502 (2020).
Fürst, S. et al. Raman investigations to identify Corallium rubrum in Iron Age jewelry and ornaments. Minerals. 6(2), 56 (2016).
Rodrigues, L. C., van den Bergh, J. C. J. M., Loureiro, M. L., Nunes, P. A. L. D. & Rossi, S. The Cost of Mediterranean Sea warming and acidification: a choice experiment among scuba divers at Medes Islands, Spain. Environ Resource Econ 63, 289–311 (2016).
Nguyen, N. B. A. et al. Towards sustainable medicinal resources through marine soft coral aquaculture: Insights into the chemical diversity and the biological potential. Mar. Drugs. 20(10), 640 (2022).
Lenz, E. A., Bramanti, L., Lasker, H. R. & Edmunds, P. J. Long-term variation of octocoral populations in St. John, US Virgin Islands. Coral Reefs 34, 1099–1109 (2015).
Lasker, H. R. et al. Resilience of octocoral forests to catastrophic storms. Sci. Rep. 10, 4286 (2020).
Coffroth, M. A. et al. What makes a winner? Symbiont and host dynamics determine Caribbean octocoral resilience to bleaching. Sci. Adv. 9, eadj6788 (2023).
Stobart, B., Teleki, K., Buckley, R., Downing, N. & Callow, M. Coral recovery at Aldabra Atoll, Seychelles: five years after the 1998 bleaching event. Philos. Trans. A Math. Phys. Eng. Sci. 363(1826), 251–5 (2005).
Reverter, M., Helber, S. B., Rohde, S., Goeij, J. M. & Schupp, P. J. Coral reef benthic community changes in the Anthropocene: Biogeographic heterogeneity, overlooked configurations, and methodology. Glob. Chang. Biol. 28, 1956–1971 (2022).
Garrabou J., Ledoux J.-B., Bensoussan N., Gómez-Gras D. & Linares C. Sliding toward the collapse of mediterranean coastal marine rocky ecosystems, in Ecosystem collapse and climate change (Cham, Switzerland: Springer), 291–324. (2021).
Falster, D. et al. AusTraits, a curated plant trait database for the Australian flora. Sci. Data 8, 254 (2021).
Madin, J. et al. An ontology for describing and synthesizing ecological observation data. Ecol. Inform. 2, 279–296 (2007).
Wallace, B.C., Small, K., Brodley, C.E., Lau, J. & Trikalinos, T.A. Deploying an interactive machine learning system in an evidence-based practice center: abstrackr. In Proc. of the ACM International Health Informatics Symposium (IHI), 819-824 (2012).
Rueden, C. T. et al. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18, 1934 (2017).
Anderson, E., Castanaro, J. & Lasker, H. R. Corrigendum: Colony growth responses of the Caribbean octocoral, Pseudopterogorgia elisabethae, to harvesting. Invertebr. Biol. 141(1) (2022).
Grigg, R. W. Growth rings: annual periodicity in two gorgonian corals. Ecology 55(4), 876–881 (1974).
Cadena, N. J. & Sánchez, J. A. Colony growth in the harvested octocoral Pseudopterogorgia acerosa in a Caribbean coral reef. Marine Ecol 31(4), 566–573 (2010).
Yoshioka, P. M. & Yoshioka, B. B. A comparison of the survivorship and growth of shallow-water gorgonian species of Puerto Rico. Mar. Ecol. Prog. Ser. 69(3), 253–260 (1991).
Rey-Villiers, N. & Sánchez, A. Can organic pollution affect the growth rate of octocorals in the Caribbean? Gayana 82(2), 166–170 (2018).
Manrique-Rodríguez, N., Bejarano-Chavarro, S. & Garzón-Ferreira, J. Crecimiento del abanico de mar Gorgonia ventalina (Linnaeus, 1758) (Cnidaria: Gorgoniidae) en el área de Santa Marta, Caribe colombiano. Bol. Investig. Mar. Costeras-INVEMAR 35(1), 77–90 (2006).
Birkeland, C. The effect of wave action on the population dynamics of Gorgonia ventalina Linnaeus. Stud. Trop. Oceanogr. 12, 115–126 (1974).
Kinzie, R. A. III Plexaura homomalla: The biology and ecology of a harvestable marine resource. Stud. Trop. Oceanogr. 12, 22–23 (1974).
Peck, L. S. & Brockington, S. Growth of the Antarctic octocoral Primnoella scotiae and predation by the anemone Dactylanthus antarcticus. Deep Sea Res. Part II: Top. Stud. Oceanogr 92, 73–78 (2013).
Matsumoto, A. K. Effects of low water temperature on growth and magnesium carbonate concentrations in the cold-water gorgonian Primnoa pacifica. Bull. Mar. Sci. 81(3), 423–435 (2007).
Andrews, A. H. et al. Age, growth and radiometric age validation of a deep-sea, habitat-forming gorgonian (Primnoa resedaeformis) from the Gulf of Alaska. Hydrobiologia 471, 101–110 (2002).
Risk, M. J., Heikoop, J. M., Snow, M. G. & Beukens, R. Lifespans and growth patterns of two deep-sea corals: Primnoa resedaeformis and Desmophyllum cristagalli. Hydrobiologia 471, 125–131 (2002).
Druffel, E. R., King, L. L., Belastock, R. A. & Buesseler, K. O. Growth rate of a deep-sea coral using 210Pb and other isotopes. Geochim. Cosmochim. Acta 54(5), 1493–1499 (1990).
Andrews, A. H. et al. Investigations of age and growth for three deep-sea corals from the Davidson Seamount off central California. In Freiwald, A., Roberts, J.M. (eds) Cold-Water Corals and Ecosystems. Erlangen Earth Conference Series. Springer, Berlin, Heidelberg.
Roark, E. B., Guilderson, T. P., Dunbar, R. B. & Ingram, B. L. Radiocarbon-based ages and growth rates of Hawaiian deep-sea corals. Mar. Ecol. Prog. Ser. 327, 1–14, https://doi.org/10.3354/meps327001 (2006).
Mortensen, P. B. & Buhl-Mortensen, L. Morphology and growth of the deep-water gorgonians Primnoa resedaeformis and Paragorgia arborea. Mar. Biol. 147, 775–788 (2005).
Aranha, R., Edinger, E., Layne, G. & Piercey, G. Growth rate variation and potential paleoceanographic proxies in Primnoa pacifica: Insights from high-resolution trace element microanalysis. Deep Sea Res. Part II: Top. Stud. Oceanogr. 99, 213–226 (2014).
Sherwood, O. A. & Edinger, E. N. Ages and growth rates of some deep-sea gorgonian and antipatharian corals of Newfoundland and Labrador. Can. J. Fish. Aquat. Sci. 66(1), 142–152 (2009).
Weinberg, S. & Weinberg, F. The life cycle of a gorgonian: Eunicella singularis (Esper, 1794). Bijdragen tot de Dierkunde 48(2), 127–140 (1979).
Gomez, C. G., Guzman, H. M., Gonzalez, A. & Breedy, O. Survival, growth, and recruitment of octocoral species (Coelenterata: Octocorallia) in Coiba National Park, Pacific Panama. Bull. Mar. Sci. 90(2), 623–650 (2014).
Munari, C., Serafin, G. & Mistri, M. Structure, growth and secondary production of two Tyrrhenian populations of the white gorgonian Eunicella singularis (Esper 1791). Estuar. Coast. Shelf Sci. 119, 162–166 (2013).
Mistri, M. & Ceccherelli, V. U. Growth and secondary production of the Mediterranean gorgonian. Paramuricea clavata. Mar. Ecol. Prog. Ser. 103, 291–291 (1994).
Coma, R., Ribes, M., Zabala, M. & Gili, J. M. Growth in a modular colonial marine invertebrate. Estuar. Coast. Shelf Sci 47(4), 459–470 (1998).
Mistri, M. & Ceccherelli, V. U. Growth of the Mediterranean gorgonian Lophogorgia ceratophyta (L., 1758). Mar. Ecol. 14(4), 329–340 (1993).
Mistri, M. Population structure and secondary production of the Mediterranean octocoral Lophogorgia ceratophyta (L. 1758). Mar. Ecol. 16(3), 181–188 (1995).
Bramanti, L., Magagnini, G., De Maio, L. & Santangelo, G. Recruitment, early survival and growth of the Mediterranean red coral Corallium rubrum (L. 1758), a 4-year study. J. Exp. Mar. Biol. Ecol. 314(1), 69–78 (2005).
Garcia-Rodriguez, M. & Mass, C. Estudio biometrico de poblaciones de coral rajo (Corallium rubrum L.) del litoral de Gerona (NE de España). Bol. Inst. Esp. Ocean 3, 61–64 (1986).
Abbiati, M., Buffoni, G., Caforio, G., Di Cola, G. & Santangelo, G. Harvesting, predation and competition effects on a red coral population. Neth. J. Sea Res. 30, 219–228 (1992).
Cerrano, C., Bavestrello, G., Cicogna, F. & Cattaneo-Vietti, R. New experiences on transplantation and red coral harvesting effects in the Ligurian Sea. In Red Coral and Other Mediterranean Octocorals, Biology and Protection (Ministero Risorse Agricole, Alimentari, Forestali, Roma), pp. 62-67 (1999).
Garrabou, J. & Harmelin, J. G. A 20‐year study on life‐history traits of a harvested long‐lived temperate coral in the NW Mediterranean: insights into conservation and management needs. J. Anim. Ecol. 71, 966–978 (2002).
Weinbauer, M. G. & Velimirov, B. Morphological variations in the Mediterranean sea fan Eunicella cavolini (Coelenterata: Gorgonacea) in relation to exposure, colony size and colony region. Bull. Mar. Sci. 56, 283–295 (1995).
Sartoretto, S. & Francour, P. Bathymetric distribution and growth rates of Eunicella verrucosa (Cnidaria: Gorgoniidae) populations along the Marseilles coast (France). Sci. Mar. 76, 349–355 (2012).
Cary, L. R. Observations upon the growth-rate and oecology of gorgonians. No. 182 (Princeton University Press, 1914).
Kinzie, R. A. The zonation of the West Indian gorgonians. Bull. Mar. Sci. 23, 95–155 (1973).
Brazeau, D. A. & Lasker, H. R. Growth rates and growth strategy in a clonal marine invertebrate, the Caribbean octocoral Briareum asbestinum. Biol. Bull. 183, 269–277 (1992).
Kinzie, R. A. III The ecology of the gorgonians (Cnidaria, Octocorallia) of Discovery Bay, Jamaica. (Yale University, 1970)
Velimirov, B. Wachstum und Altersbestimmung der Gorgonie Eunicella cavolinii. Oecologia 192, 259–272 (1975).
Sardà, J. M. G. & García, À. G. Biologia de “Paramuricea clavata” (Anthozoa: Octocorallia) a les costes catalanes. I. Creixement i característiques generals. Butlletí de la Institució Catalana d’Història Natural, 25-32 (1985).
Velimirov, B. Orientation in the sea fan Eunicella cavolinii related to water movement. Helgoland Mar. Res. 24, 163–173 (1973).
Goffredo, S. & Lasker, H. R. Modular growth of a gorgonian coral can generate predictable patterns of colony growth. J. Exp. Mar. Biol. Ecol. 336, 221–229 (2006).
Sini, M., Kipson, S., Linares, C., Koutsoubas, D. & Garrabou, J. The yellow gorgonian Eunicella cavolini: demography and disturbance levels across the Mediterranean Sea. PLoS ONE 10, e0126253 (2015).
Stone, R. P., Malecha, P. W. & Masuda, M. M. A five-year, in situ growth study on shallow-water populations of the gorgonian octocoral Calcigorgia spiculifera in the Gulf of Alaska. PLoS ONE 12, e0169470 (2017).
Andrews, A. H., Stone, R. P., Lundstrom, C. C. & DeVogelaere, A. P. Growth rate and age determination of bamboo corals from the northeastern Pacific Ocean using refined 210Pb dating. Mar. Ecol. Progr. Ser. 397, 173–185 (2009).
Viladrich, N., Gori, A. & Gili, J. M. Fast growth rate in a young colony of the Mediterranean gorgonian Eunicella singularis. Mar. Biod. 48, 951–952 (2018).
Martinez-Dios, A., Dominguez-Carrió, C., Zapata-Guardiola, R. & Gili, J. M. New insights on Antarctic gorgonians’ age, growth and their potential as paleorecords. Deep Sea Res. I Oceanogr. Res. Pap. 112, 57–67 (2016).
Sherwood, O. A., Scott, D. B., Risk, M. J. & Guilderson, T. P. Radiocarbon evidence for annual growth rings in the deep-sea octocoral Primnoa resedaeformis. Mar. Ecol. Prog. Ser. 301, 129–134 (2005).
Roark, E. B., Guilderson, T. P., Dunbar, R. B., Fallon, S. J. & Mucciarone, D. A. Extreme longevity in proteinaceous deep-sea corals. Proc. Natl. Acad. Sci. USA. 106(13), 5204–5208 (2009).
Lasker, H. R., Boller, M. L., Castanaro, J. & Sánchez, J. A. Determinate growth and modularity in a gorgonian octocoral. Biol. Bull. 205(3), 319–330 (2003).
Grigg, R. W. Fishery management of precious and stony corals in Hawaii. Fish. Bull. 74, 593–600 (1976).
Thresher, R. E., Fallon, S. J. & Townsend, A. T. A “core-top” screen for trace element proxies of environmental conditions and growth rates in the calcite skeletons of bamboo corals (Isididae). Geochim. Cosmochim. Acta 193, 75–99 (2016).
Benedetti, M. C., Priori, C., Erra, F. & Santangelo, G. Growth patterns in mesophotic octocorals: timing the branching process in the highly-valuable Mediterranean Corallium rubrum. Estuar. Coast. Shelf Sci. 171, 106–110 (2016).
Priori, C. et al. Demography of deep-dwelling red coral populations: Age and reproductive structure of a highly valued marine species. Estuar. Coast. Shelf Sci. 118, 43–49 (2013).
Santangelo, G., Carletti, E., Maggi, E. & Bramanti, L. Reproduction and population sexual structure of the overexploited Mediterranean red coral Corallium rubrum. Mar. Ecol. Prog. Ser. 248, 99–108 (2003).
Marschal, C., Garrabou, J., Harmelin, J. G. & Pichon, M. A new method for measuring growth and age in the precious red coral Corallium rubrum (L.). Coral Reefs 23, 423–432 (2004).
Gallmetzer, I., Haselmair, A. & Velimirov, B. Slow growth and early sexual maturity: bane and boon for the red coral Corallium rubrum. Estuar. Coast. Shelf Sci. 90(1), 1–10 (2010).
Santangelo, G., Bongiorni, L., Giannini, F., Abbiati, M. & Buffoni, G. Structure analysis of two red coral populations dwelling in different habitats. In: Cicogna, F., Bavestrello, G., Cattaneo-Vietti, R. (Eds.), Red Coral and Other Mediterranean Octocorals: Biology and Protection, Ministero per le Politiche Agricole, Roma, pp. 23-43 (1997)
Cattaneo-Vietti, R., Bavestrello, G. & Senes, L. Red coral from the Portofino promontory. In: Cicogna, F., Cattaneo-Vietti, R. (Eds.), Red Coral in the Mediterranean Sea: Art, History and Science, Ministero delle Risorse Agricole, Alimentari e Forestali, Roma, pp. 109-130 (1993).
de Moura Neves, B. et al. Size metrics, longevity, and growth rates in Umbellula encrinus (Cnidaria: Pennatulacea) from the eastern. Canadian Arctic. Aquat. Sci. 80, 1073, https://doi.org/10.1139/as-2018-0009 (2018).
Fabricius, K. E., Genin, A. & Benayahu, Y. Flow-dependent herbivory and growth in zooxanthellae-free soft corals. Limnol. Oceanogr. 40(7), 1290 (1995).
Boavida, J. et al. A well-kept treasure at depth: precious red coral rediscovered in Atlantic deep coral gardens (SW Portugal) after 300 years. PLoS One 11(1), e0147228, https://doi.org/10.1371/journal.pone.0147228 (2016).
Zibrowius, H., Montero, M. & Grasshoff, M. La répartition du Corallium rubrum dans l’Atlantique. Thetys 11, 163–170 (1984).
Teixidó, N., Garrabou, J. & Harmelin, J. G. Low dynamics, high longevity and persistence of sessile structural species dwelling on Mediterranean coralligenous outcrops. PLoS One 6, e23744 (2011).
Garrabou, J. Life-history traits of Alcyonium acaule and Parazoanthus axinellae (Cnidaria, Anthozoa), with emphasis on growth. Mar. Ecol. Prog. Ser. 178, 193–203 (1999).
Weinberg, S. Revision of common Octocorallia of the Mediterranean circalittoral II. Beaufortia 25(326), 131–166 (1977).
Skoufas, G., Poulicek, M. & Chintiroglou, C. C. Growth variation of Eunicella singularis (Esper, 1794) (Gorgonacea, Anthozoa). Belg. J. Zool. 130(1), 121–124 (2000).
Chimienti, G., Di Nisio, A. & Lanzolla, A. M. Size/Age Models for Monitoring of the pink sea fan Eunicella verrucosa (Cnidaria: Alcyonacea) and a case study application. J. Mar. Sci. Eng. 8(11), 951 (2020).
Rossi, S., Gili, J. M. & Garrofé, X. Net negative growth detected in a population of Leptogorgia sarmentosa: quantifying the biomass loss in a benthic soft bottom-gravel gorgonian. Mar. Ecol. 32(3), 671–680 (2011).
Pelosi, J., Eaton, K. M., Mychajliw, S., TerHorst, C. P. & Coffroth, M. A. Thermally tolerant symbionts may explain Caribbean octocoral resilience to heat stress. Coral Reefs 40(3), 699–713 (2021).
Coz, R. et al. Development of a new standardised method for sustainable monitoring of the vulnerable pink sea fan Eunicella verrucosa. Mar. Ecol. Prog. Ser. 455, 81–89 (2012).
Chang, W. L., Chi, K. J., Fan, T. Y. & Dai, C. F. Skeletal modification in response to flow during growth in colonies of the sea whip Junceella fragilis. J. Exp. Mar. Biol. Ecol. 351, 99–108 (2007).
Ghanem, R., Ben Souissi, J., Ledoux, J. B., Linares, C. & Garrabou, J. Population structure and conservation status of the white gorgonian Eunicella singularis (Esper, 1791) in Tunisian waters (Central Mediterranean). Mar. Mar. Sci. 26, 24984 (2021).
Bennecke, S., Kwasnitschka, T., Metaxas, A. & Dullo, W. C. In situ growth rates of deep-water octocorals determined from 3D photogrammetric reconstructions. Coral Reefs 35, 1471–1482 (2016).
Courtney, T. A., Guest, J. R., Edwards, A. J. & Dizon, R. M. Linear extension, skeletal density, and calcification rates of the blue coral Heliopora coerulea. Coral Reefs 40, 37–47 (2021).
Guzman, C., Atrigenio, M., Shinzato, C., Aliño, P. & Conaco, C. Warm seawater temperature promotes substrate colonization by the blue coral Heliopora coerulea. PeerJ 7, e7785 (2019).
Matsumoto, A. K. Heterogeneous and compensatory growth in Melithaea flabellifera (Octocorallia: Melithaeidae) in Japan. Hydrobiologia 527, 167–175 (2004).
Murillo, F. J. et al. Morphometry and growth of sea pen species from dense habitats in the Gulf of St. Lawrence, eastern Canada. J. Mar. Biol. Assoc. U.K. 98, 1415–1424 (2018).
Adams, R. Investigations of color, morphology and development of the sea whip Leptogorgia virgulata (Lamarck) (Cnidaria: Octocorallia: Gorgonacea). PhD thesis. Florida State University, Tallahassee, Florida, USA (1980)
Cordes, E. E., Nybakken, J. W. & VanDykhuizen, G. Reproduction and growth of Anthomastus ritteri (Octocorallia: Alcyonacea) from Monterey Bay, California, USA. Mar. Biol. 138, 491–501 (2001).
Wilson, M. T., Andrews, A. H., Brown, A. L. & Cordes, E. E. Axial rod growth and age estimation of the sea pen, Halipteris willemoesi Kölliker. Hydrobiologia 474, 1–7 (2002).
Khalesi, M. K., Beeftink, H. H. & Wijffels, R. H. Light-dependency of growth and secondary metabolite production in the captive zooxanthellate soft coral Sinularia flexibilis. Mar. Biol. 156, 1117–1125 (2009).
Thresher, R. E. Environmental and compositional correlates of growth rate in deep-water bamboo corals (Gorgonacea; Isididae). Mar. Ecol. Prog. Ser. 82, 45–52 (2009).
de Moura Neves, B. D. M., Edinger, E., Layne, G. D. & Wareham, V. E. Decadal longevity and slow growth rates in the deep-water sea pen Halipteris finmarchica (Sars, 1851) (Octocorallia: Pennatulacea): implications for vulnerability and recovery from anthropogenic disturbance. Hydrobiologia 761, 157–171 (2015).
Brodeur, R. D. Habitat-specific distribution of Pacific ocean perch (Sebastes alutus) in Pribilof Canyon, Bering Sea. Fish. Res. 50, 1–15 (2001).
Birkeland, C. Interactions between a sea pen and seven of its predators. Ecol. Monogr. 44(2), 211–232 (1974).
Vinogradov, G. M. Growth rate of the colony of a deep-water gorgonarian Chrysogorgia agassizi: in situ observations. Ophelia 53, 101–103 (2000).
Girard, F., Cruz, R., Glickman, O., Harpster, T. & Fisher, C. R. In situ growth of deep-sea octocorals after the Deepwater Horizon oil spill. Elementa 7, 349 (2019).
Walker, T. A. & Bull, G. D. A newly discovered method of reproduction in gorgonian coral. Mar. Ecol. Prog. Ser. 37, 143 (1983).
Wenker, R. P. & Stevens, B. G. Sea whip coral Leptogorgia virgulata in the Mid-Atlantic Bight: Colony complexity, age, and growth. PeerJ 8, e8372 (2020).
Mitchell, N. D., Dardeau, M. R. & Schroeder, W. W. Colony morphology, age structure, and relative growth of two gorgonian corals, Leptogorgia hebes (Verrill) and Leptogorgia virgulata (Lamarck), from the northern Gulf of Mexico. Hydrobiologia 266, 53–63 (1993).
Altvater, L. & Coutinho, R. Colonisation, competitive ability and influence of Stragulum bicolor van Ofwegen and Haddad, 2011 (Cnidaria, Anthozoa) on the fouling community in Paranaguá Bay, Southern Brazil. J. Exp. Mar. Biol. Ecol. 461, 128–136 (2015).
Goffredo, S. & Lasker, H. R. An adaptive management approach to an octocoral fishery based on the Beverton-Holt model. Coral Reefs 27, 377–385 (2008).
Humann, P. & Deloach, N. Reef coral. Identification guide: Florida, Caribbean, Bahamas. In New World Publications Inc., U.S. 276 p. (1993).
Baker, K. D. et al. Small-scale patterns in the distribution and condition of bamboo coral Keratoisis grayi, in submarine canyons on the Grand Banks, Newfoundland. Front. Mar. Sci. 6, 374 (2019).
