Abstract
The remarkable progress of deep learning in dermatological tasks has brought us closer to achieving diagnostic accuracies comparable to those of human experts. However, while large datasets play a crucial role in the development of reliable deep neural network models, the quality of data therein and their correct usage are of paramount importance. Several factors can impact data quality, such as the presence of duplicates, data leakage across train-test partitions, mislabeled images, and the absence of a well-defined test partition. In this paper, we conduct meticulous analyses of three popular dermatological image datasets: DermaMNIST, its source HAM10000, and Fitzpatrick17k, uncovering these data quality issues, measure the effects of these problems on the benchmark results, and propose corrections to the datasets. Besides ensuring the reproducibility of our analysis, by making our analysis pipeline and the accompanying code publicly available, we aim to encourage similar explorations and to facilitate the identification and addressing of potential data quality issues in other large datasets.
Similar content being viewed by others
Introduction
Skin diseases are the most common reason for clinical consultations in studied populations1, affecting almost a third of the global population2,3. The 2013 Global Burden of Disease found skin diseases to be the fourth leading cause of nonfatal disabilities globally, accounting for 41.6 million Disability Adjusted Life Years and 39.0 million Years Lost due to Disability4. In the USA alone, the healthcare cost of skin diseases was estimated to be $75 billion in 20165. Of all the skin diseases, skin cancer is a particularly concerning skin disease condition that merits special attention due to its potential seriousness. With the increased incidence rates of skin cancer over the past decades6, coupled with the projected decline in the ratio of dermatologists to populations5, automated systems for dermatological diagnosis can be immensely valuable.
Advances in deep learning (DL)-based methods for dermatological tasks have produced models that are approaching the diagnostic accuracies of experts, some even mimicking clinical approaches of hierarchical7,8,9 and differential10 diagnoses. The data-driven nature of these DL methods implies that large and diverse datasets are needed to train accurate, robust, and generalizable models. However, unlike natural computer vision datasets, medical image datasets are relatively smaller, primarily because of the large costs associated with image acquisition and annotation, legal, ethical, and privacy concerns11, and are more cost prohibitive to expand12. This is also true for skin cancer image datasets13,14, where the surge in skin image analysis research over the past decade can be attributed in part to recent publicly available datasets, most notably the datasets and challenges of the International Skin Imaging Collaboration (ISIC) and the associated HAM1000015 and BCN200016 datasets, which are primarily dermoscopic image datasets, as well as other clinical image datasets such as SD-19817, SD-26018, derm7pt19, and Fitzpatrick17k20.
Although large data sets are important for the development of reliable models, the quality of the data therein and their correct use are equally important21,22,23,24: low-quality data may result in inefficient training, inaccurate models that exhibit biases, poor generalizability and low robustness, and may negatively affect the interpretability of such models. The data quality can be affected by several factors: mislabeled images, data leakage across training and evaluation partitions, the absence of a held-out test partition, etc. The issue of data leakage, in particular, is in fact quite widespread, and a recent survey25 of 17 fields spanning 294 articles, on topics ranging from medicine and bioinformatics to information technology operations and computer security, showed that ML adoption in all these fields suffers from data leakage. In an analysis of 10 popular natural computer vision, natural language, and audio datasets, Northcutt et al.26 estimated an average label error rate of at least 3.3%. In medical image analysis domains, too, investigation into the use of machine learning best practices has found several instances of incorrect data partitioning and feature leakage between training and evaluation partitions. Oner et al.27 showed that a peer-reviewed article published in Nature Medicine using DL for histopathology image analysis suffered from data leakage by using slide-level stratification for data