Abstract
Samul-tang (SM) is a traditional multi-ingredient herbal formula that is widely used in clinical practice for treating female infertility. Despite its known therapeutic benefits, the complexity of its action mechanisms owing to the combination of multiple compounds has limited its research and integration into modern pharmacological science. To address this challenge, we identified 38 herbal compounds as the major ingredients in SM and generated their transcriptome data from aged-mice ovaries by administering these individual compounds. This dataset provides a resource for investigating the action mechanisms of SM and assist in identifying effective combinations of ingredients that can reproduce the therapeutic effects of SM.
Similar content being viewed by others
Background & Summary
Selecting the best-quality embryo is imperative for successful pregnancy in clinical in vitro fertilization and embryo transfer (IVF-ET)1,2. Additionally, oocyte quality significantly affects the quality of blastocytes obtained after IVF3,4. Therefore, the IVF-ET success rate in older women is significantly lower than that in younger women owing to a decrease in the quality and number of oocytes retrieved after ovarian stimulation5,6.
To improve the oocyte quality associated with age-related infertility, we screened various Traditional Korean Medicine prescriptions through animal experiments, specifically using an aged mouse. Among these prescriptions, only SM demonstrated the most significant improvements in oocyte quality, follicle number, embryo quality, and pregnancy rate when administered orally to 38-week-old mice for 4 weeks7. SM is a widely used traditional herbal formulation comprising equal parts of Angelicae Gigantis Radix, Cnidii Rhizoma, Rehmanniae Radix Preparata, and Paeoniae Radix8,9. Despite its therapeutic benefits, the complexity of its action mechanisms owing to the combination of multiple compounds has hindered its integration into modern pharmacological science. Therefore, this study aimed to generate a comprehensive transcriptome dataset of individual SM ingredients to facilitate a deeper understanding of their effects on ovarian function.
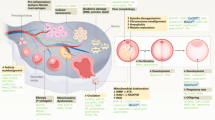
To achieve this, we conducted a study to build ovarian transcriptome data for individual ingredients of SM. As illustrated in Fig. 1a, each of the 38 compounds constituting SM or a vehicle was orally administered to 38-week-old mice for four weeks, and the ribonucleic acid (RNA) from their ovarian tissue was analyzed using Illumina RNA-Seq technology. Consequently, we obtained high-quality transcriptome data with high correlation within the group treated with the same compound (three mice per group). These results were achieved through labor-intensive experiments that involved oral administration of the compounds over four weeks, which is the approximate timeframe required for primary follicles to develop into antral follicles10. This duration allows for assessing the effectiveness of the drug treatment on follicle maturation and oocyte quality. This transcriptomic dataset can serve as a valuable resource for understanding the pharmacodynamics of natural compounds in herbal medicine for aged female mice.
Experimental design and RNA-Seq analysis pipeline. (a) Schematic of the experiment for generating the RNA sequencing (RNA-Seq) dataset following compound treatment. (b) Schematic of the experiment for obtaining a compound-induced ovarian transcriptome. A group of three aged mice (38 weeks old) were orally administered each of the 38 compounds (5 mg/kg) (OC + Compound) or a vehicle of 0.5% DMSO in DW (OC), five times a week for four weeks. Subsequently, the mice were superovulated using gonadotropins (PMSG and hCG) and total RNA was isolated from their left ovaries and subjected to RNA-Seq. (c) Flowchart depicting the key steps involved in the RNA-Seq data preprocessing pipeline for subsequent differential gene expression analysis.