Gómez-Gras, D. et al. Population collapse of habitat-forming species in the Mediterranean: a long-term study of gorgonian populations affected by recurrent marine heatwaves. Proc. R. Soc. B 288, 20212384 (2021).
Garrabou, J. et al. Re-shifting the ecological baseline for the overexploited Mediterranean red coral. Sci. Rep. 7, 42404 (2017).
Goh, N. K. C. & Chow, L. M. Growth of five species of gorgonians (Sub-Class Octocorallia) in the sedimented waters of Singapore. Mar. Ecol. 16(4), 337–346 (1995).
Hasegawa, H., Rahman, M. A., Luan, N. T., Maki, T. & Iwasaki, N. Trace elements in Corallium spp. as indicators for origin and habitat. J. Exp. Mar. Biol. Ecol. 424, 29–36 (2012).
Van Oppen, M. J. H., Mieog, J. C., Sanchez, C. A. & Fabricius, K. E. Diversity of algal endosymbionts (zooxanthellae) in octocorals: the roles of geography and host relationships. Mol. Ecol. 14, 2403–2417 (2005).
Sánchez, J. A. Black coral-octocoral distribution patterns on Imelda Bank, a deep-water reef, Colombia, Caribbean Sea. Bull. Mar. Sci. 65(1), 215–225 (1999).
Ryu, T. et al. First transcriptome assembly of the temperate azooxanthellate octocoral Eleutherobia rubra. Mar. Genomics 45, 1–4 (2019).
Kupfner Johnson, S. & Hallock, P. A review of symbiotic gorgonian research in the western Atlantic and Caribbean with recommendations for future work. Coral Reefs 39, 15–25 (2020).
Bayer, F. M. The shallow-water octocorallia of the West Indian region. Studies on the Fauna of Curaçao and Other Caribbean Islands 2(1), 1–373 (1961).
Yoshioka, B. B. Aspects of the ecology of Pseudopterogorgia americana and Pseudopterogorgia acerosa. MSc thesis, University of Puerto Rico, 105 p. (1979).
Gutiérrez‐Rodríguez, C. & Lasker, H. R. Reproductive biology, development, and planula behavior in the Caribbean gorgonian Pseudopterogorgia elisabethae. Invertebr. Biol. 123, 54–67 (2004).
Fitzsimmons-Sosa, K., Hallock, P., Wheaton, J., Hackett, K. & Callahan, M. Annual cycles of gonadal development of six common gorgonians from Biscayne National Park, Florida, USA. Caribb. J. Sci 40, 144–150 (2004).
Kahng, S. E., Benayahu, Y. & Lasker, H. R. Sexual reproduction in octocorals. Mar. Ecol. Prog. Ser. 437, 203–218 (2011).
Bastidas, C. et al. Coral mass- and split-spawning at a coastal and an offshore Venezuelan reef, southern Caribbean. Hydrobiologia 541, 101–106 (2005).
Bardales, A. T. Reproductive patterns of three species of octocorals (families Telestidae, Briareidae, Plexauridae) in the vicinity of La Parguera, Puerto Rico. MSc thesis, University of Puerto Rico, p 85. (1981).
Lasker, H. R. & Kim, K. Larval development and settlement behavior of the gorgonian coral Plexaura kuna. J Exp Mar Biol Ecol 207, 161–175 (1996).
Suzuki, H. Notes on Cornularia (Stolonifera, Alcyonaria) found in the vicinity of the Manazuru Marine Biological Laboratory. Sci. Rep. Yokohama Natl. Univ., Sect. 2, Biol. Sci., Biol. Geol. Sci. 18, 1–6 (1971).
Kahng, S. E. Ecology and ecological impact of an alien octocoral, Carijoa riisei, in Hawaii. PhD thesis, University of Hawaii, p 284. (2006).
Rees, J. T. Aspects of growth and nutrition in the octocoral Telesto riisei. PhD thesis, University of Puerto Rico (1969).
Benayahu, Y. & Loya, Y. Surface brooding in the Red Sea soft coral Parerythropodium fulvum fulvum (Forskal, 1775). Biol. Bull. 165, 353–369 (1983).
Weinberg, S. Mediterranean octocorallia: description of Clavularia carpediem n.sp. and synonymy of Clavularia crassa and C. ochracea on etho-ecological grounds. Bijdr. Dierk. 56, 232–246 (1986).
Alino, P. M. & Coll, J. C. Observations of the synchronized mass spawning and post-settlement activity of octocorals on the Great Barrier Reef, Australia: biological aspects. Bull. Mar. Sci. 45, 697–707 (1989).
Hartnoll, R. G. The annual cycle of Alcyonium digitatum. Estuar. Coast. Shelf Sci. 3, 71–75 (1975).
Gohar, H. A. F. Studies on the Xeniidae of the Red Sea. Publ. Mar. Biol. Stn. Ghadarqa, Red Sea 2, 27–118 (1940).
Bastidas, C., Benzie, J. A. H. & Fabricius, K. E. Genetic differentiation among populations of the brooding soft coral Clavularia koellikeri on the Great Barrier Reef. Mar. Ecol. Prog. Ser. 239, 115–121 (2002).
Hickson, S. The Alcyonaria & Hydrocorallinae of the Cape of Good Hope. Mar. Investig. S. Afr. 1, 67–96 (1900).
McFadden, C. S. Genetic and taxonomic relationships among Northeastern Atlantic and Mediterranean populations of the soft coral Alcyonium coralloides. Mar. Biol. 133, 171–184 (1999).
Quintanilla, E. et al. Sexual reproductive cycle of the epibiotic soft coral Alcyonium coralloides (Pallas, 1766) (Octocorallia, Alcyonacea). Aquatic Biol 18(2), 113–124 (2013).
Fiorillo, I., Rossi, S., Alvà, V., Gili, J. M. & López-González, P. J. Seasonal cycle of sexual reproduction of the Mediterranean soft coral Alcyonium acaule (Anthozoa, Octocorallia). Mar Biol 160, 719–728 (2013).
McFadden, C. S. A molecular phylogenetic analysis of reproductive trait evolution in the soft coral. Alcyonium. Evolution 55, 54–67 (2001).
Hartnoll, R.G. Reproductive strategy in two British species of Alcyonium. In: Keegan BF, Ceidigh PO, Boaden PJS (eds) Biology of benthic organisms 11th European symposium on marine biology Galway, October 1976. Pergamon Press, Oxford, p. 321–328. (1977).
McFadden, C. S. & Hochberg, F. G. Biology and taxonomy of encrusting alcyoniid soft corals in the northeastern Pacific Ocean with descriptions of two new genera (Cnidaria, Anthozoa, Octocorallia). Invertebr. Biol. 122, 93–113 (2003).
Lo Bianco, S. Notizie biologiche riguardanti specialmente il periodo di maturità sessuale degli animali del golfo di Napoli. Mitteilungen aus der Zoologischen Station zu Neapel 19, 513–761 (1909).
Sebens, K. P. The larval and juvenile ecology of the temperate octocoral Alcyonium siderium Verrill. II. Fecundity, survival, and juvenile growth. J Exp Mar Biol Ecol 72(3), 263–285 (1983).
Mercier, A. & Hamel, J. F. Contrasting reproductive strategies in three deep-sea octocorals from eastern Canada: Primnoa resedaeformis, Keratoisis ornata, and Anthomastus grandiflorus. Coral Reefs 30, 337–350 (2011).
Beazley, L. I. & Kenchington, E. L. Reproductive biology of the deep-water coral Acanella arbuscula (Phylum Cnidaria: Class Anthozoa: Order Alcyonacea), northwest Atlantic. Deep Sea Res. I 68, 92–104 (2012).
Benayahu, Y. & Loya, Y. Sexual reproduction of a soft coral: synchronous and brief annual spawning of Sarcophyton glaucum (Quoy & Gaimard, 1833). Biol. Bull. 170, 32–42 (1986).
Nobchinwong, P. & Yeemin, T. Reproduction of a soft coral, Cladiella tuberosa Tixier-Durivault (Cnidaria: Anthozoa: Alcyonacea) in coral communities of the inner and the eastern Gulf of Thailand. 10th International Coral Reef Symposium. Japanese Coral Reef Society, Okinawa, Japan, p. 369. (2004).
Yamazato, K., Sato, M. & Tamashiro, H. Reproductive biology of an alcyonacean coral, Lobophytum crassum Marenzeller. Proceedings of the 4th International Coral Reef Symposium 2, 671–678 (1981).
Benayahu, Y. Developmental episodes in reef soft corals: ecological and cellular determinants. Proceedings from the 8th International Coral Reef Symposium 2, 1213–1218 (1997).
Benayahu, Y., Weil, D. & Kleinman, M. Radiation of broadcasting and brooding patterns in coral reef alcyonaceans. in: Hoshi M, Yamashita O (eds) Advances in Invertebrate Reproduction, 5, Elsevier, Amsterdam, 323–328. (1990).
Mohammed, T. A., Ali, A. H. A., El-Komi, M. M., El-Arab, M. A. E. & Shoukr, F. A. Growth Rate Assessment of Alcyonacean Sarcophyton glaucum from Northern Hurghada, Red Sea, Egypt. Nat. Resour. 7(6), 384–398 (2016).
Fan, T.-Y., Chou, Y.-H. & Dai, C.-F. Sexual Reproduction of the Alcyonacean Lobophytum pauciflorum in Southern Taiwan. Bull. Mar. Sci. 76(1), 143–154 (2005).
Chou, Y.-H. Sexual Reproduction of Three Alcyonacean Species in Southern Taiwan. MSc thesis, National Taiwan University, p. 50 (2002).
Schleyer, M. H., Kruger, A. & Benayahu, Y. Reproduction and the Unusual Condition of Hermaphroditism in Sarcophyton glaucum (Octocorallia, Alcyoniidae) in KwaZulu-Natal, South Africa. Hydrobiologia 530/531, 399–409 (2004).
Wu, C-S. Sexual Reproduction and Population Structure of Three Sinularia Species in Nanwan Bay. MSc thesis, National Taiwan University p 44 (1994).
Slattery, M., Hines, G. A., Starmer, J. & Paul, V. J. Chemical signals in gametogenesis, spawning, and larval settlement and defense of the soft coral Sinularia polydactyla. Coral Reefs 18, 75–84 (1999).
Farrant, P. A. Gonad development and the planulae of the temperate Australian soft coral Capnella gaboensis. Mar. Biol. 92, 381–392 (1986).
Hwang, S. J. & Song, J. I. Reproductive biology and larval development of the temperate soft coral Dendronephthya gigantea (Alcyonacea: Nephtheidae). Mar. Biol. 152, 273–284 (2007).
Dahan, M. & Benayahu, Y. Reproduction of Dendronephythya hemprichi (Cnidaria: Octocorallia): year-round spawning in an azooxanthellate soft coral. Mar. Biol. 129, 573–579 (1997).
Choi, E. J. & Song, J. I. Reproductive biology of the temperate soft coral Dendronephthya suensoni (Alcyonacea: Nephtheidae). Integr. Biosci. 11, 215–225 (2007).
Sun, Z., Hamel, J. F. & Mercier, A. Planulation of deep-sea octocorals in the NW Atlantic. Coral Reefs 28, 781 (2009).
Sun, Z., Hamel, J. F. & Mercier, A. Planulation periodicity, settlement preferences and growth of two deep-sea octocorals from the northwest Atlantic. Mar. Biol. 410, 71–87 (2010).
Henry, L. A., Kenchington, E. L. & Silvaggio, A. Effects of mechanical experimental disturbance on aspects of colony responses, reproduction, and regeneration in the cold-water octocoral Gersemia rubiformis. Can. J. Zool. 81, 1691–1701 (2003).
Nørrevang, A. On a peculiar mode of germ-layer formation. The early embryology of Gersemia rubiformis (Ehrenberg 1834). Acta Zool 54, 59–74 (1973).
Ben-David-Zaslow, R. & Benayahu, Y. Competence and longevity in planulae of several species of soft corals. Mar. Ecol. Prog. Ser. 171, 235–243 (1998).
Benayahu, Y. Reproduction and developmental pathways of Red Sea Xeniidae (Octocorallia, Alcyonacea). Coral Reefs 10, 179–186 (1991).
Utinomi, H. Some xeniid alcyonarians from Japan and adjacent localities. Publ. Seto Mar. Biol. Lab. 1, 1–26 (1950).
Kruger, A., Schleyer, M. & Benayahu, Y. Reproduction in Anthelia glauca (Octocorallia: Xeniidae). I. Gametogenesis and larval brooding. Mar. Biol. 131, 423–432 (1998).
Ben-David-Zaslow, R. & Benayahu, Y. Longevity, competence and energetic content in planulae of the soft coral Heteroxenia fuscescens. J. Exp. Mar. Biol. Ecol. 206, 55–68 (1996).
Benayahu, Y., Berner, T. & Achituv, Y. Development of planulae within a mesogleal coat in the soft coral Heteroxenia fuscescens. Mar. Biol. 100, 203–210 (1989).
Benayahu, Y. & Loya, Y. Life history studies on the Red Sea soft coral Xenia macrospiculata Gohar, 1940. I. Annual dynamics of gonadal development. Biol. Bull. 166, 32–43 (1984).
Benayahu, Y. & Loya, Y. Life history studies on the Red Sea soft coral Xenia macrospiculata Gohar, 1940. II. Planulae shedding and post larval development. Biol. Bull. 166, 44–53 (1984).
Davis, S.A. Some aspects of the biology of Anthelia edmondsoni (Verrill). MSc thesis, University of Hawaii, p75, (1977).
Benayahu, Y., Achituv, Y. & Berner, T. Embryogenesis and acquisition of algal symbionts by planulae of Xenia umbellata (Octocorallia: Alcyonacea). Mar. Biol. 100, 93–101 (1988).
Brazeau, D. A. & Lasker, H. R. The reproductive cycle and spawning in a Caribbean gorgonian. Biol. Bull. 176, 1–7 (1989).
Brazeau, D. A. & Lasker, H. R. Sexual reproduction and external brooding by the Caribbean gorgonian Briareum asbestinum (Pallas). Mar. Biol. 104, 465–474 (1990).
Benayahu, Y. Reproductive cycle and developmental processes during embryogenesis of Clavularia hamra (Cnidaria, Octocorallia). Acta Zool 70, 29–36 (1989).
Excoffon, A. C., Acuña, F. H., Zamponi, M. O. & Genzano, G. N. Reproduction of the temperate octocoral Tripalea clavaria (Octocorallia: Anthothelidae) from sublittoral outcrops off Mar del Plata, Argentina. J. Mar. Biol. Assoc. UK 84, 695–699 (2004).
Chang, T. Sexual reproduction of four gorgonian corals in southern Taiwan. MSc thesis, National Sun Yet-sen University, 46 (2007).
Waller, R. G. & Baco, A. R. Reproductive morphology of three species of deep-water precious corals from the Hawaiian Archipelago: Gerardia sp., Corallium secundum, and Corallium lauuense. Bull. Mar. Sci. 81, 533–542 (2007).
Tsounis, G., Rossi, S., Aranguren, M., Gili, J. M. & Arntz, W. Effects of spatial variability and colony size on the reproductive output and gonadal development cycle of the Mediterranean red coral (Corallium rubrum L.). Mar. Biol. 148, 513–527 (2006).
Torrents, O., Garrabou, J., Marschal, C. & Harmelin, J. G. Age and size at first reproduction in the commercially exploited red coral Corallium rubrum (L.) in the Marseilles area (France, NW Mediterranean). Biol. Conserv. 121, 391–397 (2005).
Ben-Yosef, D. & Benayahu, Y. The gorgonian coral Acabaria biserialis: Life history of a successful colonizer of artificial substrata. Mar. Ecol. Prog. Ser. 163, 235–243 (1999).
Fine, M., Aluma, Y., Meroz-Fine, E., Abelson, A. & Loya, Y. Acabaria erythraea (Octocorallia: Gorgonacea) a successful invader to the Mediterranean Sea? Coral Reefs 24, 161–164 (2005).
Seo, S. Y., Hwang, S. J. & Song, J. I. Sexual reproduction of Anthoplexaura dimorpha (Gorgonacea: Octocorallia) from Munseom, Jejudo Islands, Korea. Anim. Cells Syst. 12, 231–240 (2008).
Cho, I. Y. & Song, J. I. Reproductive biology, embryogenesis and larval development of the gorgonian Calicogorgia granulosa (Gorgonacea: Plexauridae). In 11th International Coral Reef Symposium, Fort Lauderdale, FL, 97, (2008).
Beiring, E. A. & Lasker, H. R. Egg production by colonies of a gorgonian coral. Mar. Ecol. Prog. Ser. 196, 169–177 (2000).
Beiring, E. A. Life history and reproductive success of a broadcast-spawning gorgonian coral and implications for conservation biology. PhD thesis, State University of New York at Buffalo (1997).
Pakes, M. & Woollacott, R. Reproduction of the gorgonian Plexaura flexuosa in Bermuda. J. Exp. Mar. Biol. Ecol. 357, 121–127 (2008).
Gori, A., Linares, C., Rossi, S., Coma, R. & Gili, J. M. Spatial variability in reproductive cycle of the gorgonians Paramuricea clavata and Eunicella singularis (Anthozoa, Octocorallia) in the Western Mediterranean Sea. Mar. Biol. 151, 1571–1584 (2007).
Ribes, M., Coma, R., Rossi, S. & Micheli, M. Cycle of gonadal development in Eunicella singularis (Cnidaria: Octocorallia): trends in sexual reproduction in gorgonians. Invertebrate Biol 126, 307–317 (2007).
Grigg, R. W. Population dynamics of two gorgonian corals. Ecology 58, 278–290 (1977).
Coma, R., Ribes, M., Zabala, M. & Gili, J. M. Reproduction and cycle of gonadal development in the Mediterranean gorgonian Paramuricea clavata. Mar. Ecol. Prog. Ser. 117, 173–183 (1995).
Coma, R., Zabala, M. & Gili, J. M. Sexual reproductive effort in the Mediterranean gorgonian Paramuricea clavata. Mar. Ecol. Prog. Ser. 117, 185–192 (1995).
Linares, C. et al. Early life history of the Mediterranean gorgonian Paramuricea clavata: implications for population dynamics. Invertebrate Biol 127, 1–1 (2008).
Simpson, A. Reproductive morphology of two deep-water octocorals, Paramuricea placomus (Plexauridae) and Metallogorgia melanotrichos (Chrysogorgiidae). In 3rd International Symposium on Deep-Sea Corals, Rosenstiel School of Marine and Atmospheric Sciences, University of Miami, Miami, FL, 203, (2005).
Goldberg, W. M. & Hamilton, R. D. The sexual cycle in Plexaura homomalla. In Bayer, F. M. & Weinheimer, A. J. (eds), Prostaglandins From Plexaura homomalla: Ecology, Utilization and Conservation of a Major Medical Marine Resource. University of Florida Press, Coral Gables, 15, 165, (1974).
Lasker, H. R. et al. In situ rates of fertilization among broadcast-spawning gorgonian corals. Biol. Bull. 190, 45–55 (1996).
Kapela, W. & Lasker, H. R. Size-dependent reproduction in the Caribbean gorgonian Pseudoplexaura porosa. Mar. Biol. 135, 107–114 (1999).
Ferreira, M. C. C. S. Biologia reprodutiva de Phyllogorgia dilatata (Esper 1806) (Cnidaria, Anthozoa, Octocorallia) de recifes de coral de Porto Seguro. MSc thesis, Bahai, Museu Nacional/UFRJ, 82 (2009).
Beasley, S. E., Dardeau, M. R. & Schroeder, W. W. Reproductive biology of the gorgonian Leptogorgia hebes (Verrill). In Proceedings of the American Academy of Underwater Sciences, 22nd Annual Scientific Diving Symposium, (2003).
Silveira, F. L. Aspectos da biologia de Lophogorgia punicea (Milne Edwards & Haime, 1857) (Gorgonacea, Gorgoniidae) do Canal de São Sebastião, estado de São Paulo. PhD thesis, Instituto de Biociências, Universidade de São Paulo, 211 (1986).
Vermeire, M. J. Reproduction and growth of a gorgonian sea whip, Junceella fragilis, in southern Taiwan. MSc thesis, National Taiwan University, (1994).
Orejas, C. et al. Distribution and reproductive ecology of the Antarctic octocoral Ainigmaptilon antarcticum in the Weddell Sea. Mar. Ecol. Prog. Ser. 231, 101–114 (2002).
Orejas, C., Gili, J. M., López-González, P. J., Hasemann, C. & Arntz, W. E. Reproduction patterns of four Antarctic octocorals in the Weddell Sea: an interspecific, shape, and latitude comparison. Mar. Biol. 150, 551–563 (2006).
Brito, T. A. S. Taxonomic and ecological studies on Antarctic octocorals of the genus Thouarella (Octocorallia, Primoidae). PhD thesis, University of Southampton, 272 (1993).
Torrents, O. & Garrabou, J. Fecundity of red coral Corallium rubrum (L.) populations inhabiting in contrasting environmental conditions in the NW Mediterranean. Mar. Biol. 158(5), 1019–1028 (2011).
Soong, K. Reproduction and colony integration of the sea pen Virgularia juncea. Mar. Biol. 146, 1103–1109 (2005).
Satterlie, R. A. & Case, J. F. Development of bioluminescence and other effector responses in the pennatulid coelenterate Renilla köllikeri. Biol. Bull. 157(3) (1979).
Tremblay, M. È., Henry, J. & Anctil, M. Spawning and gamete follicle rupture in the cnidarian Renilla koellikeri: effects of putative neurohormones. Gen. Comp. Endocrinol. 137, 9–18 (2004).
Wilson, E. B. The development of Renilla. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 174, 723–815 (1883).
Rice, A. L., Tyler, P. A. & Paterson, G. J. L. The pennatulid Kophobelemnon stelliferum (Cnidaria: Octocorallia) in the Porcupine Seabight (North-East Atlantic Ocean). J. Mar. Biol. Assoc. UK. 72, 417–434 (1992).
Pires, D. O., Castro, C. B. & Silva, J. C. Reproductive biology of the deep-sea pennatulacean Anthoptilum murrayi (Cnidaria, Octocorallia). Mar. Ecol. Prog. Ser. 397, 103–112, https://doi.org/10.3354/meps08322 (2009).
Edwards, D. C. B. & Moore, C. G. Reproduction in the sea pen Pennatula phosphorea (Anthozoa: Pennatulacea) from the west coast of Scotland. Mar. Biol. 155, 303–314 (2008).
Edwards, D. C. B. & Moore, C. G. Reproduction in the sea pen Funiculina quadrangularis (Anthozoa: Pennatulacea) from the west coast of Scotland. Estuarine, Coastal and Shelf Sci 82(1), 161–168 (2009).
Tyler, P. A., Bronsdon, S. K., Young, C. M. & Rice, A. L. Ecology and gametogenic biology of the genus Umbellula (Pennatulacea) in the North Atlantic Ocean. Int. Rev. Gesamten Hydrobiol. Hydrograph. 80, 187–199 (1995).