partitioning instead of patient-level stratification. In a large-scale study, Bussola et al.28 showed how DL models can exhibit considerably inflated performance measures when evaluated on datasets where histopathology image patches from the same subject are present in training and validation partitions. In mammography analysis, Samala et al.29,30 showed the risks of feature leakage between training and validation partitions and how this could lead to an overly optimistic performance on the validation partition, compared to a completely held out test partition. Similar investigations have been conducted on the adverse effects of incorrect data partitioning on test performance in optical coherence tomography (OCT) image classification31, brain magnetic resonance imaging (MRI) classification32, and longitudinal brain MRI analysis33
Specific to skin image analysis, our previous work34 showed that the popular ISIC Skin Lesion Segmentation Challenge datasets from 2016 through 2018 have considerable overlap among their training partitions, and that 706 images are present in all three datasets’ training splits, a surprising discovery since ISIC 2016 only has 900 training images. Cassidy et al.35 analyzed the ISIC Skin Lesion Diagnosis Challenge datasets from 2016 to 2020, and found overlap and duplicates across these datasets. They used a duplicate removal strategy to curate new clean training, validation, and testing sets. Vega et al.36 found that a popular monkeypox skin image dataset, used in several peer-reviewed publications, contained “medically irrelevant images” and that models trained on these images did not necessarily rely on features underlying the diseases. Very recently, Groger et al.37 carried out an analysis of six dermatology skin data sets (MED-NODE, PH2, DDI, derm7pt, PAD-UFES-20, and SD-128), detecting and removing near duplicates and “irrelevant samples” from them. However, all the datasets in their study were small, with 3 of 6 datasets (MED-NODE, PH2, DDI) consisting of less than 700 images and the largest (SD-128) containing 5,619 images.
In this paper, we look at three popular and large skin image analysis datasets: the DermaMNIST dataset38,39 and its source HAM10000 dataset15 (10,015 images) and the Fitzpatrick17k dataset20 (16,577 images). We perform systematic analyses of both datasets and report instances of data duplication, data leakage across training and evaluation partitions, and data mislabeling in both datasets, fixing them where possible. Our corrected datasets and detailed analysis results are available online on Zenodo40.
Results
DermaMNIST
Released as a biomedical imaging dataset equivalent to the MNIST dataset of handwritten digits, MedMNIST consists of images from standardized biomedical imaging datasets resized to MNIST-like 28 × 28 resolution. Despite being a fairly new dataset, it has been quite popular (1009 citations as of November, 2024: 633 citations of [a more recent] 2023 paper39, and 376 of [an older] 2021 version of the paper38). The dermatological subset of MedMNIST, DermaMNIST, contains resized images from the popular “Human Against Machine with 10000 training images” (HAM10000) dataset15. HAM10000 contains 10,015 dermoscopic images of pigmented skin lesions collected from patients at two study sites in Australia and Austria, with their diagnoses confirmed by either histopathology, confocal microscopy, clinical follow-up visits, or expert consensus. The 7 disease labels in the dataset cover 95% of the lesions encountered in clinical practice15. Because of these meritorious properties, HAM10000 is a good candidate dataset for dermatological analysis, as aimed for with DermaMNIST. While the “lightweight” nature of DermaMNIST due to its “small size” is appealing for its adoption in machine learning for biomedical imaging39, the low spatial resolution (28 × 28) does not capture sufficient morphological structures of skin lesions compared to the source dataset HAM10000. Despite this, DermaMNIST has been used for a wide variety of applications in peer-reviewed publications: semi- and self-supervised learning41,42,43, federated learning44,45,46, privacy-preserving learning47,48, neural architecture search49,50, adversarially robust learning51,52, data augmentation53,54, generative modeling55, model interpretability56, AutoML57, active learning58, quantum vision transformers59, and biomedical vision-language foundation models60,61,62, as well as derivative benchmark datasets63,64. However, as we investigate below, the resulting DermaMNIST and its benchmarks suffer from serious flaws.