Methods
Compounds
To identify the chemical composition of SM, we conducted a literature search using the terms “Siwu-decoction” and “Shimotsu to,” which are Chinese and Japanese terms for SM, respectively, in reputable academic databases such as PubMed and ScienceDirect. Additionally, we searched for compounds in the four constituent herbs of SM: Angelicae Gigantis Radix (Angelica gigas Nakai), Cnidii Rhizoma (Cnidium officinale Makino), Rehmanniae Radix Preparata (Rehmannia glutinosa Liboschitz ex Steudel), and Paeoniae Radix (Paeonia lactiflora Pallas), and compiled a list of compounds in SM. To further clarify and verify the searched compounds, we performed an ultra-high performance liquid chromatography-tandem mass spectrometry (UHPLC-MS/MS) using a Dionex UltiMate 3000 system equipped with a Thermo Q-Exactive mass spectrometer (ThermoFisher Scientific, Waltham, MA, USA). SM powder, obtained from the National Institute for Korean Medicine Development (NIKOM), was dissolved in methanol at a concentration of 100 mg/ml. Subsequently, the extracted solution was processed according to the method reported in a previous study11. Data acquisition and analysis were performed using Xcalibur 4.2 and Tracefinder 4.1. A total of 33 distinct compounds were detected in SM, and their presence was confirmed by comparing the retention times and mass spectra with those listed in our established reference database12 (Supplementary Table 1). Subsequently, 38 commercially available compounds were selected by combining our findings from the literature search and the compounds identified in SM (Table 1).
Mice
Thirty-eight-week-old Balb/c female mice were purchased from Central Lab. Animal Inc. (Seoul, Korea). All mice were maintained on an ad libitum diet in a pathogen-free room under a 12 h light-dark cycle. Mice handling and experimental procedures were approved by the Korea Institute of Oriental Medicine Institutional Animal Care and Use Committee (Approval Number: 20-090). The mice were randomly divided into groups of three and orally administered either vehicle (distilled water containing 0.5% dimethyl sulfoxide (DMSO; D2650; Sigma-Aldrich, St. Louis, MO, USA)) or one of the 38 compounds at a dose of 5 mg/kg five times a week for four weeks. This dosage was selected to minimize the risk of chronic toxicity based on evidence suggesting that 5 mg/kg/day yields high predictive accuracy for the lowest observed adverse effect level in chronic-toxicity studies13. Hereafter, the vehicle-treated group is referred to as the old control (OC), and the compound-treated groups are referred to as OC + [name of compound]. The details of the compounds are listed in Table 1. Following compound administration for four weeks, the mice received an intraperitoneal injection of 5 IU of pregnant mare serum gonadotropin (PMSG; HOR-272; Prospec, Rehovot, Israel), followed 48 h later by an injection of 5 IU of human chorionic gonadotropin (hCG; HOR-250; Prospec) to create a post-ovulatory state, similar to a previous study7. Subsequently, their ovaries were removed and weighed 14–16 h post-hCG administration. The ovaries were immediately preserved in liquid nitrogen for RNA sequencing (Fig. 1b).
RNA extraction and sequencing
Following the administration of one of the 38 compounds, the left ovary (n = 3) of each post-ovulation mouse was immersed in 300 μl of TRIzol (Invitrogen, Carlsbad, CA, USA) and homogenized using a disposable grinder (Biomasher II; 890863; LMS Co., Tokyo, Japan). Total RNA was isolated using TRIzol reagent (Invitrogen). For RNA library preparation, we employed the QuantSeq FWD 3′ mRNA-Seq Library Prep Kit (Lexogen, Inc., Austria), which allows strand-specific RNA sequencing. To preserve RNA integrity and minimize RNA processing steps, DNase treatment was omitted before RNA extraction, in accordance with the manufacturer’s protocol. A total of 500 ng of RNA extracted from each sample was used as input for library preparation. In summary, an oligo-dT primer containing an Illumina-compatible sequence at its 5′ end was hybridized to the RNA, and reverse transcription was performed. Following degradation of the RNA template, second-strand synthesis was initiated using a random primer containing an Illumina-compatible linker sequence at its 5′ end. The double-stranded library was purified using magnetic beads to remove all reaction components. The library was amplified to add the complete adapter sequences required for cluster generation. The final library was purified from the polymerase chain reaction components. High-throughput sequencing was performed as single-end 75-bp sequencing using the NextSeq 500 system (Illumina, Inc., USA).