Chia, F. S. & Crawford, B. J. Some observations on gametogenesis, larval development and substratum selection of the sea pen Ptilosarcus guerneyi. Mar. Biol. 23, 73–82 (1973).
Eckelbarger, K. J., Tyler, P. A. & Langton, R. W. Gonadal morphology and gametogenesis in the sea pen Pennatula aculeata (Anthozoa: Pennatulacea) from the Gulf of Maine. Mar. Biol. 132, 677–690 (1998).
Babcock, R. C. Reproduction and development of the blue coral Heliopora coerulea (Alcyonaria: Coenothecalia). Mar. Biol. 104, 475–481 (1990).
Harii, S. & Kayanne, H. Larval dispersal, recruitment, and adult distribution of the brooding stony octocoral Heliopora coerulea on Ishigaki Island, southwestern Japan. Coral Reefs 22, 188–196 (2003).
Harii, S., Kayanne, H., Takigawa, H., Hayashibara, T. & Yamamoto, M. Larval survivorship, competency periods and settlement of two brooding corals, Heliopora coerulea and Pocillopora damicornis. Mar. Biol. 141, 39–46 (2002).
Waller, R. G., Stone, R. P., Johnstone, J. & Mondragon, J. Sexual reproduction and seasonality of the Alaskan red tree coral, Primnoa pacifica. PLOS ONE. 9(4), e90893 (2014).
Waller, R. G. et al. Phenotypic plasticity or a reproductive dead end? Primnoa pacifica (Cnidaria: Alcyonacea) in the Southeastern Alaska Region. Front. Mar. Sci. 6709 (2019).
Achituv, Y. & Benayahu, Y. Polyp dimorphism and functional, sequential hermaphroditism in the soft coral Heteroxenia fuscescens (Octocorallia). Mar. Ecol. Prog. Ser. 64, 263–269 (1990).
Maldonado, M., López-Acosta, M., Sánchez-Tocino, L. & Sitjà, C. The rare, giant gorgonian Ellisella paraplexauroides: demographics and conservation concerns. Mar. Ecol. Prog. Ser. 479, 127–141 (2013).
Achituv, Y., Benayahu, Y. & Hanania, J. Planulae brooding and acquisition of zooxanthellae in Xenia macrospiculata (Cnidaria: Octocorallia). Helgoländer Meeresuntersuchungen 46, 301–310 (1992).
Barbosa, T. M. et al. Comparisons of sexual reproduction in Carijoa riisei (Cnidaria, Alcyonacea) in South Atlantic, Caribbean, and Pacific areas. Hydrobiologia 734(1) (2014).
Mandelberg-Aharon, Y. & Benayahu, Y. Reproductive features of the Red Sea octocoral Sarcophyton auritum Verseveldt & Benayahu, 1978 are uniform within generic boundaries across wide biogeographical regions. Hydrobiologia 759, 119–132 (2015).
Torre, L. Reproductive biology of the Antarctic “sea pen” Malacobelemnon daytoni (Octocorallia, Pennatulacea, Kophobelemnidae). Polar Research 32 (2013).
Lopes, V. M. et al. Reproduction in Octocorallia: Synchronous spawning and asynchronous oogenesis in the pennatulid Veretillum cynomorium. Mar. Biol. Res. 8(9), 893–900 (2012).
Ritson-Williams, R. Surface brooding in the Caribbean gorgonian Pterogorgia anceps. Coral Reefs 29, 437 (2010).
Baillon, S., Hamel, J. F., Wareham, V. E. & Mercier, A. Seasonality in reproduction of the deep-water pennatulacean coral Anthoptilum grandiflorum. Mar. Biol. 161, 29–43 (2014).
Hwang, S. J. & Song, J. I. Sexual reproduction of the soft coral Dendronephthya castanea (Alcyonacea: Nephtheidae). Animal Cells and Systems 16(2), 135–144 (2012).
Teixidó, N., Bensoussan, N., Gori, A., Fiorillo, I. & Viladrich, N. Sexual reproduction and early life-history traits of the Mediterranean soft coral Alcyonium acaule. Mar. Ecol. 37(1), 134–144 (2015).
Coelho, M. A. & Lasker, H. R. Reproductive biology of the Caribbean brooding octocoral Antillogorgia hystrix. Invertebrate Biology 133(4), 299–313 (2014).
Cupido, R. et al. Sexual structure of a highly reproductive, recovering gorgonian population: quantifying reproductive output. Mar. Ecol. Prog. Ser. 469, 25–36 (2014).
Coma, R. & Lasker, H. R. Effects of spatial distribution and reproductive biology on in situ fertilization rates of a broadcast-spawning invertebrate. Biol. Bull. 193(1) (1997).
Rossi, S. & Gili, J. M. Reproductive features and gonad development cycle of the soft bottom-gravel gorgonian Leptogorgia sarmentosa (Esper, 1791) in the NW Mediterranean Sea. Invertebr. Reprod. Dev.t 53(4), 175–190 (2009).
Benayahu, Y. & Loya, Y. Settlement and recruitment of a soft coral: why is Xenia macrospiculata a successful colonizer? Bull. Mar. Sci. 36(1), 177–188 (1985).
De Putron, S. J. & Ryland, J. S. Effect of seawater temperature on reproductive seasonality and fecundity of Pseudoplexaura porosa (Cnidaria: Octocorallia): latitudinal variation in Caribbean gorgonian reproduction. Biol. Bull. 128(3), 213–222 (2009).
Heidari, F. & Salamat, N. Artificial reproduction of octocoral (Plumarella flabellata). Pak. Vet. J. 32(3), 403–407 (2012).
Hamel, J. F., Wareham-Hayes, V. E. & Mercier, A. Reproduction of a bathyal pennatulacean coral in the Canadian Arctic. Deep-Sea Res. I: Oceanogr. Res. Pap. 162, 103321 (2020).
Hellström, M., Kavanagh, K. D. & Benzie, J. A. H. Multiple spawning events and sexual reproduction in the octocoral Sarcophyton elegans (Cnidaria: Alcyonacea) on Lizard Island, Great Barrier Reef. Mar. Biol. 157(2), 383–392 (2010).
Rakka, M., Sampaio, I., Colaco, A. & Carreiro-Silva, M. Reproductive biology of two deep-sea octocorals in the Azores Archipelago. Deep-Sea Res. I: Oceanogr. Res. Pap. 175, 103587 (2021).
Rossin, A. M., Waller, R. G. & Forsterra, G. Reproduction of the cold-water coral Primnoella chilensis (Philippi, 1894). Continent. Shelf Res. 144, 31–37 (2017).
Grinyó, J. et al. Reproduction, energy storage and metabolic requirements in a mesophotic population of the gorgonian Paramuricea macrospina. PloS One 13, e0203308 (2018).
Fountain, C. T., Waller, R. G. & Auster, P. J. Individual and population level variation in the reproductive potential of deep-sea corals from different regions within the Gulf of Maine. Front. Mar. Sci. 6, 172 (2019).
Topçu, N. E. & Oztürk, B. Reproduction in the Mediterranean endemic gorgonian Spinimuricea klavereni (Anthozoa, Octocorallia, Plexauridae). Invertebr. Biol. 135, 13–19 (2016).
Michalek-Wagner, K. & Willis, B. Impacts of bleaching on the soft coral Lobophytum compactum. I. Fecundity, fertilization and offspring viability. Coral Reefs 19, 231–239 (2001).
Dahan, M. & Benayahu, Y. Embryogenesis, planulae longevity, and competence in the octocoral Dendronephthya hemprichi. Invertebr. Biol. 117(4), 271–280 (1998).
Nonaka, M., Nakamura, M. & Muzik, K. Sexual reproduction in precious corals (Coralliidae) collected in the Ryukyu Archipelago. Pac. Sci. 69(1), 15–46 (2015).
Luan, N. T., Rahman, M. A., Maki, T., Iwasaki, N. & Hasegawa, H. Growth characteristics and growth rate estimation of Japanese precious corals. J. Exp. Mar. Biol. Ecol. 441, 117–125 (2013).
Lasker, H. R. Asexual reproduction, fragmentation, and skeletal morphology of a plexaurid gorgonian. Mar. Ecol. Prog. Ser. 9, 261–268 (1984).
Coffroth, M. A. & Lasker, H. R. Larval paternity and male reproductive success of a broadcast-spawning gorgonian, Plexaura kuna. Mar. Biol. 131(2), 329–337 (1998).
McFadden, C. S. Contributions of sexual and asexual reproduction to population structure in the clonal soft coral, Alcyonium rudyi. Evolution 51, 112–126 (1997).
Lasker, H. R., Kim, K. & Coffroth, M. A. Production, settlement, and survival of plexaurid gorgonian recruits. Mar. Ecol. Prog. Ser. 162, 111–123 (1998).
Lasker, H. R. Reproductive strategy, larval behavior and recruitment among Caribbean octocorals. Society for Integrative and Comparative Biology Annual Meeting 51 (2011).
Rakka, M., Godinho, A., Orejas, C. & Carreiro-Silva, M. Embryo and larval biology of the deep-sea octocoral Dentomuricea aff. meteor under different temperature regimes. PeerJ 9, e11604 (2021).
Kim, S., Wild, C. & Tilstra, A. Effective asexual reproduction of a widespread soft coral: comparative assessment of four different fragmentation methods. PeerJ 10, e12589 (2022).
Excoffon, A. C., Navella, L., Acuña, F. H. & Garese, A. Oocyte production, fecundity and size at first reproduction of Tripalea clavaria (Cnidaria, Octocorallia, Anthothelidae) in the Southwest Atlantic. Zoological Studies 50(4), 434–442 (2011).
Baran, C. C. & Baria‐Rodriguez, M. V. Sexual reproduction in the soft coral Lobophytum schoedei in Bolinao-Anda Reef Complex, Pangasinan, northwestern Philippines. Invertebrate Biology 140(2), e12316 (2021).
Sun, Z., Hamel, J. F. & Mercier, A. Planulation, larval biology, and early growth of the deep-sea soft corals Gersemia fruticosa and Duva florida (Octocorallia: Alcyonacea). Invertebr. Biol. 130(2), 91–99 (2011).
Kim, E., Lasker, H. R., Coffroth, M. A. & Kim, K. Morphological and genetic variation across reef habitats in a broadcast-spawning octocoral. Hydrobiologia 530, 423–432 (2004).
Feehan, K. A. & Waller, R. G. Notes on reproduction of eight species of Eastern Pacific cold-water octocorals. J. Mar. Biol. Assoc. UK 95(4), 691–696 (2015).
Sekida, S., Iwasaki, N. & Okuda, K. Gonadal morphology and gametogenesis in Japanese red coral Corallium japonicum (Octocorallia: Alcyonacea) collected off Cape Ashizuri, Japan. Zool. Sci. 33(3), 320–336 (2016).
Tsai, S., Jhuang, Y., Spikings, E., Sung, P. J. & Lin, C. Ultrastructural observations of the early and late stages of gorgonian coral (Junceella juncea) oocytes. Tissue and Cell 46(4), 225–232 (2014).
Porcu, C. et al. Reproductive patterns in deep versus shallow populations of the precious Mediterranean gorgonian Corallium rubrum (Linnaeus, 1758) (Sardinia, Central-Western Mediterranean). Mediterr. Mar. Sci. 18(1), 64–76 (2017).
Pelosi, J. A. et al. Fine-scale morphological, genomic, reproductive, and symbiont differences delimit the Caribbean octocorals Plexaura homomalla and P. kükenthali. Coral Reefs 41, 635–653 (2021).
Gomez, C. G., Gonzalez, A. & Guzman, H. M. Reproductive traits and their relationship with water temperature in three common octocoral (Anthozoa: Octocoralia) species from the tropical eastern Pacific. Bull. Mar. Sci. 94(4), 1527–1541 (2018).
Coelho, M. A. & Lasker, H. R. Larval behavior and settlement dynamics of a ubiquitous Caribbean octocoral and its implications for dispersal. Mar. Ecol. Prog. Ser. 561, 109–121 (2016).
Gori, A. et al. Reproductive cycle and trophic ecology in deep versus shallow populations of the Mediterranean gorgonian Eunicella singularis (Cap de Creus, northwestern Mediterranean Sea). Coral Reefs 31, 823–837 (2012).
Sun, Z., Hamel, J. F., Edinger, E. & Mercier, A. Reproductive biology of the deep-sea octocoral Drifa glomerata in the Northwest Atlantic. Mar. Biol. 157, 863–873 (2010).
Tonra, K. J., Wells, C. D. & Lasker, H. R. Spawning, embryogenesis, settlement, and post-settlement development of the gorgonian Plexaura homomalla. Invertebr. Biol. 140(2), e12319 (2021).
Toh, T. C. et al. Mass brooding of the blue octocoral Heliopora coerulea on a sedimented equatorial reef. Mar. Freshw. Behav. Physiol. 49(1), 69–74 (2016).
Viladrich, N. et al. Variation in lipid and free fatty acid content during spawning in two temperate octocorals with different reproductive strategies: surface versus internal brooder. Coral Reefs 35, 1033–1045 (2016).
Page, C. A. & Lasker, H. R. Effects of tissue loss, age and size on fecundity in the octocoral Pseudopterogorgia elisabethae. J. Exp. Mar. Biol. Ecol. 434, 47–52 (2012).
Liberman, R., Shlesinger, T., Loya, Y. & Benayahu, Y. Octocoral Sexual Reproduction: Temporal Disparity Between Mesophotic and Shallow-Reef Populations. Front. Mar. Sci. 5, 445 (2018).
Betti, F., Bo, M., Di Camillo, C. G. & Bavestrello, G. Life history of Cornularia cornucopiae (Anthozoa: Octocorallia) on the Conero Promontory (North Adriatic Sea). Mar. Ecol. 33(1), 49–55 (2011).
Baillon, S., Hamel, J. F. & Mercier, A. Protracted oogenesis and annual reproductive periodicity in the deep-sea pennatulacean Halipteris finmarchica (Anthozoa, Octocorallia). Mar. Ecol. 36(4), 1364–1378 (2015).
Lasker, H. R. Vegetative reproduction in the octocoral Briareum asbestinum (Pallas). J. Exp. Mar. Biol. Ecol. 72, 157–169 (1983).
Sebens, K. P. & Miles, J. S. Sweeper tentacles in a gorgonian octocoral: morphological modification for interference competition. Biol. Bull. 175, 378–387 (1988).
Barneah, O., Weis, V. M., Perez, S. & Benayahu, Y. Diversity of dinoflagellate symbionts in Red Sea soft corals: mode of symbiont acquisition matters. Mar. Ecol. Prog. Ser. 275, 89–95 (2004).
Ramsby, B. D. & Goulet, T. L. Symbiosis and host morphological variation: Symbiodiniaceae photosynthesis in the octocoral Briareum asbestinum at ambient and elevated temperatures. Coral Reefs 38(2), 359–371 (2019).
Parrin, A. P. et al. Within-colony migration of symbionts during bleaching of octocorals. Biol. Bull. 223(2), 245–256 (2012).
Kirk, N. L., Andras, J. P., Harvell, C. D., Santos, S. R. & Coffroth, M. A. Population structure of Symbiodinium sp. associated with the common sea fan, Gorgonia ventalina, in the Florida Keys across distance, depth, and time. Mar. Biol. 156, 1609–1623 (2009).
Chong, G., Tsai, S., Wang, L. H., Huang, C. Y. & Lin, C. Cryopreservation of the gorgonian endosymbiont. Symbiodinium. Sci. Rep. 6(1), 18816 (2016).
Goulet, T. L., Shirur, K. P., Ramsby, B. D. & Iglesias-Prieto, R. The effects of elevated seawater temperatures on Caribbean gorgonian corals and their algal symbionts, Symbiodinium spp. PLoS One 12(2), e0171032 (2017).
Goulet, T. L. & Coffroth, M. A. The genetic identity of dinoflagellate symbionts in Caribbean octocorals. Coral Reefs 23, 465–472 (2004).
Andras, J. P., Kirk, N. L. & Drew Harvell, C. Range-wide population genetic structure of Symbiodinium associated with the Caribbean Sea fan coral, Gorgonia ventalina. Mol. Ecol. 20(12), 2525–2542 (2011).
Forcioli, D. et al. Symbiont diversity is not involved in depth acclimation in the Mediterranean sea whip Eunicella singularis. Mar. Ecol. Prog. Ser. 439, 57–71 (2011).
Nuzzo, G. et al. Dinoflagellate-Related Amphidinolides from the Brazilian Octocoral Stragulum bicolor. J. Nat. Prod. 79(7), 1881–1885 (2016).
Goulet, T. L., LaJeunesse, T. C. & Fabricius, K. E. Symbiont specificity and bleaching susceptibility among soft corals in the 1998 Great Barrier Reef mass coral bleaching event. Mar. Biol. 154, 795–804 (2008).
Lau, Y. W., Stokvis, F. R., Imahara, Y. & Reimer, J. D. The stoloniferous octocoral, Hanabira yukibana, gen. nov., sp. nov., of the southern Ryukyus has morphological and symbiont variation. Contrib. Zool. 88(1), 54–77 (2019).
Hannes, A. R., Barbeitos, M. & Coffroth, M. A. Stability of symbiotic dinoflagellate type in the octocoral Briareum asbestinum. Mar. Ecol. Prog. Ser. 391(1), 65–72 (2009).
Gori, A. et al. Characterization of the zooxanthellate and azooxanthellate morphotypes of the Mediterranean gorgonian Eunicella singularis. Mar. Biol. 159(7), 1485–1496 (2012).
Singh, A. P. & Mercer, E. H. Algal symbiont of a soft coral. J. Phycol. 12(4), 464–467 (1976).
Strychar, K. B., Coates, M., Sammarco, P. W., Piva, T. J. & Scott, P. T. Loss of Symbiodinium from bleached soft corals Sarcophyton ehrenbergi, Sinularia sp., and Xenia sp. J. Exp. Mar. Biol. Ecol. 320(2), 159–177 (2005).
Lau, Y. W. & Reimer, J. D. Zooxanthellate, Sclerite-Free, and Pseudopinnuled Octocoral Hadaka nudidomus gen. nov. et sp. nov. (Anthozoa, Octocorallia) from Mesophotic Reefs of the Southern Ryukyus Islands. Diversity 11(10), 176 (2019).
Plaza, P. U., Mehrotra, R., Scott, C. M. & Reimer, J. D. Rare zooxanthellate Nanipora octocoral (Helioporacea) in the Gulf of Thailand. Mar. Biodivers. 48(4), 1961–1967 (2018).
Miyazaki, Y. & Reimer, J. D. A new genus and species of octocoral with aragonite calcium-carbonate skeleton (Octocorallia, Helioporacea) from Okinawa, Japan. ZooKeys 511, 1 (2015).
McCauley, M., Banaszak, A. T. & Goulet, T. L. Species traits dictate seasonal-dependent responses of octocoral–algal symbioses to elevated temperature and ultraviolet radiation. Coral Reefs 37(3), 901–917 (2018).
Ferrier-Pagès, C. et al. Symbiotic stony and soft corals: Is their host-algae relationship really mutualistic at lower mesophotic reefs? Limnol. Oceanogr. 67(1), 261–271 (2019).
Baker, D. M. et al. Productivity links morphology, symbiont specificity, and bleaching in the evolution of Caribbean octocoral symbioses. ISME J 9(12), 2620–2629 (2015).
Goulet, T. L., Simmons, C. & Goulet, D. Worldwide biogeography of Symbiodinium in tropical octocorals. Mar. Ecol. Prog. Ser. 355(1), 45–58 (2008).
LaJeunesse, T. Diversity and community structure of symbiotic dinoflagellates from Caribbean coral reefs. Mar. Biol. 141(2), 387–400 (2002).
Santos, S. R., Gutierrez-Rodriguez, C. & Coffroth, M. Phylogenetic Identification of Symbiotic Dinoflagellates via Length Heteroplasmy in Domain V of Chloroplast Large Subunit (cp23S)—Ribosomal DNA Sequences. Mar. Biotechnol. 5(2), 130–140 (2003).
Santos, S. R., Shearer, T. L., Hannes, A. R. & Coffroth, M. A. Fine-scale diversity and specificity in the most prevalent lineage of symbiotic dinoflagellates (Symbiodinium, Dinophyceae) of the Caribbean. Mol. Ecol. 13(2), 459–469 (2004).
LaJeunesse, T. Closely related Symbiodinium spp. differ in relative dominance in coral reef host communities across environmental, latitudinal, and biogeographic gradients. Mar. Ecol. Prog. Ser. 284(1), 147–161 (2004).
Kirk, N. L., Ward, J. R. & Coffroth, M. A. Stable Symbiodinium Composition in the Sea Fan Gorgonia ventalina During Temperature and Disease Stress. Biol. Bull. 209(3), 227–234 (2005).
Barneah, O. et al. Three-party symbiosis: acoelomorph worms, corals, and unicellular algal symbionts in Eilat (Red Sea). Mar. Biol. 151(4), 1215–1223 (2007).
Santodomingo, N. et al. Diversity and distribution of azooxanthellate corals in the Colombian Caribbean. Mar. Biodivers. 43(1), 7–22 (2013).
Maia, L. F., de Oliveira, V. E., de Oliveira, M. E. R., Fleury, B. G. & de Oliveira, L. F. C. Polyenic pigments from the Brazilian octocoral Phyllogorgia dilatata Esper, 1806 characterized by Raman spectroscopy. J. Raman Spectrosc. 43(1), 161–164 (2011).
Watling, L., Rowley, S. & Guinotte, J. The World’s largest known Gorgonian. Zootaxa 3630(1), 198–199 (2013).
Imbs, A. B., Latyshev, N. A., Zhukova, N. V. & Dautova, T. N. Comparison of fatty acid compositions of azooxanthellate Dendronephthya and zooxanthellate soft coral species. Comp. Biochem. Physiol. B 148(3), 314–321 (2007).
Edgar, G. J. Australian marine life. Revised edition. The plants and animals of temperate waters. ISBN: 1877069485, 9781877069482 (2000).
Humann, P. Reef Coral Identification: Florida, Caribbean, Bahamas 3rd Edition (Reef Set (New World)). ISBN: 187834854X (1996).
Fabricius, K. E. & Alderslade, P. Soft Corals and Sea Fans: A comprehensive guide to the tropical shallow water genera of the central-west Pacific, the Indian Ocean and the Red Sea. ISBN: 0 642 322104 (2001)
Lewis, J. C. & Wallis, E. V. The function of surface sclerites in gorgonians (Coelenterata, Octocorallia). Biol. Bull. 181(2), 275–288 (1991).
Yogesh Kumar, J. S., Geetha, S., Raghunathan, C. & Sornaraj, R. Two new records of Dendronephthya octocorals (Family: Nephtheidae) from Andaman and Nicobar Islands, India. Indian J. Geo-Marine Sci 48(03), 343–348 (2019).
Utinomi, H. On some octocorals from deep waters of Prov. Tosa, Shikoku. Publ. Seto Mar. Biol. Lab. 7(1), 89–110 (1958).
Molodtsova, T. N. Deep-sea mushroom soft corals (Octocorallia: Alcyonacea: Alcyoniidae) of the Northern Mid-Atlantic Ridge. Mar. Biol. Res. 9(5-6), 488–515 (2013).