RNA-seq data preprocessing
The quality of the raw RNA-Seq FASTQ files was initially evaluated using FastQC v1.9 (Babraham Bioinformatics, UK) with default parameters. To eliminate adapter sequences and low-quality reads, the BBduk module within the BBMap toolkit (v38.95)14 was used with the following parameters: k = 13, ktrim = r, useshortkmers = t, mink = 5, qtrim = t, trimq = 10, and minlength = 20. After trimming, the cleaned reads were quality-checked (QC) again using FastQC. The trimmed reads were then aligned to the mouse reference genome build GRCm39 using the STAR aligner (v2.7.9a)15 to map their genomic positions. The aligned reads within the genome were then quantified per gene using RSEM (v1.3.3)16 in conjunction with the mouse gene annotation file (GRCm39.104.gtf). The RSEM tool was executed with the–strandedness forward option to account for the strandedness of the libraries, providing counts and transcripts per million (TPM) values for each gene. To further examine the distribution of RNA-seq reads across genomic regions (exonic, intronic, and intergenic), the BAM files generated from the STAR alignment were processed using the QualiMap (v2.2.1) RNA-seq module with the–outformat PDF:HTML option. Following individual QC steps, MultiQC (v1.11)17 was used to summarize and visualize the quality metrics across all 124 RNA-seq samples (Fig. 1c). MultiQC was used to aggregate data from FastQC and QualiMap, providing an integrated view of quality and alignment statistics to evaluate sample quality and processing consistency.
Differential gene expression analysis
The RNA-Seq read count data were normalized using the voom function of the limma package18 in R (v4.2.2) to account for technical variability. Subsequently, a linear model was fitted using the lmFit function, followed by the application of the eBayes function to compute moderated t-statistics to assess the significance of differential expression between the compound- and vehicle-treated groups. This approach yielded three key metrics per gene: log2 fold change, t-statistic, and adjusted p-value (false discovery rate, FDR). Thereafter, differentially expressed genes (DEGs) were identified by applying a stringent FDR threshold of <0.05 (Fig. 1c).
Data Records
The raw sequencing reads (FASTQ format, compressed with gzip) and processed gene expression data (expected read count and TPM) for 124 samples were deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database under accession number GSE26400219. The detailed information for each sample, including the production methodology, utilized strain, and treatment parameters (dosage and duration), is summarized in Supplementary Table 2.
Technical Validation
RNA quality and integrity
The nucleic acid purity of the isolated RNA samples was assessed by measuring the optical density (OD) ratio at ultraviolet wavelengths of 260 and 280 nm (OD 260/280) using the NanoDrop ND-2000 Spectrophotometer (ThermoFisher Scientific, DE, USA). All samples exhibited high purity, with an OD 260/280 ratio (mean ± SD) of 1.89 ± 0.06 (Fig. 2a and Supplementary Figure 1a), indicating minimal protein or DNA contamination. High-resolution analysis of RNA integrity was performed using the Agilent 2100 Bioanalyzer (Agilent Technologies, Amstelveen, Netherlands). The analysis revealed a high degree of RNA integrity, as evidenced by the 28S/18S ribosomal RNA (rRNA) ratio of 1.69 ± 0.15 (Fig. 2b and Supplementary Figure 1b) and RNA integrity number (RIN) of 9.36 ± 0.38 (Fig. 2c and Supplementary Figure 1c). These results confirmed the quality and integrity of the isolated RNA, suitable for downstream RNA sequencing analysis (Supplementary Table 3).