Cairns, S. D., Stone, R. P., Moon, H. W. & Lee, J. H. Primnoidae (Octocorallia: Calcaxonia) from the Emperor Seamounts, with notes on Callogorgia elegans (Gray, 1870). Pac. Sci. 72(1), 125–142 (2018).
Lam, K. & Morton, B. Soft corals, sea fans, gorgonians (Octocorallia: Alcyonacea) and black and wire corals (Ceriantipatharia: Antipatharia) from submarine caves in Hong Kong with a checklist of local species and a description of a new species of Paraminabea. J. Nat. Hist. 42(9-12), 749–780 (2008).
Horvath, E. A. A review of gorgonian coral species (Cnidaria, Octocorallia, Alcyonacea) held in the Santa Barbara Museum of Natural History research collection: focus on species from Scleraxonia, Holaxonia, and Calcaxonia – Part I: Introduction, species of Scleraxonia and Holaxonia (Family Acanthogorgiidae). ZooKeys 860, 183 (2019).
Moore, K. M., Alderslade, P. & Miller, K. J. A taxonomic revision of Anthothela (Octocorallia: Scleraxonia: Anthothelidae) and related genera, with the addition of new taxa, using morphological and molecular data. Zootaxa 4304(1), 14 (2017).
Sánchez, J. A. Systematics of the bubblegum corals (Cnidaria: Octocorallia: Paragorgiidae) with description of new species from New Zealand and the Eastern Pacific. Zootaxa 1014(1), 1–72 (2005).
Lamare, V. & Weinberg, S. Alcyonium acaule Marion, 1878. DORIS: Données d’Observations pour la Reconnaissance et l’Identification de la faune et la flore Subaquatiques. http://doris.ffessm.fr/ref/specie/91 (2021).
Perrier, P., André, F. & Pean, M. Alcyonium coralloides (Pallas, 1766). DORIS: Données d’Observations pour la Reconnaissance et l’Identification de la faune et la flore Subaquatiques. http://doris.ffessm.fr/ref/specie/100 (2021).
Perrin, M., Ader, D. & Prouzet, A. Alcyonium digitatum Linnaeus, 1758. DORIS: Données d’Observations pour la Reconnaissance et l’Identification de la faune et la flore Subaquatiques. http://doris.ffessm.fr/ref/specie/267 (2021).
Le Granché, P. & Müller, Y. Alcyonium glomeratum (Hassal, 1843). DORIS: Données d’Observations pour la Reconnaissance et l’Identification de la faune et la flore Subaquatiques. http://doris.ffessm.fr/ref/specie/271 (2020).
Le Granché, P. Alcyonium hibernicum (Renouf, 1931). DORIS: Données d’Observations pour la Reconnaissance et l’Identification de la faune et la flore Subaquatiques. http://doris.ffessm.fr/ref/specie/4311 (2022).
André, F., La Riviere, M. & Pean, M. Alcyonium palmatum Pallas, 1766. DORIS: Données d’Observations pour la Reconnaissance et l’Identification de la faune et la flore Subaquatiques. http://doris.ffessm.fr/ref/specie/1485 (2021).
Fey, L. & Maran, V. Gersemia rubiformis (Ehrenberg, 1834). DORIS: Données d’Observations pour la Reconnaissance et l’Identification de la faune et la flore Subaquatiques. http://doris.ffessm.fr/ref/specie/1581 (2022).
Weinberg, S. & Corolla, J.-P. Maasella edwardsi (de Lacaze-Duthiers, 1888). DORIS: Données d’Observations pour la Reconnaissance et l’Identification de la faune et la flore Subaquatiques. http://doris.ffessm.fr/ref/specie/288 (2020).
Girard, P., Weinberg, S. & Maran, V. Paralcyonium spinulosum Delle Chiaje, 1822. DORIS: Données d’Observations pour la Reconnaissance et l’Identification de la faune et la flore Subaquatiques. http://doris.ffessm.fr/ref/specie/2725 (2022).
Girard, P., Müller, Y. & Maran, V. Annella mollis (Nutting, 1910). DORIS: Données d’Observations pour la Reconnaissance et l’Identification de la faune et la flore Subaquatiques. http://doris.ffessm.fr/ref/specie/2253 (2020).
Girard, P., Müller, Y. & Maran, V. Annella reticulata (Ellis & Solander, 1786). DORIS: Données d’Observations pour la Reconnaissance et l’Identification de la faune et la flore Subaquatiques. http://doris.ffessm.fr/ref/specie/3778 (2022).
Reguieg, A., Kupfer, M., Dumas, J. & Sittler, A.-P. Corallium rubrum (Linnaeus, 1758). DORIS: Données d’Observations pour la Reconnaissance et l’Identification de la faune et la flore Subaquatiques. http://doris.ffessm.fr/ref/specie/308 (2020).
Oms, R., Goyeau, A. & Prouzet, A. Erythropodium caribaeorum (Duchassaing & Michelotti, 1860). DORIS: Données d’Observations pour la Reconnaissance et l’Identification de la faune et la flore Subaquatiques. http://doris.ffessm.fr/ref/specie/1275 (2021).
Dumas, J., Ader, D., Maran, V. & Sittler, A.-P. Eunicella cavolini (Koch, 1887). DORIS: Données d’Observations pour la Reconnaissance et l’Identification de la faune et la flore Subaquatiques. http://doris.ffessm.fr/ref/specie/247 (2020).
Dumas, J., Maran, V., Ader, D. & Huet, S. Eunicella singularis (Esper, 1791). DORIS: Données d’Observations pour la Reconnaissance et l’Identification de la faune et la flore Subaquatiques. http://doris.ffessm.fr/ref/specie/239 (2022).
Dumas, J., Maran, V., Ader, D. & Huet, S. Eunicella verrucosa (Pallas, 1766). DORIS: Données d’Observations pour la Reconnaissance et l’Identification de la faune et la flore Subaquatiques. http://doris.ffessm.fr/ref/specie/235 (2022).
Prouzet, A. & Philippot, V. Iciligorgia schrammi Duchassaing, 1870. DORIS: Données d’Observations pour la Reconnaissance et l’Identification de la faune et la flore Subaquatiques. http://doris.ffessm.fr/ref/specie/2821 (2021).
Dumas, J., Ader, D., Maran, V. & Huet, S. Leptogorgia sarmentosa (Esper, 1789). DORIS: Données d’Observations pour la Reconnaissance et l’Identification de la faune et la flore Subaquatiques. http://doris.ffessm.fr/ref/specie/253 (2022).
Girard, P., Müller, Y. & Maran, V. Melithaea caledonica Grasshoff, 1999. DORIS: Données d’Observations pour la Reconnaissance et l’Identification de la faune et la flore Subaquatiques. http://doris.ffessm.fr/ref/specie/3757 (2017).
Prouzet, A. & Dumas, J. Melithaea ochracea (Linnaeus, 1758). DORIS: Données d’Observations pour la Reconnaissance et l’Identification de la faune et la flore Subaquatiques. http://doris.ffessm.fr/ref/specie/3756 (2021).
Dumas, J., Maran, V., Ader, D. & Huet, S. Paramuricea clavata (Risso, 1826). DORIS: Données d’Observations pour la Reconnaissance et l’Identification de la faune et la flore Subaquatiques. http://doris.ffessm.fr/ref/specie/223 (2022).
Jeglot, S. & Weinberg, S. Cavernularia pusilla (Philippi, 1835). DORIS: Données d’Observations pour la Reconnaissance et l’Identification de la faune et la flore Subaquatiques. http://doris.ffessm.fr/ref/specie/4690 (2022).
Vassard, E., Sittler, A.-P. & Reguieg, A. Pennatula rubra (Ellis, 1761). DORIS: Données d’Observations pour la Reconnaissance et l’Identification de la faune et la flore Subaquatiques. http://doris.ffessm.fr/ref/specie/930 (2020).
Jeglot, S., Arguel, M.-J. & Weinberg, S. Pteroeides griseum (Bohadsch, 1761). DORIS: Données d’Observations pour la Reconnaissance et l’Identification de la faune et la flore Subaquatiques. http://doris.ffessm.fr/ref/specie/4537 (2020).
Deschomets, Y., Jeglot, S., Weinberg, S. & Didierlaurent, S. Veretillum cynomorium (Pallas, 1766). DORIS: Données d’Observations pour la Reconnaissance et l’Identification de la faune et la flore Subaquatiques. http://doris.ffessm.fr/ref/specie/844 (2022).
Jeglot, S., Arguel, M.-J. & Weinberg, S. Virgularia mirabilis (Müller, 1776). DORIS: Données d’Observations pour la Reconnaissance et l’Identification de la faune et la flore Subaquatiques. http://doris.ffessm.fr/ref/specie/4538 (2020).
Dumas, J., André, F. & Huet, S. Clavularia crassa (Milne Edwards, 1848). DORIS: Données d’Observations pour la Reconnaissance et l’Identification de la faune et la flore Subaquatiques. http://doris.ffessm.fr/ref/specie/841 (2022).
André, F. & Weinberg, S. Cornularia cornucopiae (Pallas, 1766). DORIS: Données d’Observations pour la Reconnaissance et l’Identification de la faune et la flore Subaquatiques. http://doris.ffessm.fr/ref/specie/3343 (2021).
André, F. & Weinberg, S. Sarcodictyon catenatum Forbes, 1847. DORIS: Données d’Observations pour la Reconnaissance et l’Identification de la faune et la flore Subaquatiques. http://doris.ffessm.fr/ref/specie/3320 (2021).
Larkin, M. F. et al. The rapid decline of an Endangered temperate soft coral species. Estuar. Coast. Shelf Sci. 107364, 1–9, https://doi.org/10.1016/j.ecss.2021.107364 (2021).
IUCN. The IUCN Red List of Threatened Species. Version 2019–2. Downloaded on 1 September 2019. Online resource. www.iucnredlist.org (2019)
Trowbridge, C. D. et al. Shallow subtidal octocorals in an Irish marine reserve. Mar. Biodiv. 46, 879–887, https://doi.org/10.1007/s12526-016-0450-0 (2016).
Horvath, E. A. & Stone, R. P. Another unusual new gorgonian (Anthozoa: Octocorallia: Plexauridae) from the Aleutian Islands of Alaska. Zootaxa 4524(1), 112–120, https://doi.org/10.11646/zootaxa.4524.1.8 (2018).
Cordeiro, R. & McFadden, C. World List of Octocorallia. In O. Bánki, Y. et al. Catalogue of Life Checklist (ver. (09/2022)) https://doi.org/10.48580/dfqc-3d2 (2022).
Fabricius, K. E. Octocorallia. In D. Hopley (Ed.), Encyclopedia of Modern Coral Reefs. Encyclopedia of Earth Sciences Series (pp. 1–6). Springer, Dordrecht https://doi.org/10.1007/978-90-481-2639-2_35 (2011).
Halász, A. N. N. A., McFadden, C. S., Toonen, R. & Benayahu, Y. Re-description of type material of Xenia Lamarck, 1816 (Octocorallia: Xeniidae). Zootaxa 4652(2), 101–112, https://doi.org/10.11646/zootaxa.4652.2.1 (2019).
Picton, B. E. & Morrow, C. C. Cervera cf. atlantica (Johnson, 1861). [In] Encyclopedia of Marine Life of Britain and Ireland. Online identification guide. https://www.habitas.org.uk/marinelife/species.asp?item=D10110 (2016).
Picton, B. E. & Morrow, C. C. Sarcodictyon catenatum Forbes, 1847. [In] Encyclopedia of Marine Life of Britain and Ireland. Online identification guide. http://www.habitas.org.uk/marinelife/species.asp?item=D10170 (2016).
Picton, B. E. & Morrow, C. C. Alcyonium digitatum Linnaeus, 1758. [In] Encyclopedia of Marine Life of Britain and Ireland. Online identification guide. http://www.habitas.org.uk/marinelife/species.asp?item=D10240 (2016).
Picton, B. E. & Morrow, C. C. Alcyonium glomeratum (Hassall, 1843). [In] Encyclopedia of Marine Life of Britain and Ireland. Online identification guide. http://www.habitas.org.uk/marinelife/species.asp?item=D10250 (2016).
Picton, B. E. & Morrow, C. C. Alcyonium hibernicum (Renouf, 1931). [In] Encyclopedia of Marine Life of Britain and Ireland. Online identification guide. http://www.habitas.org.uk/marinelife/species.asp?item=D10300 (2016).
Picton, B. E. & Morrow, C. C. Swiftia pallida Madsen, 1970. [In] Encyclopedia of Marine Life of Britain and Ireland. Online identification guide. http://www.habitas.org.uk/marinelife/species.asp?item=D10370 (2016).
Picton, B. E. & Morrow, C. C. Eunicella verrucosa (Pallas, 1766). [In] Encyclopedia of Marine Life of Britain and Ireland. Online identification guide. http://www.habitas.org.uk/marinelife/species.asp?item=D10430 (2016).
Picton, B. E. & Morrow, C. C. Funiculina quadrangularis (Pallas, 1766). [In] Encyclopedia of Marine Life of Britain and Ireland. Online identification guide. http://www.habitas.org.uk/marinelife/species.asp?item=D10500 (2016).
Picton, B. E. & Morrow, C. C. Virgularia mirabilis (O F Müller, 1776). [In] Encyclopedia of Marine Life of Britain and Ireland. Online identification guide. http://www.habitas.org.uk/marinelife/species.asp?item=D10560 (2016).
Picton, B. E. & Morrow, C. C. Pennatula phosphorea Linnaeus, 1758. [In] Encyclopedia of Marine Life of Britain and Ireland. Online identification guide. http://www.habitas.org.uk/marinelife/species.asp?item=D10670 (2016).
van Ofwegen, L. P. Towards a revision of the Nephtheidae (Coelenterata: Octocorallia). PhD thesis, Faculty of Science, Leiden University (2007).
Grossowicz, M., Shemesh, E., Martinez, S., Benayahu, Y. & Tchernov, D. New evidence of Melithaea erythraea colonization in the Mediterranean. Estuar. Coast. Shelf Sci. 106652, 1–10 (2020).
van Ofwegen, L. P. M. Coelenterata: Anthozoa) from the Indian Ocean and the Malay Archipelago. Zool. Verh. 39, 1–57 (1987).
van Ofwegen, L. P. O. Wrightella coccinea (Ellis & Solander, 1786) and Wrightella stiasnyi spec. nov. (Anthozoa: Gorgonacea: Melithaeidae). Zool. Meded. 63(3), 27–34 (1989).
van Ofwegen, L. P. Notes on the Keroeididae (Anthozoa: Gorgonacea) collected in 1963 and 1964 by the International Indian Ocean Expedition of the R/V “Anton Bruun”, in the Indian Ocean, with the description of a new species, Lignella hartogi spec. nov. Zool. Meded. 63(13), 163–168 (1990).
van Ofwegen, L. P. Notes on Octocorallia from the Laccadives (SW India). Zool. Meded. 65(9), 143–154 (1991).
van Ofwegen, L. P. & Benayahu, Y. Notes on Alcyonacea (Octocorallia) from Tanzania. Zool. Meded. 66(6), 139–154 (1992).
Verseveldt, J. & van Ofwegen, L. P. New and redescribed species of Alcyonium Linnaeus, 1758 (Anthozoa: Alcyonacea). Zool. Meded. 66(7), 155–181 (1992).
van Ofwegen, L. P. & Vennam, J. Results of the Rumphius Biohistorical Expedition to Ambon (1990). Part 3. The Alcyoniidae (Octocorallia: Alcyonacea). Zool. Meded. 68(14), 135–158 (1994).
van Ofwegen, L. P. Octocorallia from the Bismarck Sea (part II). Zool. Meded. 70(13), 207–215 (1996).
Vennam, J. & van Ofwegen, L. P. Soft corals (Coelenterata: Octocorallia: Alcyonacea) from the Laccadives (SW India), with a re-examination of Sinularia gravis Tixier-Durivault, 1970. Zool. Meded. 70(29), 437–452 (1996).
Casas, C., Ramil, F. & van Ofwegen, L. P. Octocorallia (Cnidaria: Anthozoa) from the Scotia Arc, South Atlantic Ocean. I. The genus Alcyonium Linnaeus, 1758. Zool. Meded. 71(26), 299–311 (1997).
van Ofwegen, L. P. & Schleyer, M. H. Corals of the South-west Indian Ocean V. Leptophyton benayahui gen. nov. & spec. nov. (Cnidaria, Alcyonacea) from deep reefs off Durban and on the KwaZulu-Natal south coast, South Africa. Oceanographic Research Institute Investigational Report No. 71, 1–12 (1997).
Benayahu, Y., van Ofwegen, L. P. & Alderslade, P. A. A case study of variation in two nominal species of Sinularia (Coelenterata: Octocorallia), S. brassica May, 1898, and S. dura (Pratt, 1903), with a proposal for their synonymy. Zool. Verhand. 323, 277–309 (1998).
van Ofwegen, L. P. Lobophytum jasparsi spec. nov. from Indonesia (Coelenterata: Octocorallia: Alcyonacea). Zool. Meded. 73(8), 177–185 (1999).
van Ofwegen, L. P., Goh, N. K. C. & Chou, L. M. The Melithaeidae (Coelenterata: Octocorallia) of Singapore. Zool. Meded. 73(19), 285–304 (2000).
van Ofwegen, L. P. Sinularia vanderlandi spec. nov. (Octocorallia: Alcyonacea) from the Seychelles. Zool. Verhand. 334, 103–114 (2001).
van Ofwegen, L. P. & Hoeksema, B. W. Astrogorgia bayeri, a new gorgonian octocoral species from South Sulawesi (Coelenterata: Octocorallia: Plexauridae). Bull. Biol. Soc. Washington 10, 66–70 (2001).
van Ofwegen, L. P. Status of knowledge of the Indo Pacific soft coral genus Sinularia May, 1898 (Anthozoa: Octocorallia). Proc. 9th Int. Coral Reef Symp., Bali, Indonesia, 23–27 October 2000, 1, 167–171 (2002).
Ocaña, O. & van Ofwegen, L. P. Two new genera and one new species of stoloniferous octocorals (Anthozoa: Clavulariidae). Zool. Verh. 345, 269–274 (2003).
Hermanlimianto, M. I. Y. T. & van Ofwegen, L. P. A new species of Astrogorgia (Coelenterata: Octocorallia: Plexauridae) from Bali. Zool. Meded. 80(4), 103–108 (2006).
Stokvis, F. R. & van Ofwegen, L. P. New and redescribed encrusting species of Alcyonium from the Atlantic Ocean (Octocorallia: Alcyonacea: Alcyoniidae). Zool. Meded. 80(4), 165–183 (2006).
van Ofwegen, L. P. & Benayahu, Y. A new genus of paralcyoniid soft corals (Octocorallia, Alcyonacea, Paralcyoniidae) from the Indo-West Pacific. Galaxea, J. Jap. Coral Reef Soc 8(1), 25–37 (2006).
van Ofwegen, L. P., Häussermann, V. & Försterra, G. A new genus of soft coral (Octocorallia: Alcyonacea: Clavulariidae) from Chile. Zootaxa 1219(1), 47–57 (2006).
Manuputty, A. E. W. & van Ofwegen, L. P. The genus Sinularia (Octocorallia: Alcyonacea) from Ambon and Seram (Moluccas, Indonesia). Zool. Meded. 81(11), 187–216 (2007).
van Ofwegen, L. P. & Alderslade, P. A. A new species of Alertigorgia (Coelenterata: Octocorallia: Anthothelidae) from the Indo Malayan region. Zool. Meded. 81(13), 241–249 (2007).
van Ofwegen, L. P., Häussermann, V. & Försterra, G. The genus Alcyonium (Octocorallia: Alcyonacea: Alcyoniidae) in Chile. Zootaxa 1607(1), 1–19 (2007).
van Ofwegen, L. P. The genus Sinularia (Octocorallia: Alcyonacea) from Bremer and West Woody Islands (Gulf of Carpentaria, Australia). Zool. Meded. 82(16), 131–165 (2008).
van Ofwegen, L. P. The genus Sinularia (Octocorallia: Alcyonacea) at Palau, Micronesia. Zool. Meded. 82(51), 631–735 (2008).
Benayahu, Y. & van Ofwegen, L. P. New species of Sarcophyton and Lobophytum (Octocorallia: Alcyonacea) from Hong Kong. Zool. Meded. 83(30), 863–876 (2009).
Samimi Namin, K. & van Ofwegen, L. P. The second observation of a live Trimuricea species (Octocorallia: Plexauridae). Coral Reefs 28(2), e517 (2009).
Payri, C. E. & de Forges, R. B. Compendium of Marine Species From New Caledonia. Report No. Institute of Research for Development (2007).
Dautova, T. N., van Ofwegen, L. P. & Savinkin, O. V. New species of the genus Sinularia (Octocorallia: Alcyonacea) from Nha Trang Bay, South China Sea, Vietnam. Zool. Med. 84(5), 47–91 (2010).
Samimi Namin, K. & van Ofwegen, L. P. An overview of Bebryce (Cnidaria, Octocorallia, Plexauridae) species with tiny rosettes, with the description of a new species from the Gulf of Oman. Zoosyst 32(3), 479–493 (2010).
van Ofwegen, L. P. & McFadden, C. S. A new family of octocorals (Anthozoa: Octocorallia) from Cameroon waters. J. Nat. Hist. 44(1), 23–29 (2010).
Opresko, D. M. & Bayer, F. M. Rediscovery of the enigmatic coelenterate Dendrobrachia (Octocorallia: Gorgonacea) with descriptions of two new species. Trans. R. Soc. S. Aust. 115, 1–19 (1991).
Benayahu, Y. & van Ofwegen, L. P. New species of the genus Sinularia (Octocorallia: Alcyonacea) from Singapore with notes on the occurrence of other species of the genus. Raffles Bull. Zool. 59(2), 117–125 (2011).
Benayahu, Y. & Chow, L. M. On some Octocorallia (Cnidaria: Anthozoa: Alcyonacea) from Singapore, with description of a new Cladiella species. Raffles Bull. Zool. 58(1), 1–13 (2010).
Benayahu, Y. & van Ofwegen, L. P. New species of octocorals (Coelenterata: Anthozoa) from the Penghu archipelago, Taiwan. Zool. Stud. 50(3), 350–362 (2011).
Samimi-Namin, K., van Ofwegen, L. P., Wilson, S. C. & Claereboudt, M. R. The first in situ and shallow-water observation of the genus Pseudothelogorgia (Octocorallia: Keroeididae). Zool. Stud. 50(2), e265 (2011).
van Ofwegen, L. P. & Haddad, M. A. A probably invasive new genus and new species of soft coral (Octocorallia: Alcyonacea: Clavulariidae) from Brazil. Zootaxa 3107(1), 38–46 (2011).
Benayahu, Y. & van Ofwegen, L. P. Octocorals (Cnidaria, Anthozoa) from Reunion, with a description of two new species of the genus Sinularia May, 1898 and notes on the occurrence of other species. Zoosyst 34(4), 673–699 (2012).