Quality of RNA-Seq data
The quality of the raw RNA-Seq reads generated from the 124 samples was evaluated using FastQC, a software suite that provides comprehensive quality reports. These reports encompass metrics such as the base-wise quality scores and guanine-cytosine (GC) content distribution. Subsequently, MultiQC was used to consolidate the individual FastQC reports into a single unified file for efficient data exploration. The overall quality of the sequencing reads was high, as evidenced by a mean Phred quality score of >30 (>99.9% base-call accuracy) across all sequenced positions (Fig. 3a). To obtain clean data, adapter sequences and low-quality bases with Phred scores under 20 were eliminated from the reads using the BBduk module of the BBMap toolkit. The GC content of the trimmed reads revealed a normal distribution with an average of 47.66 ± 1.50% (Fig. 3b), further supporting the absence of substantial contamination within the sequencing data. Subsequent alignment of the cleaned reads with the reference mouse genome (GRCm38) yielded a median total mapped read count of 18,017,346 reads ± 1,454,114 (Fig. 3c and Supplementary Table 4). Notably, all samples, except for a single outlier, exhibited uniquely mapped read percentages exceeding 80% (Fig. 3d and Supplementary Figure 2a). This outlier, corresponding to an OC sample treated with chlorogenic acid, exhibited a uniquely mapped read percentage of 51.36%. Owing to this significant deviation, this sample was excluded from further analyses to maintain data integrity.
RNA-Seq read quality assessments. (a) Average Phred quality scores across all base positions for the raw RNA-Seq data obtained from all 124 samples. Most bases across all samples exhibit Phred scores exceeding 30, demonstrating high overall read quality. (b) Distribution of the average GC content in the trimmed RNA-Seq reads of all 124 samples. A normal distribution of the GC content is typically observed in randomly generated libraries. The distribution shown aligns with this expectation, suggesting unbiased library preparation. (c) Bar plot summarizing the uniquely mapped, multi-mapped, and unmapped reads for each sample library. Uniquely mapped reads represent those that map to a single location in the reference genome, whereas multi-mapped reads represent those that map to multiple locations. Unmapped reads are those that failed to map to the reference genome. (d) Distribution of the percentage of uniquely mapped reads among the total reads across all 124 samples. A higher percentage of uniquely mapped reads generally indicates high-quality mapping. Although most samples exhibit high percentages of uniquely mapped reads (>80%), one sample exhibits a lower percentage (51.36%), as indicated by the red arrow. This outlier was excluded from further analysis.
We used QualiMap to further assess the distribution of RNA-seq reads across various genomic regions, including exonic, intronic, and intergenic areas. This analysis revealed that, on average (±standard deviation), 68.2 ± 3.75% of reads aligned to exonic regions, 24.6 ± 2.63% to intronic regions, and 7.23 ± 1.53% to intergenic regions (Supplementary Figure 2b and Supplementary Table 5). This high proportion of exonic alignments (~70%) surpasses the typical ~64% observed in standard RNA-seq experiments20, indicating the high quality and robustness of our dataset for quantitative gene expression analyses. This distribution supports the suitability of our dataset for reliable downstream analysis, with minimal interference from non-coding regions.
Biological reproducibility
Through the RNA-Seq analysis, we identified 20,009 protein-coding genes across the 124 samples. To assess the reproducibility of our data, we evaluated gene expression patterns between replicate samples under each experimental condition. Each replicate was independently subjected to RNA extraction, library preparation, and sequencing. The Pearson correlation coefficient between replicate control samples of OC mice was 0.98 ± 0.01 (Fig. 4a). Additionally, the Pearson correlation coefficients for OC samples subjected to the same compound treatment (0.99 ± 0.02) were higher than those subjected to different compound treatments (0.98 ± 0.01) (Fig. 4a), indicating a high degree of biological replicability.
Correlation coefficients of gene expression between biological replicates. (a)Heatmap of Pearson correlation coefficients between 10 replicates of OC (vehicle-treated) samples. (b) Distribution of Pearson correlation coefficients for OC samples treated with the same compound (yellow) versus those treated with different compounds (gray). The median Pearson correlation for each distribution is indicated with a dotted line.