Benayahu, Y. et al. Diversity, distribution, and molecular systematics of octocorals (Coelenterata: Anthozoa) of the Penghu Archipelago, Taiwan. Zool. Stud. 51(8), 1529–1548 (2012).
van Ofwegen, L. P. & Benayahu, Y. Two new species and a new record of the genus Sinularia (Octocorallia, Alcyonacea) from the Penghu archipelago, Taiwan. Zool. Stud. 51(3), 283–398 (2012).
Breedy, O., van Ofwegen, L. P. & Vargas, S. A new family of soft corals (Anthozoa, Octocorallia, Alcyonacea) from the aphotic tropical eastern Pacific waters revealed by integrative taxonomy. Syst. Biodivers. 10(3), 351–359 (2012).
McFadden, C. S. & van Ofwegen, L. P. Stoloniferous octocorals (Anthozoa, Octocorallia) from South Africa, with descriptions of a new family of Alcyonacea, a new genus of Clavulariidae, and a new species of Cornularia (Cornulariidae). Invertebr. Syst. 26(4), 331–356 (2012).
Samimi-Namin, K. & van Ofwegen, L.P. The octocoral fauna of the Gulf. In: Riegl BM, Purkis SJ (Eds) Coral Reefs of the Gulf: Adaptation to Climatic Extremes (Springer, Dordrecht, 2012), pp. 225–252.
McFadden, C. S. & van Ofwegen, L. P. A revision of the soft coral genus Eunephthya Verrill, 1869 (Anthozoa: Octocorallia: Nephtheidae), with a description of four new species from South Africa. Zootaxa 3485(1), 1–25 (2012).
McFadden, C. S. & van Ofwegen, L. P. A second, cryptic species of the soft coral genus Incrustatus (Anthozoa: Octocorallia: Clavulariidae) from Tierra del Fuego, Argentina, revealed by DNA barcoding. Helgol. Mar. Res. 67(1), 137–147 (2013).
McFadden, C. S. & van Ofwegen, L. P. Molecular phylogenetic evidence supports a new family of octocorals and a new genus of Alcyoniidae (Octocorallia, Alcyonacea). ZooKeys 346, 59–83 (2013).
Dautova, T. N. & Savinkin, O. V. New data on soft corals (Cnidaria: Octocorallia: Alcyonacea) from Nha Trang Bay, South China Sea. Zootaxa 2027, 1–27 (2009).
van Ofwegen, L. P., Benayahu, Y. & McFadden, C. S. Sinularia leptoclados (Ehrenberg, 1834) (Cnidaria: Octocorallia) re-examined. ZooKeys 272, 29–59 (2013).
Bryce, M., Poliseno, A., Alderslade, P. & Vargas, S. Digitate and capitate soft corals (Cnidaria: Octocorallia: Alcyoniidae) from Western Australia with reports on new species and new Australian geographical records. Zootaxa 3963(2), 160–200 (2015).
Reijnen, B. T., McFadden, C. S., Hermanlimianto, Y. T. & van Ofwegen, L. P. A molecular and morphological exploration of the generic boundaries in the family Melithaeidae (Coelenterata: Octocorallia) and its taxonomic consequences. Mol. Phylogenet. Evol. 70, 383–401 (2014).
Alderslade, P. New subfamilies and a new genus and species of Melithaeidae (Coelenterata: Octocorallia: Alcyonacea) with comparative data on the structure of both melithaeid and subergorgiid axes. Zootaxa 1199(1), 19–47 (2006).
van Ofwegen, L. P. & Hermanlimianto, M. I. Y. T. Siboga plexaurids (Coelenterata: Octocorallia) re-examined. Zool. Med. 88(3), 19–58 (2014).
van Ofwegen, L. P., Aurelle, D. & Sartoretto, S. A new genus of soft coral (Cnidaria, Octocorallia) from the Republic of Congo (Pointe-Noire Region). ZooKeys 462, 1–10 (2014).
Matsumoto, A. K. & van Ofwegen, L. P. Melithaeidae of Japan (Octocorallia, Alcyonacea) re-examined with descriptions of 11 new species. ZooKeys 522, 1–127 (2015).
Bayer, F. M. & van Ofwegen, L. P. The type specimens of Bebryce (Cnidaria, Octocorallia, Plexauridae) re-examined, with emphasis on the sclerites. Zootaxa 4083(3), 301–358 (2016).
Devictor, S. T. & Morton, S. L. Identification guide to the shallow water (0–200 m) octocorals of the South Atlantic Bight. Zootaxa 2599(1), 1–62 (2010).
Deichmann, E. The Alcyonaria of the western part of the Atlantic Ocean. Memoirs of the Museum of Comparative Zoölogy, at Harvard College, Cambridge, Mass 53, 1–308 (1936). 37.
Matsumoto, A. K. & van Ofwegen, L. P. The genus Bebryce (Cnidaria, Octocorallia, Plexauridae) at Japan, with descriptions of three new species. Zootaxa 587, 1–20 (2016).
Matsumoto, A. K. & van Ofwegen, L. P. Species of Elasmogorgia and Euplexaura (Cnidaria, Octocorallia) from Japan with a discussion about the genus Filigella. Zootaxa 589, 1–21 (2016).
Samimi-Namin, K. & van Ofwegen, L. P. Overview of the genus Briareum (Cnidaria, Octocorallia, Briareidae) in the Indo-Pacific, with the description of a new species. Zookeys 557, 1–44 (2016).
Samimi-Namin, K. & van Ofwegen, L. P. A revision of Trimuricea Gordon, 1926 (Cnidaria: Octocorallia: Plexauridae) with the description of six new species. Zootaxa 4105(1), 1–44 (2016).
Samimi-Namin, K., van Ofwegen, L. P. & McFadden, C. S. A new species of Melithaea (Anthozoa, Octocorallia, Melithaeidae) from the Oman Sea, off Oman. Zookeys 623, 15–29 (2016).
van Ofwegen, L. P. The genus Litophyton Forskål, 1775 (Octocorallia, Alcyonacea, Nephtheidae) in the Red Sea and the western Indian Ocean. Zookeys 567, 1–128 (2016).
van Ofwegen, L. P., McFadden, C. S. & Benayahu, Y. Sinularia polydactyla (Ehrenberg, 1834) (Cnidaria, Octocorallia) re-examined, with the description of a new species. Zookeys 581, 71–126 (2016).
Benayahu, Y., McFadden, C. S., Shoham, E. & van Ofwegen, L. P. Search for mesophotic octocorals (Cnidaria, Anthozoa) and their phylogeny. II. A new zooxanthellate species from Eilat, northern Red Sea. ZooKeys 676, 1–12 (2017).
McFadden, C. S. & van Ofwegen, L. P. Revisionary systematics of the endemic soft coral fauna (Octocorallia: Alcyonacea: Alcyoniina) of the Agulhas Bioregion, South Africa. Zootaxa 4363(4), 451–488 (2017).
Benayahu, Y., van Ofwegen, L. P. & McFadden, C. S. Evaluating the genus Cespitularia Milne Edwards & Haime, 1850 with descriptions of new genera of the family Xeniidae (Octocorallia, Alcyonacea). ZooKeys 754, 63–101 (2018).
Benayahu, Y. et al. The octocorals of Dongsha Atoll (South China Sea): an iterative approach to species identification using classical taxonomy and molecular barcodes. Zoological Studies 57, 1–50 (2018).
Lau, Y. W., Stokvis, F. R., van Ofwegen, L. P. & Reimer, J. D. Stolonifera from shallow waters in the north-western Pacific: a description of one new genus and two new species within the Arulidae (Anthozoa, Octocorallia). ZooKeys 790, 1–19 (2018).
Tuti, Y. & van Ofwegen, L.P. Gorgonians in Indonesian Waters (Media Sains National, Bogor, Indonesia, 2018), 89 pp.
Matsumoto, A. K., van Ofwegen, L. P. & Bayer, F. M. A revision of the genus Calcigorgia (Cnidaria, Octocorallia, Acanthogorgiidae) with the description of three new species. Zootaxa 4571(1), 1–27 (2019).
van Ofwegen, L. P. The genus Litophyton Forskål, 1775 (Octocorallia, Alcyonacea, Nephtheidae) from Australia. Zootaxa 4764(1), 1–131 (2020).
Verselveldt, J. & Alderslade, P. Description of types and other alcyonacean material (Coelenterata: Octocorallia) in the Australian Museum, Sydney. Rec. Aust. Mus. 34(15), 619–647 (1982).
Benayahu, Y., van Ofwegen, L. P., Ruiz Allais, J. P. & McFadden, C. S. Revisiting the type of Cespitulariastolonifera Gohar, 1938 leads to the description of a new genus and a species of the family Xeniidae (Octocorallia, Alcyonacea). Zootaxa 4964(2), 330–344 (2021).
Breedy, O., van Ofwegen, L. P., McFadden, C. S. & Murillo-Cruz, C. Rhodolitica on rhodoliths: a new stoloniferan genus (Anthozoa, Octocorallia, Alcyonacea). ZooKeys 1032, 63–77 (2021).
Shuler, A.J. & Etnoyer, P.J. in Alcyonacean Octocorals of the Pinnacle Trend: A Photo-Identification Guide (2020).
Kükenthal, W. Gorgonaria. Wissenschaftliche Ergebnisse der Deutschen Tiefsee-Expedition auf dem Dampfer “Valdivia”, 1898–1899. 13(2), 946. (1919).
Sánchez, J.A. et al. Gorgonian Corals. In Mesophotic Coral Ecosystems (ed. Loya, Y., Puglise, K. A. & Bridge, T. C.) (Springer, 2019).
Kessel, G. M. et al. Dead man’s fingers point to new taxa: two new genera of New Zealand soft corals (Anthozoa, Octocorallia) and a revision of Alcyonium aurantiacum Quoy & Gaimard, 1833. Eur. J. Taxon. 837, 1–85 (2022).
López-González, P. J., Gili, J. M. & Williams, G. C. New records of Pennatulacea (Anthozoa: Octocorallia) from the African Atlantic coast, with description of a new species and a zoogeographic analysis. Sci. Mar. 65(1), 59–74 (2001).
Breedy, O. & Guzmán, H. M. New species of the gorgonian genus Pacifigorgia (Coelenterata: Octocorallia: Gorgoniidae) from Pacific Panama. Zootaxa 541, 1–18 (2004).
Pérez, C. D. & Zamponi, M. O. New records of octocorals (Cnidaria, Anthozoa) from the southwestern Atlantic Ocean, with zoogeographic considerations. Zootaxa 630(0), 1–12 (2004).
Dueñas, L. F., Alderslade, P. & Sánchez, J. A. Molecular systematics of the deep-sea bamboo corals (Octocorallia: Isididae: Keratoisidinae) from New Zealand with descriptions of two new species of Keratoisis. Mol. Phylogenet. Evol. 74, 15–28 (2014).
Bayer, F. M. A new isidid octocoral (Anthozoa: Gorgonacea) from New Caledonia, with descriptions of other new species from elsewhere in the Pacific Ocean. Proc. Biol. Soc. Wash. 103(1), 205–228 (1990).
Alderslade, P. & McFadden, C. S. A new genus and species of the family Isididae (Coelenterata: Octocorallia) from a CMAR Biodiversity study, and a discussion on the subfamilial placement of some nominal isidid genera. Zootaxa 3154(1), 21–39 (2012).
Watling, L. & France, S. C. A new genus and species of bamboo coral (Octocorallia: Isididae: Keratoisidinae) from the New England Seamounts. Bull. Peabody Mus. Nat. Hist. 52(2), 209–220 (2011).
Cairns, S. D. A new species of Chrysogorgia (Anthozoa: Octocorallia) from the Antarctic. Proc. Biol. Soc. Wash. 115, 217–222 (2002).
Bryce, M. & Wilson, N. G. A new genus with two new capitate species of dimorphic soft corals (Octocorallia: Alcyoniidae) from north-western Australia. Invertebr. Syst. 33(1), 71–88 (2019).
Williams, G. C. New taxa and revisionary systematics of alcyonacean octocorals from the Pacific coast of North America (Cnidaria, Anthozoa). ZooKeys 283, 1–75 (2013).
Williams, G. C. & Breedy, O. A new species of gorgonian octocoral from the mesophotic zone off the central coast of California, Eastern Pacific with a key to related regional taxa (Anthozoa, Octocorallia, Alcyonacea). Proc. Calif. Acad. Sci. Ser. 4 65(6), 143–158 (2019).
Horvath, E. A. A review of gorgonian coral species (Cnidaria, Octocorallia, Alcyonacea) held in the Santa Barbara Museum of Natural History research collection: focus on species from Scleraxonia, Holaxonia, Calcaxonia - Part II: species of Holaxonia, families Gorgoniidae and Plexauridae. ZooKeys 860, 183–208 (2019).
Breedy, O. & Guzman, H. A revision of the genus Leptogorgia Milne Edwards & Haime, 1857 (Coelenterata: Octocorallia: Gorgoniidae) in the eastern Pacific. Zootaxa 1419, 1–90 (2007).
Cairns, S. D. Studies on western Atlantic Octocorallia (Coelenterata: Anthozoa) Part 1: The genus Chrysogorgia Duchassaing & Michelotti, 1864. Proc. Biol. Soc. Wash. 114(3), 746–787 (2001).
Cairns, S. D. & Bayer, F. M. Studies on western Atlantic Octocorallia (Coelenterata: Anthozoa). Part 2: The genus Callogorgia Gray, 1858. Proc. Biol. Soc. Wash. 115(4), 840–867 (2002).
Cairns, S. D. & Bayer, F. M. Studies on western Atlantic Octocorallia (Coelenterata: Anthozoa). Part 3. The genus Narella Gray, 1870. Proc. Biol. Soc. Wash. 116(3), 617–648 (2003).
Cairns, S. D. & Bayer, F. M. Studies on western Atlantic Octocorallia (Coelenterata: Anthozoa). Part 4. The genus Paracalyptrophora Kinoshita, 1908. Proc. Biol. Soc. Wash. 117(1), 114–139 (2004).
Cairns, S. D. & Bayer, F. M. Studies on western Atlantic Octocorallia (Coelenterata: Anthozoa). Part 5: the genera Plumarella Gray, 1870; Acanthoprimnoa, n. gen.; and Candidella Bayer, 1954. Proc. Biol. Soc. Wash. 117(4), 447–487 (2004).
Cairns, S. D. Studies on western Atlantic Octocorallia (Coelenterata: Anthozoa). Part 6. The genera Primnoella Gray, 1858; Thouarella Gray, 1870; Dasystenella Versluys, 1906. Proc. Biol. Soc. Wash. 119(2), 161–194 (2006).
Cairns, S. D. Studies on western Atlantic Octocorallia (Gorgonacea: Ellisellidae). Part 7: The genera Riisea Duchassaing & Michelotti, 1860 and Nicella Gray, 1870. Proc. Biol. Soc. Wash. 120(1), 1–38 (2007).
Cairns, S. D. Studies on western Atlantic octocorallia (Gorgonacea: Primnoidae). Part 8: New records of Primnoidae from the New England and corner rise seamounts. Proc. Biol. Soc. Wash. 120(3), 243–263 (2007).
Castro, C.-B. A new species of Plexaurella Valenciennes, 1855 (Coelenterata, Octocorallia), from the Abrolhos Reefs, Bahia, Brazil. Rev. Bras. Ecol. 49(2), 597–603 (1989).
Dautova, T.N. Two new species of deep-water Calcigorgia gorgonians (Anthozoa: Octocorallia) from the Kurile Islands, Sea of Okhotsk, with a review of distinctive characters of the known species of the genus. Eur. J. Taxon. 408 (2018).
López-González, P. J., Gili, J. M. & Williams, G. C. On some veretillid pennatulaceans from the eastern Atlantic and western Pacific Oceans (Anthozoa: Octocorallia), with a review of the genus Cavernularia, and descriptions of new taxa. J. Zool. 250(2), 201–216 (2006).
Williams, G. C. The pennatulacean genus Cavernularia Valenciennes (Octocorallia: Veretillidae). Zool. J. Linn. Soc. 95(4), 285–310 (1989).
Soler-Hurtado, M. D. M. & López-González, P. J. Two new gorgonian species (Anthozoa: Octocorallia: Gorgoniidae) from Ecuador (Eastern Pacific). Mar. Biol. Res. 8(4), 380–387 (2012).
Breedy, O., Guzman, H. M. & Vargas, S. A revision of the genus Eugorgia Verrill, 1868 (Coelenterata: Octocorallia: Gorgoniidae). Zootaxa 2151(1), 1–7 (2009).
López-González, P. J. & Gili, J. M. A new species of Nidalia Gray, 1835 from Mid-Atlantic seamounts (Octocorallia, Alcyonacea, Nidaliidae). Helgol. Mar. Res. 62, 389–392 (2008).
Conti-Jerpe, I. E. & Freshwater, D. W. Hedera caerulescens (Alcyonacea: Alcyoniidae), a new genus and species of soft coral from the temperate North Atlantic: invasive in its known range? Invert. Syst. 31(6), 723–733 (2017).
Perez, C. D. New records of octocorals (Cnidaria: Anthozoa) from the Brazilian coast. Aquat. Biol. 13(3), 203–214 (2011).
Xu, Y., Zhan, Z. & Xu, K. Morphology and phylogenetic analysis of five deep-sea golden gorgonians (Cnidaria, Octocorallia, Chrysogorgiidae) in the Western Pacific Ocean, with the description of a new species. ZooKeys 989, 1 (2020).
Bayer, F. M., Cairns, S. D., Cordeiro, R. T. & Pérez, C. D. New records of the genus Callogorgia (Anthozoa: Octocorallia) in the western Atlantic, including the description of a new species. J. Mar. Biol. Assoc. U.K. 95(5), 905–911 (2015).
Cairns, S. D. Review and five new Alaskan species of the deep-water octocoral Narella (Octocorallia: Primnoidae). Syst. Biodivers. 5(4), 391–407 (2007).
Herrera, S., Baco, A. & Sánchez, J. A. Molecular systematics of the bubblegum coral genera (Paragorgiidae, Octocorallia) and description of a new deep-sea species. Mol. Phylogenet. Evol. 55(1), 123–135 (2010).
Bayer, F. M. A new species of the gorgonacean genus Bebryce (Coelenterata: Octocorallia) from Papua New Guinea. Bull. Mar. Sci. 54(2), 546–553 (1994).
Williams, G. C. A new species of stoloniferous octocoral (Cnidaria: Alcyonacea) from the south-western Indian Ocean. J. Nat. Hist. 21(1), 207–218 (2007).
López González, P. J., Gili, J. M. & Orejas, C. A new primnoid genus (Anthozoa: Octocorallia) from the Southern Ocean. Sci. Mar. 66(4), 383–397 (2002).
López-González, P. J. & Gili, J. M. A new octocoral genus (Cnidaria: Anthozoa) from Antarctic waters. Polar Biol 23, 452–458 (2000).
Bayer, F. M. New Primnoid Gorgonians (Coelenterata: Octocorallia) from Antarctic Waters. Bull. Mar. Sci. 58(2), 511–530 (1996).
Etnoyer, P. J. A new species of Isidella bamboo coral (Octocorallia: Alcyonacea: Isididae) from northeast Pacific Seamounts. Proc. Biol. Soc. Wash. 121(4), 541–553 (2009).
Cordeiro, R. A review of the genus Trichogorgia (Cnidaria, Octocorallia), including descriptions of new species. Zootaxa 4706(4) (2019).
Zapata-Guardiola, R. & López-González, P. J. Redescription of Thouarella brucei Thomson and Ritchie, 1906 (Cnidaria: Octocorallia: Primnoidae) and description of two new Antarctic primnoid species. Zootaxa 2616(1), 48–68 (2010).
Verseveldt, J. & Cohen, J. Some new species of Octocorallia from the Gulf of Elat (Red Sea). Isr. J. Ecol. Evol. 20(1), 53–67 (1971).
López-González, P. J. A new gorgonian genus from deep-sea Antarctic waters (Octocorallia, Alcyonacea, Plexauridae). Helgol. Mar. Res. 60, 1–6 (2006).
Enrichetti, F., Bavestrello, G., Coppari, M., Betti, F. & Bo, M. Placogorgia coronata first documented record in Italian waters: Use of trawl bycatch to unveil vulnerable deep-sea ecosystems. Aquat. Conserv.: Mar. Freshw. Ecosyst. 28(5), 1123–1138 (2018).
Imahara, Y. Two new paralcyoniid octocorals from Japan (Anthozoa: Alcyonacea). Zootaxa 3652(4) (2013).
Cairns, S. D. Deep-Water Octocorals (Cnidaria, Anthozoa) from the Galápagos and Cocos Islands. Part 1: Suborder Calcaxonia. ZooKeys 729, 1 (2018).
Hernández, O., Gomez-Gutiérrez, J. & Sánchez, C. Three new species of the sea fan genus Leptogorgia (Octocorallia, Gorgoniidae) from the Gulf of California, Mexico. ZooKeys 1017, 1 (2020).
Breedy, O., Rouse, G. W., Stabbins, A., Cortes, J. & Cordes, E. E. New records of Swiftia (Cnidaria, Anthozoa, Octocorallia) from off the Pacific Costa Rican margin, including a new species from methane seeps. Zootaxa 4671(3) (2019).
Cordeiro, R. Callogorgia lucaya sp. nov., a new species of deep-sea Primnoidae (Anthozoa: Octocorallia) from the western Atlantic. Zootaxa 4441(3), 529–536 (2018).
Zapata-Guardiola, R. & López-González, P. J. Designation of Thouarella abies Broch, 1965 as the type species of the subgenus Fannyella (Scyphogorgia) Cairns and Bayer, 2009, and description of a new genus for Stenella (Dasystenella) liouvillei Gravier, 1913 (Octocorallia: Primnoidae). J. Nat. Hist. 44(33–34), 1995–2013 (2013).
Perez, C. D. First record of Leptogorgia punicea (Milne-Edwards & Haime) (Cnidaria, Octocorallia) from Maranhão State, Brazil. Rev. Bras. Zool. 22, 810–811 (2005).
Bayer, F. M. & Stefani, J. A new species of Chrysogorgia (Octocorallia: Gorgonacea) from New Caledonia, with descriptions of some other species from the western Pacific. Proc. Biol. Soc. Wash. 101, 257–279 (1988).
Veena, S. & Kaladharan, P. The first record of Cavernulina orientalis (Thomson & Simpson, 1909) (Octocorallia: Pennatulacea: Veretillidae) from the Bay coast of Visakhapatnam, Andhra Pradesh. Zootaxa 3204, 61–64 (2012).
Breedy, O. & Guzman, H. M. Leptogorgia ignita, a new shallow-water coral species (Octocorallia: Gorgoniidae) from the tropical eastern Pacific. J. Mar. Biol. Assoc. U. K. 88, 893–899 (2008).
Bayer, F. M. A new species of Leptogorgia from the eastern Pacific (Coelenterata: Octocorallia: Holaxonia). Proc. Biol. Soc. Wash. 113, 609–616 (2000).
Bayer, F. M. Narella nuttingi, a new gorgonacean octocoral of the family Primnoidae (Anthozoa) from the eastern Pacific. Proc. Biol. Soc. Wash. 110, 511–519 (1997).
Cairns, S. D. & Häussermann, V. A new species of Thouarella from Chilean Patagonia. Spixiana 44, 1–8 (2021).
Richards, Z. T. et al. Integrated evidence reveals a new species in the ancient blue coral genus Heliopora (Octocorallia). Sci. Rep. 8, 15875 (2018).
Perez, C. D. & Cordeiro, R. Ideogorgia laurae, an uncommon new octocoral species (Alcyonacea: Keroeididae) from a newly established Marine Protected Area at Burdwood Bank, Argentina. Polar Biol 43, 63–69 (2020).
Imahara, Y. et al. Two new species of the genus Chironephthya (Octocorallia, Alcyonacea, Nidaliidae, Siphonogorgiinae) from the Gulf of Thailand. Zootaxa 4780(2), 324–340 (2020).
Koido, T., Imahara, Y. & Fukami, H. Xenia konohana sp. nov. (Cnidaria, Octocorallia, Alcyonacea), a new soft coral species in the family Xeniidae from Miyazaki, Japan. ZooKeys 1085, 29 (2022).
Breedy, O. & Guzman, H. M. A new species of the genus Eugorgia (Cnidaria: Octocorallia: Gorgoniidae) from mesophotic reefs in the eastern Pacific. Bull. Mar. Sci. 89, 735–743 (2013).
Zapata-Guardiola, R. & Lopez-Gonzalez, P. J. Two new gorgonian genera (Octocorallia: Primnoidae) from Southern Ocean waters. Polar Biol 33, 313–320 (2010).
Alderslade, P. & McFadden, C. S. Pinnule-less polyps: a new genus and new species of Indo-Pacific Clavulariidae and validation of the soft coral genus Acrossota and the family Acrossotidae (Coelenterata: Octocorallia). Zootaxa 1400, 27–44 (2007).
Williams, G. C. A new species of the octocorallian genus Alcyonium (Anthozoa: Alcyonacea) from southern Africa, with a revised diagnosis of the genus. J. Nat. Hist. 20, 53–63 (2007).
Nieves, B. & Perez, C. D. A new species of Sclerobelemnon Kölliker, 1872 from Brazil (Octocorallia: Pennatulacea: Kophobelemnidae). Cah. Biol. Mar. 53, 429–434 (2012).
Rajendra, S., Raghunathan, C. & Chandra, K. New record of Sarcophyton cornispiculatum Verseveldt, 1971 (Octocorallia: Alcyonacea: Alcyoniidae) in India, from the Andaman Islands. Eur. Zool. J. 84, 167–171 (2017).
Watling, L. A new genus of bamboo coral (Octocorallia: Isididae) from the Bahamas. Zootaxa 3918(2), 239–249 (2015).
Mastrototaro, F., Chimienti, G., Capezutto, F., Carlucci, R. & Williams, G. First record of Protoptilum carpenteri (Cnidaria: Octocorallia: Pennatulacea) in the Mediterranean Sea. Ital. J. Zool. 82, 61–68 (2014).
Cairns, S. D. & Cordeiro, R. A new genus and species of golden coral (Anthozoa, Octocorallia, Chrysogorgiidae) from the Northwest Atlantic. ZooKeys 668, 1–15 (2017).
Breedy, O., Guzman, H. M., Murillo, C. & Vargas, S. A new species of the genus Psammogorgia (Cnidaria: Anthozoa: Octocorallia) from the Hannibal Bank in Pacific Panama. Bull. Mar. Sci. 96, 169–180 (2019).
Sartoretto, S. & Zibrowius, H. Note on new records of living Scleractinia and Gorgonaria between 1700 and 2200 m depth in the western Mediterranean Sea. Mar. Biodiv. 48, 689–694 (2018).
López-González, P. J. & Williams, G. C. A new deep-sea pennatulacean (Anthozoa: Octocorallia: Chunellidae) from the Porcupine Abyssal Plain (NE Atlantic). Helgol. Mar. Res. 65, 309–318 (2011).
Bayer, F. M. & Grasshoff, M. Two new species of the gorgonacean genus Ctenocella (Coelenterata: Anthozoa, Octocorallia) from deep reefs in the western Atlantic. Bull. Mar. Sci. 56, 625–652 (1995).
Bayer, F. M. & Muzik, K. M. A new solitary octocoral, Taiaroa tauhou gen. et sp. nov. (Coelenterata: Protoalcyonaria) from New Zealand. J. R. Soc. N. Z. 6, 499–515 (1976).
Bayer, F. M. New species of Calyptrophora (Coelenterata: Octocorallia: Primnoidae) from the western part of the Atlantic Ocean. Proc. Biol. Soc. Wash. 114, 367–380 (2001).
Williams, G. C. & Alderslade, P. Three new species of pennatulacean octocorals with the ability to attach to rocky substrata (Cnidaria: Octocorallia: Virgulariidae). Zootaxa 3001(1), 33–48 (2011).
Vafidis, D. & Koukoufas, A. Crassophyllum thessalonicae sp. n. (Octocorallia, Pennatulacea), from the Aegean Sea. Zool. Scr. 20, 201–205 (1991).
Slattery, M., McClintock, J. B. & Bowser, S. S. Deposit feeding: a novel mode of nutrition in the Antarctic colonial soft coral Gersemia antarctica. Mar. Ecol. Prog. Ser. 149, 299–304 (1997).
Bayer, F. M. & Cairns, S. D. A new genus of the scleraxonian family Coralliidae (Octocorallia: Gorgonacea). Proc. Biol. Soc. Wash. 116, 222–228 (2003).
Benayahu, Y. & Fabricius, K. E. On some Octocorallia (Alcyonacea) from Hong Kong, with description of a new species, Paraminabea rubeusa. Pac. Sci. 64, 285–296 (2010).
Rajendra, S. & Raghunathan, C. A new record of soft coral, Sarcophyton subviride Tixier-Durivault, 1958 (Cnidaria: Anthozoa: Octocorallia) from Great Nicobar Island, India. Natl. Acad. Sci. Lett. 45, 145–148 (2022).
Horvath, E. A. An unusual new “sea fan” from the northeastern Pacific Ocean (Cnidaria: Octocorallia: Gorgoniidae). Proc. Biol. Soc. Wash. 124, 45–52 (2011).
Breedy, O. & Guzman, H. A revision of the genus Muricea Lamouroux, 1821 (Anthozoa, Octocorallia) in the eastern Pacific. Part I: Eumuricea Verrill, 1869 revisited. ZooKeys 537, 1–32 (2015).
Breedy, O. & Guzman, H. A revision of the genus Muricea Lamouroux, 1821 (Anthozoa, Octocorallia) in the eastern Pacific. Part II. ZooKeys 581, 1–69 (2016).
Pante, E. & France, S. C. Pseudochrysogorgia bellona n. gen., n. sp.: a new genus and species of chrysogorgiid octocoral (Coelenterata, Anthozoa) from the Coral Sea. Zoosystema 32, 595–612 (2010).
Ocaña, O. & Çinar, M. E. Descriptions of two new genera, six new species and three new records of Anthozoa (Cnidaria) from the Sea of Marmara. J. Nat. Hist. 52, 2243–2282 (2018).
Bayer, F. M. Thelogorgia, a new genus of gorgonacean octocorals, with descriptions of four new species from the western Atlantic. Bull. Mar. Sci. 49, 506–537 (1991).
Dautova, T. N. New species of deep-water Calcigorgia gorgonians (Anthozoa: Octocorallia) from the Sea of Okhotsk, with a re-diagnosis and a taxonomic review of the genus. Eur. J. Taxon. 549, 1–30 (2019).
López-González, P. J. Scytalium herklotsi sp. nov. (Anthozoa, Octocorallia, Pennatulacea), the first Atlantic species in the genus Scytalium Herklots, 1858. Mar. Biodiv. 51, 62 (2021).
Xu, Y., Zhan, Z. & Xu, K. Morphological and molecular characterization of five species, including three new species of golden gorgonians (Cnidaria: Octocorallia) from seamounts in the western Pacific. Biology 10, 588 (2021).
Risaro, J. A., Williams, G., Pereyra Villaurreta, M. D. R. & Lauretta, D. M. Umbellula pomona sp. nov., a new sea pen from Mar del Plata Submarine Canyon (Cnidaria: Octocorallia: Pennatulacea). Eur. J. Taxon. 720, 121–143 (2020).
Bayer, F. M. & Stefani, J. New and previously known taxa of isidid octocorals (Coelenterata: Gorgonacea), partly from Antarctic waters. Proc. Biol. Soc. Wash. 100, 937–991 (1987).
Alderslade, P. & McFadden, C. S. A new sclerite-free genus and species of Clavulariidae (Coelenterata: Octocorallia). Zootaxa 3104(1), 64–68 (2011).
Guzman, H. M. & Breedy, O. Pacifigorgia marviva (Anthozoa: Octocorallia) a new species from Coiba National Park, Pacific Panama. J. Mar. Biol. Assoc. U.K. 92, 693–698 (2011).
Lau, Y. W. & Reimer, J. D. A first phylogenetic study on stoloniferous octocorals off the coast of Kota Kinabalu, Sabah, Malaysia, with the description of two new genera and five new species. ZooKeys 872, 127 (2019).
Breedy, O. & Guzman, H. A new species of Leptogorgia (Cnidaria: Anthozoa: Octocorallia) from Golfo Dulce, Pacific Costa Rica. Zootaxa 3182(1), 65–68 (2012).
Breedy, O. & Guzman, H. A new species of alcyonacean octocoral from the Galápagos Archipelago. J. Mar. Biol. Assoc. U.K. 85, 801–807 (2005).
Arantes, R. C. & Loiola, L. L. New records of Primnoidae (Cnidaria: Octocorallia) in Brazilian deep waters. Deep Sea Res. Part II Top. Stud. Oceanogr 99, 103–112 (2014).
Cerino, N. & Lauretta, D. M. Armadillogorgia albertoi sp. nov.: a new primnoid from the Argentinean deep sea. Zootaxa 3741(3), 369–376 (2013).
Breedy, O., Abeytia, R. & Guzman, H. M. A new species of Leptogorgia (Cnidaria: Anthozoa: Octocorallia) from the Mexican Pacific coast. Bull. Mar. Sci. 88, 319–325 (2012).
Zapata-Guardiola, R. Four new species of Thouarella (Anthozoa: Octocorallia: Primnoidae) from Antarctic waters. Sci. Mar. 74, 131–146 (2010).
Breedy, O. & Guzman, H. M. A new Muricea species (Cnidaria, Anthozoa, Octocorallia) from the eastern tropical Pacific. ZooKeys 629, 1–10 (2016).
Janes, M. P., McFadden, C. S. & Chanmethakul, T. A new species of Ovabunda (Octocorallia, Xeniidae) from the Andaman Sea, Thailand, with notes on the biogeography of this genus. ZooKeys 431, 1 (2014).
Williams, G. C. A new genus of dimorphic soft coral from the south-western fringe of the Indo-Pacific (Octocorallia: Alcyoniidae). J. Zool. 221(1), 21–35 (1990).
Kumar, J. S. Y., Geetha, S., Raghunathan, C. & Sornaraj, R. New record of Melithaea retifera (Lamarck, 1816) from Andaman and Nicobar Island, India. Indian J. Geo-Mar. Sci. 48(10), 1516–1520 (2019).
Breedy, O. & Guzman, H. M. A new alcyonacean species (Cnidaria: Anthozoa: Octocorallia) from a seamount in the tropical Pacific Ocean. Bull. Mar. Sci. 94(4), 1515–1524 (2018).
Soler Hurtado, M. D. M., Machordom, A., Muñoz, J. & López González, P. J. New records of the genera Leptogorgia, Pacifigorgia, and Eugorgia (Octocorallia: Gorgoniidae) from Ecuador, with a description of a new species. Sci. Mar. 80(3), 369–394 (2016).
López González, P. J. & Gili, J. M. Rosgorgia inexspectata, new genus and species of Subergorgiidae (Cnidaria, Octocorallia) from off the Antarctic Peninsula. Polar Biol 24, 122–126 (2001).
López-González, P. J. & Williams, G. C. A new genus and species of sea pen (Octocorallia: Pennatulacea: Stachyptilidae) from the Antarctic Peninsula. Invertebr. Syst. 16(6), 919–929 (2002).
López-González, P. J., Grinyó, J. & Gili, J. M. Rediscovery of Cereopsis studeri Koch, 1891, a forgotten Mediterranean soft coral species, and its inclusion in the genus Nidalia Gray, 1835 (Octocorallia, Alcyonacea, Nidaliidae). Mar. Biol. Res. 8(7), 594–604 (2012).
Zapata-Guardiola, R., López-González, P. J. & Gili, J. M. A review of the genus Mirostenella Bayer, 1988 (Octocorallia: Primnoidae) with a description of a new subgenus and species. Helgol. Mar. Res. 67, 229–240 (2013).
Williams, G. C. Revision of the gorgonian genus Simpsonella (Octocorallia: Chrysogorgiidae) from the western margin of the Indo-Pacific, with the description of a new species from southeastern Africa. Zool. J. Linnean Soc. 105(4), 377–405 (1992).
Breedy, O., Williams, G. C. & Guzman, H. M. Two new species of gorgonian octocorals from the Tropical Eastern Pacific Biogeographic Region (Cnidaria, Anthozoa, Gorgoniidae). ZooKeys 350, 75 (2013).
Guzman, H. M. & Breedy, O. Leptogorgia christiae (Octocorallia: Gorgoniidae) a new shallow water gorgonian from Pacific Panama. J. Mar. Biol. Assoc. U.K. 88(4), 719–722 (2008).
Bayer, F. M. Two new species of the alcyonacean genus Protodendron (Octocorallia: Alcyoniidae) from the Indian Ocean off Natal. Bull. Mar. Sci. 57(2), 301–312 (1995).
Saucier, E. H., Sajjadi, A. U. Z. I. T. A. & France, S. C. A taxonomic review of the genus Acanella (Cnidaria: Octocorallia: Isididae) in the North Atlantic Ocean, with descriptions of two new species. Zootaxa 4323(3), 359–390 (2017).
Castro, C. B. A new species of Heterogorgia Verrill, 1868 (Coelenterata, Octocorallia) from Brazil with comments on the type species of the genus. Bull. Mar. Sci. Miami 47(2), 411–420 (1990).
Tu, T. H., Altuna, A. & Jeng, M. S. Coralliidae (Anthozoa: Octocorallia) from the INDEMARES 2010 expedition to north and northwest Spain (northeast Atlantic), with delimitation of a new species using both morphological and molecular approaches. Zootaxa 3926(3), 301–328 (2015).
Tu, T. H., Dai, C. F. & Jeng, M. S. Taxonomic revision of Coralliidae with descriptions of new species from New Caledonia and the Hawaiian Archipelago. Mar. Biol. Res. 12(10), 1003–1038 (2016).
Bayer, F. M. A new genus of alcyonacean coral (Anthozoa: Octocorallia) from Caribbean waters. Bull. Mar. Sci. 56, 620–624 (1995).
Bayer, F. M. A new gorgonacean octocoral from Jamaica. Bull. Mar. Sci. 23(2), 387–398 (1973).
Zapata-Guardiola, R. & Lopez-Gonzalez, P. J. Two new species of Antarctic gorgonians (Octocorallia: Primnoidae) with a redescription of Thouarella laxa Versluys, 1906. Helgol. Mar. Res. 64, 169–180 (2009).
Olvera, U., Hernández, O., Sánchez, C. & Gómez-Gutiérrez, J. Two new endemic species of Gorgoniidae (Cnidaria, Anthozoa, Octocorallia) from Revillagigedo Archipelago, Mexico. Zootaxa 4442(4), 523–538 (2018).
Viada, S. T. & Cairns, S. D. A new species of Nicella (Anthozoa: Octocorallia) from the western Atlantic. Proc. Biol. Soc. Wash. 120(2), 227–231 (2007).
Breedy, O. & Guzman, H. M. Revision of the genus Adelogorgia Bayer, 1958 (Cnidaria: Anthozoa: Octocorallia) with the description of three new species. Zootaxa 4369(3), 327–348 (2018).
Breedy, O. & Guzman, H. M. A new species of Leptogorgia (Coelenterata: Octocorallia: Gorgoniidae) from the shallow waters of the eastern Pacific. Zootaxa 899(1), 1–11 (2005).
Bayer, F. M. A new species of the gorgonacean genus Narella (Anthozoa: Octocorallia) from Hawaiian waters. Proc. Biol. Soc. Wash. 108, 147–152 (1995).
López González, P. J. A new calcaxonian genus and family for Trichogorgia utinomii Cordeiro, 2019 (Octocorallia, Alcyonacea): new records of a scleriteless gorgonian species from Antarctica. Mar. Biodivers. 50, 1–13 (2020).
Cairns, S. D. & Bayer, F. M. A review of the genus Primnoa (Octocorallia: Gorgonacea: Primnoidae), with the description of two new species. Bull. Mar. Sci. 77(2), 225–256 (2005).
Haussermann, V., Forsterra, G. & Cairns, S. D. New record of the primnoid gorgonian Primnoella delicatissima Kukenthal. 1909 for Chilean waters. Spixiana 39(2), 147–148 (2016).
Núñez-Flores, M., Gomez-Uchida, D. & López-González, P. J. Molecular systematics of Thouarella (Octocorallia: Primnoidae) with the description of three new species from the Southern Ocean based on combined molecular and morphological evidence. Invertebr. Syst. 35(6), 655–674 (2021).
Tu, T. H. & Dai, C. F. A new species of anthothelid octocoral (Cnidaria, Alcyonacea) discovered on an algal reef of Taiwan. ZooKeys 1089, 37 (2021).
López-González, P. J. & Briand, P. A new scleraxonian genus from Josephine Bank, north-eastern Atlantic (Cnidaria, Octocorallia). Hydrobiologia 482, 97–105 (2002).
Verseveldt, J. New species of Sinularia (Octocorallia: Alcyonacea) from Madagascar. Israel J. Zool. 19(3), 165–168 (1970).
Ubare, V., Mohan, P. M., Muriganantham, M. & Nagarjuna, P. New record of Cladiella pachyclados (Klunzinger, 1877) at Port Blair, South Andaman, Andaman and Nicobar Islands, India. Indian J. Geo-Mar. Sci. 44(11), 1787–1789 (2015).
Marqués, A. C. S. J. & Castro, C. B. Muricea (Cnidaria, Octocorallia) from Brazil, with description of a new species. Bull. Mar. Sci. 56(1), 161–172 (1995).
Li, Y., Zhan, Z. & Xu, K. Paragorgia papillata sp. nov., a new bubblegum coral (Octocorallia: Paragorgiidae) from a seamount in the tropical Western Pacific. J. Oceanol. Limnol. 39(5), 1758–1766 (2021).
Williams, G. C. The deep-sea pennatulacean genus Porcupinella – with the description of a new species from Tasmania (Anthozoa, Octocorallia, Chunellidae). ZooKeys 1019, 1 (2021).
Breedy, O., Guzman, H. M., Murillo-Cruz, C. & Vargas, S. Discovery of a new species of Leptogorgia Milne Edwards & Haime, 1857 (Anthozoa: Octocorallia: Gorgoniidae) from eastern tropical Pacific mesophotic reefs. Mar. Biodivers. 51(6), 95 (2021).
Williams, G. C. A new genus and species of enigmatic gorgonian coral from the Ryukyu Archipelago, northwestern Pacific, with a discussion of calcaxonian systematics (Cnidaria, Anthozoa, Octocorallia). Zootaxa 4701(5), 1–11 (2019).
Williams, G. C. A new genus and species of pennatulacean octocoral from equatorial West Africa (Cnidaria, Anthozoa, Virgulariidae). ZooKeys 546, 39 (2015).
López-González, P. J., Gili, J. M. & Fuentes, V. A new species of shallow-water sea pen (Octocorallia: Pennatulacea: Kophobelemnidae) from Antarctica. Polar Biol 32, 907–914 (2009).
Williams, G. C. Four new species of southern African octocorals (Cnidaria: Alcyonacea), with a further diagnostic revision of the genus Alcyonium Linnaeus, 1758. Zool. J. Linnean Soc. 92(1), 1–26 (1988).
Altuna, Á. & López-González, P. J. Description of two new species of bathyal Primnoidae (Octocorallia: Alcyonacea) from the Porcupine Bank (northeastern Atlantic). Zootaxa 4576(1), 1–10 (2019).
Benayahu, Y. & McFadden, C. S. A new genus of soft coral of the family Alcyoniidae (Cnidaria, Octocorallia) with re-description of a new combination and description of a new species. ZooKeys 84, 1–10 (2011).
Halász, A., McFadden, C. S., Aharonovich, D., Toonen, R. & Benayahu, Y. A revision of the octocoral genus Ovabunda Alderslade, 2001 (Anthozoa, Octocorallia, Xeniidae). ZooKeys 373, 1–10 (2014).
Bayer, F. M. The gorgonacean genus Arthrogorgia (Octocorallia: Primnoidae). Proc. Biol. Soc. Wash. 109, 605–628 (1996).
Zapata-Guardiola, R. & López-González, P. J. Revision and redescription of the species previously included in the genus Amphilaphis Studer and Wright in Studer, 1887 (Octocorallia: Primnoidae). Sci. Mar. 76(2), 357–380 (2012).
Tu, T. H., Dai, C. F. & Jeng, M. S. Phylogeny and systematics of deep-sea precious corals (Anthozoa: Octocorallia: Coralliidae). Mol. Phylogenetics Evol. 84, 173–184 (2015).
Breedy, O. & Cortés, J. Morphology and taxonomy of a new species of Leptogorgia (Cnidaria: Octocorallia: Gorgoniidae) in Cocos Island National Park, Pacific Costa Rica. Proc. Biol. Soc. Wash. 124(2), 62–69 (2011).
Cordeiro, R. T., Cairns, S. D. & Perez, C. D. A revision of the genus Radicipes Stearns, 1883 (Anthozoa: Octocorallia: Chrysogorgiidae). Zootaxa 4319(1), 1–26 (2017).
Breedy, O., Camps-Castellà, J., Försterra, G. & Häussermann, V. Taxonomic status and geographic distribution of Phycogorgia fucata (Valenciennes 1846) (Octocorallia: Gorgoniidae). Mar. Biol. Res. 17(7-8), 625–633 (2021).
Cairns, S. D., Cordeiro, R. T., Xu, Y., Zhan, Z. & Alderslade, P. A new family and two new genera of calcaxonian octocoral, including a redescription of Pleurogorgia militaris (Cnidaria: Octocorallia: Chrysogorgiidae) and its placement in a new genus. Invertebr. Syst. 35(3), 282–297 (2021).
Opresko, D. M. Abundance and distribution of shallow-water gorgonians in the area of Miami, Florida. Bull. Mar. Sci. 23(3), 535–558 (1973).
Borgstein, N., Beltrán, D. M. & Prada, C. Variable growth across species and life stages in Caribbean reef octocorals. Front. Mar. Sci. 7, 483 (2020).
Benayahu, Y. & Perkol-Finkel, S. Soft corals (Octocorallia: Alcyonacea) from southern Taiwan. I. Sarcophyton nanwanensis sp. nov. (Octocorallia: Alcyonacea). Zool. Stud. 43(3), 537–547 (2004).