Compound-specific effects on gene expression
Differential gene expression analysis was performed to investigate the effects of the 38 compounds on the cellular transcriptome. An FDR cutoff of <0.05 was employed to identify significant DEGs for each compound, and substantial heterogeneity was observed in the number of DEGs elicited by these compounds (Fig. 5). Rehmannioside D, chlorogenic acid, and ferulic acid induced a larger transcriptional response than other compounds. Notably, the effects of approximately half of the tested compounds on gene expression were relatively modest.
Usage Notes
This dataset is a collection of transcriptome profiles of ovaries of old mice following the administration of 38 individual herbal compounds found in SM. Researchers can leverage this resource to elucidate the underlying action mechanisms of these compounds on ovarian function, potentially leading to the identification and development of novel therapeutic targets. Additionally, the dataset can be utilized to identify the optimal combinations of these herbal ingredients to improve ovarian function. This analysis was achieved by integrating complementary transcriptome data generated in our previous study7 encompassing data from aged murine ovaries treated with SM and control young mice, which were processed using the same Quant-seq platform. The data have been deposited in the gene expression omnibus (GEO) under accession numbers GSE26441321 and GSE26400219.
Code availability
All software used to analyze the RNA-Seq data, along with their respective parameters, are described in the Methods section. Unless otherwise specified, the default parameter settings recommended by the developers were used. Researchers are encouraged to cite this paper when utilizing the RNA-Seq data uploaded to the GEO.
References
Vaegter, K. K. et al. Which factors are most predictive for live birth after in vitro fertilization and intracytoplasmic sperm injection (IVF/ICSI) treatments? Analysis of 100 prospectively recorded variables in 8,400 IVF/ICSI single-embryo transfers. Fertil Steril 107, 641–648 e642, https://doi.org/10.1016/j.fertnstert.2016.12.005 (2017).
Amini, P. et al. Factors Associated with In Vitro Fertilization Live Birth Outcome: A Comparison of Different Classification Methods. Int J Fertil Steril 15, 128–134, https://doi.org/10.22074/IJFS.2020.134582 (2021).
Catala, M. G., Izquierdo, D., Rodriguez-Prado, M., Hammami, S. & Paramio, M. T. Effect of oocyte quality on blastocyst development after in vitro fertilization (IVF) and intracytoplasmic sperm injection (ICSI) in a sheep model. Fertil Steril 97, 1004–1008, https://doi.org/10.1016/j.fertnstert.2011.12.043 (2012).
Braga, D. P. et al. Influence of oocyte dysmorphisms on blastocyst formation and quality. Fertil Steril 100, 748–754, https://doi.org/10.1016/j.fertnstert.2013.05.021 (2013).
Moghadam, A. R. E., Moghadam, M. T., Hemadi, M. & Saki, G. Oocyte quality and aging. JBRA Assist Reprod 26, 105–122, https://doi.org/10.5935/1518-0557.20210026 (2022).
Wang, P. et al. The association between the number of oocytes retrieved and cumulative live birth rate in different female age strata. Sci Rep 13, 14516, https://doi.org/10.1038/s41598-023-41842-7 (2023).
Kim, J. & You, S. Effect of samul-tang on female fertility via RAS signaling pathway in ovaries of aged mice. Aging (Albany NY) 13, 14829–14842, https://doi.org/10.18632/aging.203150 (2021).
Choi, J. et al. Current clinical practice status of Korean medicine for managing female infertility: A cross-sectional survey. Complement Ther Clin Pract 40, 101175, https://doi.org/10.1016/j.ctcp.2020.101175 (2020).
Liao, Y. H. et al. Traditional Chinese Medicine Treatment Associated with Female Infertility in Taiwan: A Population-Based Case-Control Study. Evid Based Complement Alternat Med 2020, 3951741, https://doi.org/10.1155/2020/3951741 (2020).
Zheng, W. et al. Two classes of ovarian primordial follicles exhibit distinct developmental dynamics and physiological functions. Hum Mol Genet 23, 920–928, https://doi.org/10.1093/hmg/ddt486 (2014).