Jahajeeah, D., Bhoyroo, V. & Ranghoo-Sanmukhiya, M. An assessment of soft coral community (Octocorallia; Alcyonacea) around Mauritius and Rodrigues Islands — New records of soft corals. Reg. Stud. Mar. Sci. 47, 101976 (2021).
Breedy, O. & Guzman, H. M. A revision of the genus Heterogorgia Verrill, 1868 (Anthozoa: Octocorallia: Plexauridae). Zootaxa 2995(1), 27–44 (2011).
Breedy, O. & Guzman, H. M. A revision of the genus Psammogorgia Verrill, 1868 (Cnidaria, Anthozoa, Octocorallia) in the tropical eastern Pacific Ocean. ZooKeys 961, 1 (2020).
Breedy, O. & Guzman, H. M. A new species of alcyonacean octocoral from the Peruvian zoogeographic region. J. Mar. Biol. Assoc. UK 94(3), 493–498 (2014).
Xu, Y., Li, Y., Zhan, Z. & Xu, K. Morphology and phylogenetic analysis of two new deep-sea species of Chrysogorgia (Cnidaria, Octocorallia, Chrysogorgiidae) from Kocebu Guyot (Magellan seamounts) in the Pacific Ocean. ZooKeys 881, 91 (2019).
Cairns, S. D. & Taylor, M. L. An illustrated key to the species of the genus Narella (Cnidaria, Octocorallia, Primnoidae). ZooKeys 822, 1 (2019).
Sánchez, J. A. Systematics of the candelabrum gorgonian corals (Eunicea Lamouroux; Plexauridae; Octocorallia; Cnidaria). Zool. J. Linn. Soc. 157(2), 237–263 (2009).
Benayahu, Y. Notes on some tropical octocorals at the zoological museum, university of Copenhagen, Denmark (Cnidaria: Octocorallia). Raffles Bull. Zool. 61(1), 7–11 (2013).
Cairns, S. D. & Wirshing, H. H. Phylogenetic reconstruction of scleraxonian octocorals supports the resurrection of the family Spongiodermidae (Cnidaria, Alcyonacea). Invertebr. System. 29(4), 345–368 (2015).
Goh, N. K. C. & Chow, L. M. An annotated checklist of the gorgonians (Anthozoa: Octocorallia) of Singapore, with a discussion of gorgonian diversity in the Indo-West Pacific. Raffles Bull. Zool. 44(2), 435–459 (2013).
Breedy, O. & Guzman, H. M. A revision of the genus Pacifigorgia (Coelenterata: Octocorallia: Gorgoniidae). Proc. Biol. Soc. Wash. 115, 782–839 (2002).
Bayer, F. M. Mirostenella articulata, A remarkable new genus and species of primnoid octocoral (Gorgonacea) with uncalcified axial nodes. Proc. Biol. Soc. Wash 101, 251–256 (1988).
Sánchez, J. A. A new genus of Atlantic octocorals (Octocorallia: Gorgoniidae): Systematics of gorgoniids with asymmetric sclerites. J. Nat. Hist. 41(9-12), 493–509 (2007).
Cairns, S. D. & Wirshing, H. H. A phylogenetic analysis of the Primnoidae (Anthozoa: Octocorallia: Calcaxonia) with analyses of character evolution and a key to the genera and subgenera. BMC Evol. Biol. 18, 1–20 (2018).
García-Cárdenas, F. J., Drewery, J. & López-González, P. J. Resurrection of the sea pen genus Ptilella Gray, 1870 and description of Ptilella grayi n. sp. from the NE Atlantic (Octocorallia: Pennatulacea). Sci. Mar. 83(3), 261–276 (2019).
Bayer, F. M. & Stefani, J. Isididae (Gorgonacea) de Nouvelle-Caledonie–Nouvelle cle des genres de la famille. Bull. Mus. natl. hist. nat. Paris 4(9 A)(1), 47–106 (1987).
Williams, G. C. The aberrant and monotypic soft coral genus Malacacanthus Thomson, 1910 (Octocorallia: Alcyoniidae) endemic to southern Africa. J. Nat. Hist. 21(6), 1337–1346 (2007).
Cordeiro, R. T., Castro, C. B. & Perez, C. D. Deep-water octocorals (Cnidaria: Octocorallia) from Brazil: Family Chrysogorgiidae Verrill, 1883. Zootaxa 4058(1), 81–100 (2015).
Moore, K., Alderslade, P. & Miller, K. A taxonomic revision of the genus Primnoisis Studer [& Wright], 1887 (Coelenterata: Octocorallia: Isididae) using morphological and molecular data. Zootaxa 4075(1), 1–141 (2016).
Li, Y., Zhan, Z. & Xu, K. Establishment of Alloptilella splendida gen. et sp. nov. and resurrection of Scytalium veneris (Thomson & Henderson, 1906), two sea pens (Cnidaria: Pennatulacea) from seamounts in the tropical Western Pacific. J. Oceanol. Limnol. 39(5), 1790–1804 (2021).
Saucier, E. H., France, S. C. & Watling, L. Toward a revision of the bamboo corals: Part 3, deconstructing the family Isididae. Zootaxa 5047(3), 247–272 (2021).
Kushida, Y. & Reimer, J. D. Molecular phylogeny and diversity of sea pens (Cnidaria: Octocorallia: Pennatulacea) with a focus on shallow water species of the northwestern Pacific Ocean. Mol. Phylogenet. Evol. 131, 233–244 (2019).
García-Cárdenas, F. J., Núñez-Flores, M. & López-González, P. J. Molecular phylogeny and divergence time estimates in pennatulaceans (Cnidaria: Octocorallia: Pennatulacea). Sci. Mar. 84(4), 317–330 (2020).
Bayer, F. M. The identity of Fannyella rossii Gray, J. E. (Coelenterata, Octocorallia). Proc. Biol. Soc. Wash. 103, 773–783 (1990).
Williams, G. C. & Chen, J. Y. Resurrection of the octocorallian genus Antillogorgia for Caribbean species previously assigned to Pseudopterogorgia, and a taxonomic assessment of the relationship of these genera with Leptogorgia (Cnidaria, Anthozoa, Gorgoniidae). Zootaxa 3505(1), 39–52 (2012).
Williams, G. C. Living genera of sea pens (Coelenterata: Octocorallia: Pennatulacea): illustrated key and synopses. Zool. J. Linn. Soc. 113(2), 93–140 (1995).
Benayahu, Y., Ekins, M., van Ofwegen, L. P., Samimi-Namin, K. & McFadden, C. S. On some encrusting Xeniidae (Octocorallia): Re-examination of the type material of Sansibia flava (May, 1898) and a description of new taxa. Zootaxa, 5093(4) (2022).
Poliseno, A. et al. An integrated morphological–molecular approach reveals new insights on the systematics of the octocoral Telestula humilis (Thomson, 1927) (Octocorallia: Alcyonacea: Clavulariidae). Invertebr. Syst. 35(3), 261–281 (2021).
Li, Y., Zhan, Z. & Xu, K. Morphology and molecular phylogeny of Paragorgia rubra sp. nov. (Cnidaria: Octocorallia), a new bubblegum coral species from a seamount in the tropical Western Pacific. Chin. J. Oceanol. Limnol. 35(4), 803–814 (2017).
McFadden, C. S., Alderslade, P., van Ofwegen, L. P., Johnsen, H. & Rusmevichientong, A. Phylogenetic relationships within the tropical soft coral genera Sarcophyton and Lobophytum (Anthozoa, Octocorallia). Invertebr. Biol. 125(4), 288–305 (2006).
Benayahu, Y., McFadden, C. S. & Shoham, E. Search for mesophotic octocorals (Cnidaria, Anthozoa) and their phylogeny: I. A new sclerite-free genus from Eilat, northern Red Sea. ZooKeys 680, 1 (2017).
Watling, L. A review of the genus Iridogorgia (Octocorallia: Chrysogorgiidae) and its relatives, chiefly from the North Atlantic Ocean. J. Mar. Biol. Assoc. U.K. 87(2), 393–402 (2007).
Pante, E., Saucier, E. H. & France, S. C. Molecular and morphological data support reclassification of the octocoral genus. Isidoides. Invertebr. Syst. 27(4), 365–378 (2013).
Taylor, M. L., Cairns, S. D., Agnew, D. J. & Rogers, A. D. A revision of the genus Thouarella Gray, 1870 (Octocorallia: Primnoidae), including an illustrated dichotomous key, a new species description, and comments on Plumarella Gray, 1870 and Dasystenella, Versluys, 1906. Zootaxa 3602(1), 1–105 (2013).
Li, Y., Zhan, Z. & Xu, K. Morphology and Molecular Phylogenetic Analysis of Deep-Sea Purple Gorgonians (Octocorallia: Victorgorgiidae) from Seamounts in the Tropical Western Pacific, with Description of Three New Species. Front. Mar. Sci. 7, 701 (2020).
Williams, G. C. A comparison of the stoloniferous octocorallian genera Scyphopodium, Stereotelesto, Bathytelesto and Rhodelinda, with the description of a new species from southeastern Africa (Anthozoa, Chavulariidae). J. Zool. 219(4), 621–635 (1989).
Chen, C. C. & Chang, K.-H. Gorgonacea (Coelenterata: Anthozoa: Octocorallia) of southern Taiwan. Bull. Inst. Zool. Acad. Sin. 30(3), 149–182 (1991).
Xu, Y., Zhan, Z. & Xu, K. Morphology and molecular phylogeny of three new deep-sea species of Chrysogorgia (Cnidaria, Octocorallia) from seamounts in the tropical Western Pacific Ocean. PeerJ 8, e8832 (2020).
Verseveldt, J. A. Octocorallia) from the Red Sea, with a discussion of a new Sinularia species from Ceylon. J. Ecol. Evol. 23(1), 1–37 (2013).
d’Hondt, M. J. Some Octocorallia (Coelenterata) from Australia. Mar. Freshwater Res. 28(2), 241–259 (1977).
Taylor, M. L. & Rogers, A. D. Primnoidae (Cnidaria: Octocorallia) of the SW Indian Ocean: new species, genus revisions and systematics. Zool. J. Linn. Soc. 181(1), 70–97 (2017).
Breedy, O., Cairns, S. D. & Haeussermann, V. A new alcyonacean octocoral (Cnidaria, Anthozoa, Octocorallia) from Chilean fjords. Zootaxa 3919(2), 327–334 (2015).
d’Hondt, M. J. & Tixier-Durivault, A. Clavularia steveninoae n. sp. new Octocorallia stolonifera from Mediterranean Sea. Cah. Biol. Mar. 26, 585–592 (1975).
Baker, K. D. et al. Distributional patterns of deep-sea coral assemblages in three submarine canyons off Newfoundland, Canada. Mar. Ecol. Prog. Ser. 445, 235–249 (2012).
Kumar, J. S. Y., Geetha, S., Raghunathan, C. & Sornaraj, R. New report of Melithaea delicata Hickson, 1905 (subclass: Octocorallia) from Little Andaman Island, India. Indian J. Geo Mar. Sci. 48(9), 1344–1350 (2019).
Cairns, S. D. Primnoidae (Cnidaria: Octocorallia: Calcaxonia) of the Okeanos Explorer expeditions (CAPSTONE) to the central Pacific. Zootaxa 4532(1), 1–43 (2018).
Cairns, S. D. & Bayer, F. M. A generic revision and phylogenetic analysis of the Primnoidae (Cnidaria: Octocorallia). Smithson. Contr. Zool. 629, 1–79 (2009).
Prada, C., Schizas, N. V. & Yoshioka, P. M. Phenotypic plasticity or speciation? A case from a clonal marine organism. BMC Evol. Biol. 8, 47 (2008).
Weinberg, S. Transplantation experiments with Mediterranean gorgonians. Bijdr. Dierk. 49(1), 31–41 (1979).
Terribile, K., Evans, J., Knittweis, L. & Schembri, P. J. Maximising MEDITS: Using data collected from trawl surveys to characterise the benthic and demersal assemblages of the circalittoral and deeper waters around the Maltese Islands (Central Mediterranean). Reg. Stud. Mar. Sci. 3, 163–175 (2016).
Mastrototaro, F. et al. A facies of Kophobelemnon (Cnidaria, Octocorallia) from Santa Maria di Leuca coral province (Mediterranean Sea). Mar. Ecol. 34(3), 313–320 (2013).
Buhl-Mortensen, P. & Buhl-Mortensen, L. Diverse and vulnerable deep-water biotopes in the Hardangerfjord. Mar. Biol. Res. 10(3), 253–267 (2013).
Bo, M. et al. Deep Coral Oases in the South Tyrrhenian Sea. PLoS One 7(11), e49870 (2012).
López González, P. J. & Gili, J. M. Two new dimorphic soft-coral species (Anthozoa: Octocorallia) from Antarctica. Hydrobiologia 544, 143–153 (2005).
Cúrdia, J. et al. Spatial and depth-associated distribution patterns of shallow gorgonians in the Algarve coast (Portugal, NE Atlantic). Helgol. Mar. Res. 67, 521–534 (2012).
Shoham, E. & Benayahu, Y. Higher species richness of octocorals in the upper mesophotic zone in Eilat (Gulf of Aqaba) compared to shallower reef zones. Coral Reefs 36(1), 71–81 (2017).
Mejía-Quintero, K. & Chasqui, L. Octocorals and Antipatharians in the Mesophotic Rocky Reefs of Colombian Pacific (Eastern Tropical Pacific). Front. Mar. Sci. 7, 311 (2020).
Beyayahu, Y. et al. Coral Reefs of the World. in Mesophotic Coral Ecosystems Vol. 12 (eds. Loya, Y., Puglise, K. & Bridge, T.) Ch. 38 (Springer, Cham, 2019).
Xu, Y., Zhan, Z. & Xu, K. Morphology and phylogeny of Chrysogorgia pinniformis sp. nov. and C. varians sp. nov., two golden corals from the Caroline seamounts in the tropical Western Pacific Ocean. J. Oceanol. Limnol. 39(5), 1767–1789 (2021).
Kushida, Y. & Reimer, J. D. Description of the sea pen Calibelemnon hinoenma sp. nov. from shallow waters in southern Japan. Mar. Biodivers. 50(6), 107 (2020).
Tu, T. H., Dai, C. F. & Jen, M. S. Precious corals (Octocorallia: Coralliidae) from the northern West Pacific region with descriptions of two new species. Zootaxa 3395(1), 1–17 (2012).
Cordeiro, R. T. S., McFadden, C. S., Sanchez, J. A. & Pérez, C. D. Revision of the genus Plexaurella Kölliker, 1865 (Anthozoa: Octocorallia) and resurrection of Plexaurellidae Verrill, 1912 new rank. Invert. Syst. 35(8), 892–921 (2021).
Drake, J. et al. Sclerites of the soft coral Ovabunda macrospiculata (Xeniidae) are predominantly the metastable CaCO3 polymorph vaterite. UCLA (2021).
López-González, P. J. & Cunha, M. R. Two new species of Dendrobrachia Brook, 1889 (Cnidaria: Octocorallia: Dendrobrachiidae) from the north-eastern Atlantic and western Mediterranean. Sci. Mar. 74(3), 423–434 (2010).
Tursch, B. & Tursch, A. The soft coral community on a sheltered reef quadrat at Laing Island (Papua New Guinea). Mar. Biol. 68, 321–332 (1982).
Dautova, T. N. Deep-water Octocorallia (Cnidaria: Anthozoa) of the temperate Northern Pacific: Notes on the distribution and new bathyal-abyssal taxa from the Sea of Okhotsk. Deep Sea Res. II: Topical Stud. Oceanogr 154, 74–86 (2018).
Bayer, F. M. The helioporacean octocoral Epiphaxum: Recent and fossil. A monographic iconography. Stud. Trop. Oceanogr. 15, 1–8 (1992).
Gómez, C. E. et al. Responses of the tropical gorgonian coral Eunicea fusca to ocean acidification conditions. Coral Reefs 34, 451–460 (2014).
Cocito, S. et al. Nutrient acquisition in four Mediterranean gorgonian species. Mar. Ecol. Prog. Ser. 473, 179–188 (2013).
Ribes, M., Coma, R. & Gili, J. M. Heterogeneous feeding in benthic suspension feeders: The natural diet and grazing rate of the temperate gorgonian Paramuricea clavata (Cnidaria: Octocorallia) over a year cycle. Mar. Ecol. Prog. Ser. 183, 125–137 (1999).
Lewis, J. C. Feeding behaviour and feeding ecology of the Octocorallia (Coelenterata: Anthozoa). J. Zool. 196(3), 371–384 (1982).
Lasker, H. R. A comparison of the particulate feeding abilities of three species of gorgonian soft coral. Mar. Ecol. Prog. Ser. 5, 61–67 (1981).
Acuña, F. H., Excoffon, A. C., Zamponi, M. O. & Genzano, G. Feeding habits of the temperate octocoral Tripalea clavaria (Studer, 1878) (Octocorallia, Gorgonaria, Anthothelidae) from sublittoral outcrops off Mar del Plata, Argentina. Belg. J. Zool. 134(1), 65–66 (2004).
Farrant, P. A., Borowitzka, M. A., Hinde, R. & King, R. J. Nutrition of the temperate Australian soft coral Capnella gaboensis. Mar. Biol. 95, 575–581 (1987).
Coma, R., Gili, J. M., Zabala, M. & Riera, T. Feeding and prey capture cycles in the aposymbiontic gorgonian Paramuricea clavata. Mar. Ecol. Prog. Ser. 115, 257–270 (1994).
Leal, M. C. et al. Coral feeding on microalgae assessed with molecular trophic markers. Mol. Ecol. 23(15), 3870–3876 (2014).
Coma, R., Llorente-Llurba, E., Serrano, E., Gili, J. M. & Ribes, M. Natural heterotrophic feeding by a temperate octocoral with symbiotic zooxanthellae: A contribution to understanding the mechanisms of die-off events. Coral Reefs 34, 549–560 (2015).
Slattery, M. & McClintock, J. B. Population structure and feeding deterrence in three shallow-water Antarctic soft corals. Mar. Biol. 122, 461–470 (1995).
Orejas, C., Gili, J., López-González, P. J. & Arntz, W. Feeding strategies and diet composition of four Antarctic cnidarian species. Polar Biol. 24, 620–627 (2001).
Lin, M., Liao, C. & Dai, C. Modeling the effects of satiation on the feeding rate of a colonial suspension feeder, Acanthogorgia vegae, in a circulating system under lab conditions. Zool. Stud. 41(4), 355–365 (2002).
Ribes, M., Coma, R. & Gili, J. M. Heterotrophic feeding by gorgonian corals with symbiotic zooxanthellae. Limnol. Oceanogr. 43(6), 1170–1179 (1998).
Lin, M. & Dai, C. The effects of flow on feeding of three gorgonians from southern Taiwan. J. Exp. Mar. Biol. Ecol. 173(1), 57–69 (1993).
Sorokin, Y. Biomass, metabolic rates and feeding of some common reef zoantharians and octocorals. Mar. Freshwater Res. 42(6), 729–741 (1991).
Ribes, M., Coma, R. & Rossi, S. Natural feeding of the temperate asymbiotic octocoral-gorgonian Leptogorgia sarmentosa (Cnidaria: Octocorallia). Mar. Ecol. Prog. Ser. 254, 141–150 (2003).
Lasker, H. R., Gottfried, M. D. & Coffroth, M. A. Effects of depth on the feeding capabilities of two octocorals. Mar. Biol. 73, 73–78 (1983).
Best, B. A. Passive suspension feeding in a sea pen: Effects of ambient flow on volume flow rate and filtering efficiency. Biol. Bull. 175(3), 332–342 (1988).
Fabricius, K. E., Benayahu, Y. & Genin, A. Herbivory in asymbiotic soft corals. Science 268(5207), 90–92 (1995).
Grossowicz, M. & Benayahu, Y. Occurrence and survivorship of azooxanthellate octocorals reflect recruitment preferences and depth distribution. Hydrobiologia 759, 85–93 (2015).
Weinberg, S. Autecology of shallow-water Octocorallia from Mediterranean rocky substrata, I. The Banyuls area. Bijdragen tot de Dierkunde 49(1), 1–15 (1979).
Chimienti, G. Vulnerable forests of the pink sea fan Eunicella verrucosa in the Mediterranean Sea. Diversity 12(5), 176 (2020).
Topçu, N. E. & Öztürk, B. First insights into the demography of the rare gorgonian Spinimuricea klavereni in the Mediterranean Sea. Mar. Ecol. 37(5), 1154–1160 (2016).
Piccinetti, C. C. et al. Herbivory in the soft coral Sinularia flexibilis (Alcyoniidae). Sci. Rep. 6(1), 22679 (2016).
Roushdy, H. M. & Hansen, V. K. Filtration of phytoplankton by the octocoral Alcyonium digitatum L. Nature 190(4776), 649–650 (1961).
Lapointe, A. & Watling, L. Bamboo corals from the abyssal Pacific: Bathygorgia. Proc. Biol. Soc. Wash. 128(2), 125–136 (2015).
Reimer, J. D. et al. Unexpected high abundance of aragonite-forming Nanipora (Octocorallia: Helioporacea) at an acidified volcanic reef in southern Japan. Mar. Biodivers. 51(1), 19 (2021).
Sánchez, J. A. et al. Corals in the mesophotic zone (40–115 m) at the Barrier Reef Complex from San Andrés Island (Southwestern Caribbean). Front. Mar. Sci. 6, 536 (2019).
Sanchez-Tocino, L. et al. Pruning treatment: A possible method for improving the conservation status of Ellisella paraplexauroides Stiasny, 1936 (Anthozoa, Alcyonacea) population in the Chafarinas Islands. Medit. Mar. Sci. 18(3), 385–392 (2017).
Giusti, M. et al. Habitat preference of Viminella flagellum (Alcyonacea: Ellisellidae) in relation to bathymetric variables in southeastern Sardinian waters. Cont. Shelf Res. 138, 41–50 (2017).
Araya, J. F., Araya, M. E., Mack, M. & Aliaga, J. A. On the presence of Distichoptilum gracile Verrill, 1882 (Octocorallia: Pennatulacea), in the southeastern Pacific. Mar. Biodivers. 48, 1637–1641 (2018).
Cúrdia, J. et al. Diversity and abundance of invertebrate epifaunal assemblages associated with gorgonians are driven by colony attributes. Coral Reefs 34, 611–624 (2015).
Brazeau, D. A. & Harvell, C. D. Genetic structure of local populations and divergence between growth forms in a clonal invertebrate, the Caribbean octocoral Briareum asbestinum. Mar. Biol. 119, 53–60 (1994).
Toledo-Hernández, C. et al. The role of sclerites in the defense against pathogens of the sea fan Gorgonia ventalina (Octocorallia). J. Exp. Mar. Biol. Ecol. 483, 20–24 (2016).
Rezaei, Z., Movahedinia, A., Salamat, N. & Ranjbar, M. S. Details on the histomorphological characteristics of marine sea pen, Virgularia gustaviana, from the Persian Gulf. Turk. J. Vet. Anim. Sci. 45(4), 735–745 (2021).
Linares, C., Doak, D. F., Coma, R., Díaz, D. & Zabala, M. Life history and viability of a long-lived marine invertebrate: the octocoral Paramuricea clavata. Ecology 88(4), 918–928 (2007).