Lee, A., Yang, H., Kim, T., Ha, H. & Hwang, Y. H. Identification and pharmacokinetics of bioavailable anti-resorptive phytochemicals after oral administration of Psoralea corylifolia L. Biomed Pharmacother 144, 112300, https://doi.org/10.1016/j.biopha.2021.112300 (2021).
Lee, A. et al. Hydroethanolic Extract of Fritillariae thunbergii Bulbus Alleviates Dextran Sulfate Sodium-Induced Ulcerative Colitis by Enhancing Intestinal Barrier Integrity. Nutrients 15, https://doi.org/10.3390/nu15122810 (2023).
Li, X., Zhang, Y., Chen, H. N., Li, H. H. & Zhao, Y. prediction of chronic toxicity with chemical category approaches. Rsc Adv 7, 41330–41338, https://doi.org/10.1039/c7ra08415c (2017).
Bushnell, B. BBMap: a fast, accurate, splice-aware aligner. (2014).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21, https://doi.org/10.1093/bioinformatics/bts635 (2013).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323, https://doi.org/10.1186/1471-2105-12-323 (2011).
Ewels, P., Magnusson, M., Lundin, S. & Kaller, M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048, https://doi.org/10.1093/bioinformatics/btw354 (2016).
Law, C. W., Chen, Y., Shi, W. & Smyth, G. K. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15, R29, https://doi.org/10.1186/gb-2014-15-2-r29 (2014).
You, S., Shin, h., Kim, J. & Lee, H. Transcriptome profiling of ovaries in aged mice administered with individual ingredients of Samul-tang, http://identifiers.org/geo/GSE264002 (2024).
Corley, S. M., Troy, N. M., Bosco, A. & Wilkins, M. R. QuantSeq 3′ Sequencing combined with Salmon provides a fast, reliable approach for high throughput RNA expression analysis. Sci Rep 9, 18895, https://doi.org/10.1038/s41598-019-55434-x (2019).
You, S., Kim, J. & Lee, H. Transcriptome profiling of ovaries in aged mice administered with Samul-tang, http://identifiers.org/geo/GSE264413 (2024).
Su, S. et al. Chemical fingerprinting and quantitative constituent analysis of Siwu decoction categorized formulae by UPLC-QTOF/MS/MS and HPLC-DAD. Chin Med 8, 5, https://doi.org/10.1186/1749-8546-8-5 (2013).
Zhu, M. et al. Identification of chemical constituents in SiWu decoction by UHPLC-DAD-TOF/MS. Acta Chromatographica 26, 517–537 (2014).
Kang, Y. Y., Kim, J. Y., Song, J. & Mok, H. Enhanced intracellular uptake and stability of umbelliferone in compound mixtures from Angelica gigas in vitro. J Pharmacol Sci 140, 8–13, https://doi.org/10.1016/j.jphs.2019.02.010 (2019).
Won, T.-H., Ryu, S.-K., Kang, S.-S. & Shin, J.-H. Simultaneous analysis of bioactive metabolites from Rehmannia glutinosa by HPLC-DAD-MS/MS. Natural Product Sciences 16, 116–122 (2010).
Zheng, X. et al. Comparative Elucidation of Age, Diameter, and “Pockmarks” in Roots of Paeonia lactiflora Pall. (Shaoyao) by Qualitative and Quantitative Methods. Front Plant Sci 12, 802196, https://doi.org/10.3389/fpls.2021.802196 (2021).
Duan, X. et al. Analysis of the active components and metabolites of Taohong Siwu decoction by using ultra high performance liquid chromatography quadrupole time-of-flight mass spectrometry. J Sep Sci 43, 4131–4147, https://doi.org/10.1002/jssc.202000498 (2020).
Takiyama, M., Matsumoto, T., Sanechika, S. & Watanabe, J. Pharmacokinetic study of traditional Japanese Kampo medicine shimotsuto used to treat gynecological diseases in rats. J Nat Med 75, 361–371, https://doi.org/10.1007/s11418-020-01474-x (2021).
Zhou, J. et al. Rehmannia glutinosa (Gaertn.) DC. polysaccharide ameliorates hyperglycemia, hyperlipemia and vascular inflammation in streptozotocin-induced diabetic mice. J Ethnopharmacol 164, 229–238, https://doi.org/10.1016/j.jep.2015.02.026 (2015).
Kim, M. R., Abd El-Aty, A. M., Kim, I. S. & Shim, J. H. Determination of volatile flavor components in danggui cultivars by solvent free injection and hydrodistillation followed by gas chromatographic-mass spectrometric analysis. J Chromatogr A 1116, 259–264, https://doi.org/10.1016/j.chroma.2006.03.060 (2006).
Jeong, S. Y. et al. Quantitative analysis of marker compounds in Angelica gigas, Angelica sinensis, and Angelica acutiloba by HPLC/DAD. Chem Pharm Bull (Tokyo) 63, 504–511, https://doi.org/10.1248/cpb.c15-00081 (2015).
Feng, W. et al. A new megastigmane from fresh roots of Rehmannia glutinosa. Acta Pharmaceutica sinica B 3, 333–336 (2013).
Hwang, Y. H., Cho, W. K., Jang, D., Ha, J. H. & Ma, J. Y. High-performance liquid chromatography determination and pharmacokinetics of coumarin compounds after oral administration of Samul-Tang to rats. Pharmacogn Mag 10, 34–39, https://doi.org/10.4103/0973-1296.126656 (2014).
Lee, H. Y., Lee, E. J., Kim, J. S., Lee, J.-H. & Kang, S. S. Phytochemical Studies on Rehmanniae Radix Preparata. Kor. J. Pharmacogn [Lexical characteristics of Korean language] 42, 117-126 (2011).
Kim, S. H. et al. Chemical constituents isolated from Paeonia lactiflora roots and their neuroprotective activity against oxidative stress in vitro. Journal of Enzyme Inhibition and Medicinal Chemistry 24, 1138–1140 (2009).
He, D. Y. & Dai, S. M. Anti-inflammatory and immunomodulatory effects of paeonia lactiflora pall., a traditional chinese herbal medicine. Front Pharmacol 2, 10, https://doi.org/10.3389/fphar.2011.00010 (2011).
Wang, Z. J. et al. Simultaneous quantification of active components in the herbs and products of Si-Wu-Tang by high performance liquid chromatography-mass spectrometry. J Pharm Biomed Anal 50, 232–244, https://doi.org/10.1016/j.jpba.2009.04.001 (2009).
Chen, Y., Li, Q. & Qiu, D. The Dynamic Accumulation Rules of Chemical Components in Different Medicinal Parts of Angelica sinensis by GC-MS. Molecules 27, https://doi.org/10.3390/molecules27144617 (2022).
Acknowledgements
This research was funded by the research program of the Korea Institute of Oriental Medicine (KSN20233305 and KSN1722122). We would like to thank Editage (www.editage.co.kr) for English language editing.
Author information
Authors and Affiliations
Contributions
H.S., J.K. and S.Y. conceived and designed the experiments. H.S. conducted the chromatographic profiling and validated the compound. J.K. and S.Y. compiled the list of SM compounds. H.S. and J.K. collected tissue samples and constructed the transcriptome. M.P. discussed the results and provided feedback. H.J. contributed to the validation of transcriptome data. M.L. performed the preprocessing of raw data. S.C. reviewed the manuscript and provided feedback. H.L. conceived the validation of transcriptome data. H.S., H.L. and S.Y. drafted the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Shin, H., Kim, J., Park, M. et al. Transcriptome profiling of aged-mice ovaries administered with individual ingredients of Samul-tang. Sci Data 12, 81 (2025). https://doi.org/10.1038/s41597-025-04370-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41597-025-04370-9