Ambroso, S. et al. Pristine populations of habitat-forming gorgonian species on the Antarctic continental shelf. Sci. Rep. 7(1), 12251 (2017).
Chimienti, G., Aguilar, R., Maiorca, M. & Mastrototaro, F. A newly discovered forest of the whip coral Viminella flagellum (Anthozoa, Alcyonacea) in the Mediterranean Sea: A non-invasive method to assess its population structure. Biology 11(1), 39 (2021).
Morato, T. et al. Dense cold-water coral garden of Paragorgia johnsoni suggests the importance of the Mid-Atlantic Ridge for deep-sea biodiversity. Ecol. Evol. 11(23), 16426–16433 (2021).
Palma, M. et al. SfM-based method to assess gorgonian forests (Paramuricea clavata (Cnidaria, Octocorallia). Remote Sens 10(7), 1154 (2018).
Rowley, S. J., Pochon, X. & Watling, L. Environmental influences on the Indo–Pacific octocoral Isis hippuris Linnaeus 1758 (Alcyonacea: Isididae): Genetic fixation or phenotypic plasticity? PeerJ 3, e1128 (2015).
Sánchez, J. A. & Lasker, H. R. Patterns of morphological integration in marine modular organisms: supra-module organization in branching octocoral colonies. Proc. R. Soc. Lond. B Biol. Sci. 270(1528), 2039–2044 (2003).
Keith, D. E. Shallow-water gorgonians (Octocorallia) of Roatan, Honduras. Bull. Mar. Sci. 50(1), 212–226 (1992).
Grinyó, J. et al. Diversity, distribution and population size structure of deep Mediterranean gorgonian assemblages (Menorca Channel, Western Mediterranean Sea). Prog. Oceanogr. 145, 42–56 (2016).
Wells, C. D. & Tonra, K. J. Polyp bailout and reattachment of the abundant Caribbean octocoral Eunicea flexuosa. Coral Reefs 40(1), 27–30 (2021).
Pastorino, G. & Ituarte, C. Rhodelinda gardineri (Gohar, 1940) (Coelenterata: Anthozoa) a stoloniferous octocoral from Patagonian shallow waters. Bull. Mar. Sci. 58(1), 196–202 (1992).
Matsumoto, A. K., Iwase, F., Imahara, Y. & Namikawa, H. Bathymetric distribution and biodiversity of cold-water octocorals (Coelenterata: Octocorallia) in Sagami Bay and adjacent waters of Japan. Bull. Mar. Sci. 81, 231–251 (2007).
Lasker, H. R., Kiho, K. & Coffroth, M. A. Reproductive and genetic variation among Caribbean gorgonians: the differentiation of Plexaura kuna, new species. Bull. Mar. Sci. 58(1), 277–288 (1996).
Lira, A. K. et al. Trophic ecology of the octocoral Carijoa riisei from littoral of Pernambuco, Brazil. I. Composition and spatio-temporal variation of the diet. J. Mar. Biol. Assoc. U.K. 89(1), 89–99 (2009).
Williams, G. C. Biogeography of the octocorallian coelenterate fauna of southern Africa. Biol. J. Linn. Soc. 46(4), 351–401 (2008).
Chimienti, G., Tursi, A. & Mastrototaro, F. Biometric relationships in the red sea pen Pennatula rubra (Cnidaria: Pennatulacea). In 2018 IEEE International Workshop on Metrology for the Sea; Learning to Measure Sea Health Parameters (MetroSea) (pp. 212-216). IEEE (2018, October).
Pierdomenico, M. et al. Effects of trawling activity on the bamboo-coral Isidella elongata and the sea pen Funiculina quadrangularis along the Gioia Canyon (Western Mediterranean, southern Tyrrhenian Sea). Prog. Oceanogr. 169, 214–226 (2018).
Migné, A., Davoult, D., Janquin, M. A. & Kupka, A. Biometrics, carbon and nitrogen content in two cnidarians: Urticina felina and Alcyonium digitatum. J. Mar. Biol. Assoc. U.K. 76(3), 595–602 (1996).
Grigg, R. W. Orientation and growth form of sea fans. Limnol. Oceanogr. 17(2), 185–192 (1972).
Fabricius, K., Yahel, G. & Genin, A. In situ depletion of phytoplankton by an azooxanthellate soft coral. Limnol. Oceanogr. 43(2), 354–356 (2003).
Tsounis, G., Rossi, S., Gili, J. M. & Arntz, W. Population structure of an exploited benthic cnidarian: the case study of red coral (Corallium rubrum L.). Mar. Biol. 149, 1059–1070 (2006).
Carlier, A. et al. Trophic relationships in a deep Mediterranean cold-water coral bank (Santa Maria di Leuca, Ionian Sea). Mar. Ecol. Prog. Ser. 397, 125–137 (2009).
Rahman, M. A. & Oomori, T. Structure, crystallization and mineral composition of sclerites in the alcyonarian coral. J. Cryst. Growth 310(15), 3528–3534 (2008).
Carugati, L. et al. Deep-dwelling populations of Mediterranean Corallium rubrum and Eunicella cavolini: Distribution, demography, and co-occurrence. Biology 11(2), 333 (2022).
Linares, C. et al. Immediate and delayed effects of a mass mortality event on gorgonian population dynamics and benthic community structure in the NW Mediterranean Sea. Mar. Ecol. Prog. Ser. 305, 127–137 (2005).
Rowley, S. J. Acclimatory capacity of the gorgonian Isis hippuris Linnaeus, 1758 to environmental change in SE Sulawesi, Indonesia. J. Exp. Mar. Biol. Ecol. 500, 73–88 (2018).
Lasker, H. R. Zooxanthella densities within a Caribbean octocoral during bleaching and non-bleaching years. Coral Reefs 22(1), 23–26 (2003).
Linares, C., Coma, R., Garrabou, J., Díaz, D. & Zabala, M. Size distribution, density and disturbance in two Mediterranean gorgonians: Paramuricea clavata and Eunicella singularis. J. Appl. Ecol. 45(2), 688–699 (2008).
Kipson, S. et al. Population structure and conservation status of the red gorgonian Paramuricea clavata (Risso, 1826) in the Eastern Adriatic Sea. Mar. Ecol. 36(4), 982–993 (2014).
Betti, F., Bo, M., Bava, S., Faimali, M. & Bavestrello, G. Shallow-water sea fans: the exceptional assemblage of Leptogorgia sarmentosa (Anthozoa: Gorgoniidae) in the Genoa harbour (Ligurian Sea). Eur. Zool. J. 85(1), 290–298 (2018).
Edmunds, P. J. High ecological resilience of the sea fan Gorgonia ventalina during two severe hurricanes. PeerJ 8, e10315 (2020).
Lacharité, M. & Metaxas, A. Early life history of deep-water gorgonian corals may limit their abundance. PLoS One 8(6), e65394 (2013).
Grinyó, J. et al. Morphological features of the gorgonian Paramuricea macrospina on the continental shelf and shelf edge (Menorca Channel, Western Mediterranean Sea). Mar. Biol. Res. 14(1), 30–40 (2017).
Davis, T. R., Harasti, D. & Smith, S. D. Extension of Dendronephthya australis soft corals in tidal current flows. Mar. Biol. 162, 2155–2159 (2015).
Velimirov, B. & King, J. Calcium uptake and net calcification rates in the octocoral Eunicella papillosa. Mar. Biol. 50, 349–358 (1979).
Velimirov, B. & Böhm, E. L. Calcium and magnesium carbonate concentrations in different growth regions of gorgonians. Mar. Biol. 35, 269–275 (1976).
Couillard, C. M., Sainte‐Marie, B. & Dionne, H. Late maturity and evidence for female biennial spawning in the sea pen Pennatula aculeata (Anthozoa, Pennatulacea) in eastern Canada. Invertebr. Biol. 140(3), 259–267 (2021).
Tsounis, G., Edmunds, P. J., Bramanti, L., Gambrel, B. & Lasker, H. R. Variability of size structure and species composition in Caribbean octocoral communities under contrasting environmental conditions. Mar. Biol. 165(3), 58 (2018).
Lewis, J. C., Barnowski, T. F. & Telesnicki, G. J. Characteristics of carbonates of gorgonian axes (Coelenterata, Octocorallia). Biol. Bull. 183(2), 207–220 (1992).
de Moura Neves, B., Edinger, E. & Wareham Hayes, V. Morphology and composition of the internal axis in two morphologically contrasting deep-water sea pens (Cnidaria: Octocorallia). J. Nat. Hist. 51(37-38), 2241–2259 (2017).
Ambroso, S. et al. Spatial distribution patterns of the soft corals Alcyonium acaule and Alcyonium palmatum in coastal bottoms (Cap de Creus, northwestern Mediterranean Sea). Mar. Biol. 160(7), 1567–1580 (2013).
Boller, M. L., Swain, T. D. & Lasker, H. R. Skeletal morphology and material properties of a fragmenting gorgonian coral. Mar. Ecol. Prog. Ser. 228, 131–139 (2002).
Doughty, C. L., Quattrini, A. M. & Cordes, E. E. Insights into the population dynamics of the deep-sea coral genus Paramuricea in the Gulf of Mexico. Deep Sea Res. Part II: Top. Stud. Oceanogr 99, 98–106 (2014).
Weinbauer, M. G. & Vellmirov, B. Calcium, magnesium and strontium concentrations in the calcite sclerites of Mediterranean gorgonians (Coelenterata: Octocorallia). Estuar. Coast. Shelf Sci. 64(4), 635–641 (2005).
Taubner, I., Böhm, F., Eisenhauer, A., Garbe-Schönberg, D. & Erez, J. Uptake of alkaline earth metals in Alcyonarian spicules (Octocorallia). Geochim. Cosmochim. Acta 83, 187–197 (2012).
Bayer, F. M. The mineral component of the axis and holdfast of some gorgonacean octocorals (Coelenterata: Anthozoa), with special reference to the family Gorgoniidae. Proc. Biol. Soc. Wash. 114(1), 66–77 (2001).
Taninaka, H. et al. Phylogeography of blue corals (Genus Heliopora) across the Indo-West Pacific. Front. Mar. Sci. 8, 714662 (2021).
Allemand, D. & Bénazet‐Tambutté, S. Dynamics of calcification in the Mediterranean red coral, Corallium rubrum (Linnaeus) (Cnidaria, Octocorallia). J. Exp. Zool. 276(4), 270–279 (1996).
Rodriguez-Lanetti, M., Marquez, L. M. & Losada, F. Changes in gorgonian morphology along a depth gradient at Isla Alcatraz, San Esteban National Park, Venezuela. Bull. Mar. Sci. 73(2), 257–270 (2003).
Gori, A. et al. Size and spatial structure in deep versus shallow populations of the Mediterranean gorgonian Eunicella singularis (Cap de Creus, northwestern Mediterranean Sea). J. Exp. Zool. 316(5), 320–330 (2011).
Castro, C. B., Medeiros, M. S. & Loiola, L. L. Octocorallia (Cnidaria: Anthozoa) from Brazilian reefs. J. Nat. Hist. 44(13), 817–830 (2010).
West, J. M., Harvell, C. D. & Walls, A. M. Morphological plasticity in a gorgonian coral (Briareum asbestinum) over a depth cline. Mar. Ecol. Prog. Ser. 94, 61–67 (1993).
Pica, D., Calcinai, B., Poliseno, A., Trainito, E. & Cerrano, C. Distribution and phenotypic variability of the Mediterranean gorgonian Paramuricea macrospina (Cnidaria: Octocorallia). Eur. Zool. J. 85(1), 37–44 (2018).
Bastari, A., Pica, D., Ferretti, F., Micheli, F. & Cerrano, C. Sea pens in the Mediterranean Sea: habitat suitability and opportunities for ecosystem recovery. ICES J. Mar. Sci. 75(5), 1881–1892 (2018).
Quintanilla, E., Madurell, T., Wilke, T. & Sánchez, J. A. Dynamic interplay of ENSO events and local hydrodynamic parameters drives demography and health status of gorgonian sea fan populations on a remote tropical eastern Pacific island. Front. Mar. Sci. 6, 694 (2019).
Breedy, O. & Cortés, J. Shallow water gorgonians (Octocorallia: Gorgoniidae) from the North Pacific of Costa Rica. Rev. Biol. Trop. 62(4), 20032 (2015).
Bastidas, C., Fabricius, K. E. & Willis, B. L. Demographic aspects of the soft coral Sinularia flexibilis leading to local dominance on coral reefs. In Coelenterate biology 2003: trends in research on Cnidaria and Ctenophora (pp. 433-441). Springer Netherlands (2004).
Namin, K. S. & Van Ofwegen, L. P. Some shallow water octocorals (Coelenterata: Anthozoa) of the Persian Gulf. Zootaxa 2058(1), 1–52 (2009).
Grillo, M. C., Goldberg, W. M. & Allemand, D. Skeleton and sclerite formation in the precious red coral Corallium rubrum. Mar. Biol. 117, 119–128 (1993).
Santangelo, G. et al. Patterns of variation in recruitment and post-recruitment processes of the Mediterranean precious gorgonian coral Corallium rubrum. J. Exp. Mar. Biol. Ecol. 411, 7–13 (2012).
Quattrini, A. M., Gómez, C. E. & Cordes, E. E. Environmental filtering and neutral processes shape octocoral community assembly in the deep sea. Oecologia 183(1), 221–236 (2017).
Farrant, P. A. Population dynamics of the temperate Australian soft coral Capnella gaboensis. Mar. Biol. 96, 401–407 (1987).
Russo, A. R. Ecological observations on the gorgonian sea fan Eunicella cavolinii in the Bay of Naples. Mar. Ecol. Prog. Ser. 24, 155–159 (1985).
Benayahu, Y. Lobe Variation in Sinularia nanolobata Verseveldt, 1977 (Cnidaria: Alcyonacea). Bull. Mar. Sci. 63(1), 229–240 (1998).
Jeyasuria, P. & Lewis, J. C. Mechanical properties of the axial skeleton in gorgonians. Coral Reefs 5, 213–219 (1987).
Muzik, K. & Wainwright, S. Morphology and Habitat of Five Fijian Sea Fans. Bull. Mar. Sci. 27(2), 308–337 (1977).
Sebens, K. P. Water flow and coral colony size: Interhabitat comparisons of the octocoral Alcyonium siderium. Proc. Natl. Acad. Sci. 81(17), 5473–5477 (1984).
Esford, L. E. & Lewis, J. C. Stiffness of Caribbean gorgonians (Coelenterata, Octocorallia) and Ca/Mg content of their axes. Mar. Ecol. Prog. Ser. 67(2), 189–200 (1990).
Imahara, Y. Nephtheid octocorals from Suruga Bay, the Pacific Coast of Central Japan. Annot. Zool. Japonenses 50(3), 164–173 (1977).
Dautova, T. N. Gorgonians (Anthozoa: Octocorallia) of the northwestern Sea of Japan. Russ. J. Mar. Biol. 33, 297–304 (2007).
Schuhmacher, H. Morphologische und ökologische Anpassungen von Acabaria-Arten (Octocorallia) im Roten Meer an verschiedene Formen der Wasserbewegung. Helgol. Wiss. Meeresunters. 25, 461–472 (1973).
Baillon, S., English, M., Hamel, J. F. & Mercier, A. Comparative biometry and isotopy of three dominant pennatulacean corals in the Northwest Atlantic. Acta Zool 97(4), 475–493 (2016).
Weinberg, S. Revision of the common Octocorallia of the Mediterranean circalittoral. I. Gorgonacea. Beaufortia 24(313), 63–104 (1976).
Weinberg, S. Revision of the common Octocorallia of the Mediterranean circalittoral II. Alcyonacea. Beaufortia 25(326), 131–166 (1977).
Weinberg, S. Revision of the common Octocorallia of the Mediterranean circalittoral. III. Stolonifera. Beaufortia 27(338), 139–176 (1978).
Verseveldt, J. Octocorallia from North-Western Madagascar (Part II). Zool. Verh. 117(1), 1–73 (1971).
Cairns, S. D. Review of Octocorallia (Cnidaria: Anthozoa) from Hawai’i and adjacent seamounts. Part 3: Genera Thouarella, Plumarella, Callogorgia, Fanellia, and Parastenella. Pac. Sci. 64(3), 413–440 (2010).
Alderslade, P. A new soft coral genus (Coelenterata: Octocorallia) from Palau. Rec. Mus. Art Gall. Northern Territory 18, 1–8 (2002).
Canessa, M., Bavestrello, G. & Trainito, E. Leptogorgia sarmentosa (Anthozoa: Octocorallia) in NE Sardinia (Mediterranean Sea): distribution and growth patterns. Mar. Biodivers. 53(1), 13 (2023).
Grasshoff, M. & Bargibant, G. Les gorgones des récifs coralliens de Nouvelle-Calédonie, 335 pp (IRD, Paris, 2001).
Horvarth, E. A. A review of gorgonian coral species (Cnidaria, Octocorallia, Alcyonacea) held in the Santa Barbara Museum of Natural History research collection: focus on species from Scleraxonia, Holaxonia, Calcaxonia – Part III: Suborder Holaxonia continued, and suborder Calcaxonia. ZooKeys 860, 183 (2019).
Bo, M. et al. Overview of the conservation status of Mediterranean anthozoa. Report No. IUCN International Union for Conservation of Nature (2017).
OBIS. Ocean Biodiversity Information System. Intergovernmental Oceanographic Commission of UNESCO. Available from: https://obis.org/ (June 6, 2022) (2019).
Bastidas, C., Benzie, J. A. H., Uthicke, S. & Fabricius, K. E. Genetic differentiation among populations of a broadcast spawning soft coral, Sinularia flexibilis, on the Great Barrier Reef. Mar. Biol. 138, 517–525 (2001).
Hwang, S. J. & Soong, J. I. Reproductive biology of temperate soft corals (Alcyonacea: Nephtheidae) in Korean waters. 11th International Coral Reef Symposium, Fort Lauderdale, FL, p. 98 (2008).
Pratt, E. M. The Alcyonaria of the Maldives. Part II. The genera Sarcophytum, Lobophytum, Sclerophytum, and Alcyonium. In Fauna of the Maldive and Laccadive Archipelago 2, 503–509 (1903).
Manuel, R. L. Synopses of the British Fauna: British Anthozoa, 2nd edn (Academic Press, London, 1988) 241 pp
Munro, C. D. & Munro, L. Eunicella verrucosa: investigating growth and reproduction from a population ecology perspective. PHMS Newsletter 13, 29–30 (2003).
Grange, K. R. et al. Phylum Cnidaria. In: Cook, S. D. C. (ed.) New Zealand Coastal Marine Invertebrates 1, 137–247 (Canterbury University Press, Christchurch, 2010).
Hickson, S. J. The Alcyonaria of the Cape of Good Hope. Part II. Marine Investigations in South Africa 3, 211–239 (1904).
Weinberg, S. Trichogorgia insulaeuropensis spec. nov., a new gorgonian from the Southern Indian Ocean (Coelenterata: Octocorallia: Alcyonacea: Chrysogorgiidae). Zool. Meded. 87, 405–412 (2013).
Bayer, F. M. & Muzik, K. M. New genera and species of the holaxonian family Chrysogorgiidae (Octocorallia: Gorgonacea). Zool. Meded. 50(5), 65–90 (1976).
Clarke, F. W. & Wheeler, W. C. The inorganic constituents of marine invertebrates (2nd edn, revised and enlarged). Prof. Pap. U.S. Geol. Surv. No. 124 (1922).
Choi, Y. W. & Kim, J. H. Mortality and growth of the soft coral, Dendronephthya gigantea in Jejudo Island, Korea. J. Korean Soc. Oceanogr. 13(4), 342–347 (2008).
Williams, G. C. New taxa of octocorals (Anthozoa: Octocorallia) from the northeastern Pacific Ocean. Proc. California Acad. Sci. 56(5), 53–65 (2005).
Ledger, P. W. & Franc, S. Calcification of the collagenous axial skeleton of Veretillum cynomorium pall. (Cnidaria: Pennatulacea). Cell Tissue Res 192, 249–266 (1978).
Provoost, P. & Bosch, S. “robis: R Client to access data from the OBIS API.” Ocean Biogeographic Information System. Intergovernmental Oceanographic Commission of UNESCO. R package version 1.0.0, https://cran.r-project.org/package=robis (2017).
RStudio Team. RStudio Desktop IDE (Version 2023.06.0 + 421). PBC https://posit.co/products/open-source/rstudio/ (2023).
Zizka et al. CoordinateCleaner: standardized cleaning of occurrence records from biological collection databases. Methods in Ecology and Evolution, 7, R package version 3.0.1, https://github.com/ropensci/CoordinateCleaner (2019).
Byrnes J. meowR: marine ecoregions of the world in R, 0.6.2, https://github.com/jebyrnes/meowR. (2016)
McFadden, C., Cordeiro, R., Williams, G. & van Ofwegen, L. World List of Octocorallia. In O. Bánki, Y. et al., Catalogue of Life Checklist (ver. (05/2024)) https://doi.org/10.48580/dg6lk-3d2 (2024).
WoRMS Editorial Board. World Register of Marine Species. Available from https://www.marinespecies.org at VLIZ. Accessed 2024-06-07. https://doi.org/10.14284/170 (2024).
Gómez-Gras et al. OctocoralTraits: a global database of trait information for octocoral species (2.2). Zenodo https://doi.org/10.5281/zenodo.14228404 (2024).
Wilkinson, M. D. et al. The FAIR guiding principles for scientific data management and stewardship. Sci. Data 3 (2016).
Spalding, M. D. et al. Marine ecoregions of the world: a bioregionalization of coastal and shelf areas. BioScience 57(7), 573-583 (2007).
Acknowledgements
D.G.G. acknowledges the funding of Margarita Salas 2021–2023 postdoctoral Fellowship program (Ministerio de Universidades). CL gratefully acknowledges the financial support by ICREA under the ICREA Academia program. D.G.G., N.V., A.G., C.L. & Y.Z. are part of the Marine Conservation research group –MedRecover (2021 SGR 01073) from the ‘Generalitat de Catalunya’. JSM acknowledges funding from the National Science Foundation (1948946).
Author information
Authors and Affiliations
Contributions
D.G.G.: Conceptualization, methodology, compilation of trait data, software, investigation, formal analysis, preparation of original draft, and visualization, project administration, funding acquisition. C.L.: Conceptualization, methodology, validation, draft review and editing, supervision. J.S.M.: Conceptualization, methodology, software, validation, draft review and editing, supervision. C.M.: Compilation of trait data, methodology, draft review, and editing, K.F.: Compilation of trait data, methodology, draft review, and editing, A.G.: Compilation of trait data, methodology, draft review, and editing. N.V.: Compilation of trait data, methodology, draft review, and editing, Y.Z.: Compilation of trait data, methodology, draft review, and editing, J.G.: Compilation of trait data, methodology, draft review, and editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gómez-Gras, D., Linares, C., Viladrich, N. et al. The Octocoral Trait Database: a global database of trait information for octocoral species. Sci Data 12, 82 (2025). https://doi.org/10.1038/s41597-024-04307-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41597-024-04307-8