Data Leakage
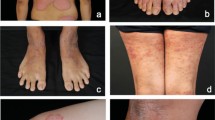
A caveat of HAM10000, despite its rather large size, is that it contains multiple images of the same lesion captured either from different viewing angles or at different magnification levels (Fig. 1(a)), i.e., the number of lesions with unique lesion IDs (HAM_xxx) is smaller than the number of images with unique image IDs (ISIC_xxx). We visualize the frequency counts of lesions and how many images of the same lesion are present in HAM10000 in Fig. 1(b) and observe that the 10,015 images are in fact derived from only 7,470 unique lesions, and 1,956 of these lesion IDs (~26.18%) contains 2 or more images: 1,423 lesions have 2 images, 490 lesions have 3 images, 34 lesions have 4 images, 5 lesions have 5 images, and 6 lesions have 4 images each. Unfortunately, this was not accounted for when preparing train-valid-test splits for DermaMNIST. The DermaMNIST dataset is released as a pre-processed NumPy array, so while the dataset does not contain filenames of the images, we confirmed this by contacting the authors to obtain the exact training-validation-testing split filenames. Therefore, there is considerable data leakage of images of the same lesion across partitions. Fig. 1(a) shows 2 examples where images of the same lesion are present in the training, validation, and testing partitions. This issue is quite pervasive across DermaMNIST, and our analysis found the following overlaps across partitions: train-test: 886 images (641 lesions), train-valid: 440 images (332 lesions); valid-test: 128 images (113 lesions); train-valid-test: 51 images (40 lesions) (Fig. 1(c)). Such data leakage naturally raises concerns on the reliability of the DermaMNIST benchmarks and related studies.
DermaMNIST analyses: (a,b) show instances of and reasons for the data leakage, and (c) visualizes how the three datasets: DermaMNIST, DermaMNIST-C, and DermaMNIST-E differ in their partition composition, yet have similarly proportionate diagnosis distributions. Images from DermaMNIST are licensed under CC BY-NC 4.038,39. Best viewed online.
We correct this data leakage by simply moving all images of a lesion ID present in the train partition from valid and test partitions back to the train partition. We choose to do this (i.e., moving the images of a lesion to the training set) instead of removing the images completely, to ensure that images of the same lesion are in one partition while also not discarding any images. Although this has the undesirable side effect of increasing the training partition size at the cost of reduced validation and testing partition sizes, it fixes the data leakage issue by ensuring there is no overlap across partitions.
Visualizing how DermaMNIST’s incorrect resizing operation leads to loss of information. DermaMNIST’s approach (top row) to generating 224 × 224 images results in visibly pixelated images. Our approach (bottom row), used for both DermaMNIST-C and DermaMNIST-E, retains much more detailed information. Images from DermaMNIST are licensed under CC BY-NC 4.038,39. Best viewed online.
Next, we examine the accuracy of HAM10000’s metadata. While the images in HAM10000 and their labels have been collected from clinical sites and the dataset itself has been widely adopted for online challenges65,66 and human-in-the-loop evaluations67,68, we found that some images with different lesion IDs are in fact duplicates and should have been assigned the same lesion ID. Therefore, for a systematic analysis, we use fastdup69, an open-source Python library for analyzing visual datasets at scale, and calculate inter-image embedding similarity scores \({\mathscr{S}}({x}_{i},{x}_{j})\) for all \(\left(\begin{array}{c}10,015\\ 2\end{array}\right)\) pairs of images (xi, xj). Figure 3 visualizes, as a confusion matrix, the following four scenarios that are possible when comparing duplication checks based on the metadata with duplicates detected using fastdup followed by visual human confirmation:
-
“Confirmed duplicates”: image pairs where both images share the same lesion IDs in the metadata and are indeed images of the same lesion.
-
“True non-duplicates”: image pairs where both images differ in their lesion IDs and are images of different lesions.
-
“Missed duplicates”: image pairs where both images differ in their lesion IDs but actually belong to the same lesion.
-
“False duplicates”: image pairs where both images share the same lesion IDs in but actually belong to the same lesion.
Visualizing the four scenarios that a pair of images from HAM10000 can be assigned to in duplicate detection, based on the metadata and the fastdup-based duplicate detection followed by manual review. “Confirmed duplicates”, as the name suggests, are pairs that are images of the same lesion, indicated by the same lesion IDs in the metadata. Similarly, “True non-duplicates” are pairs of images that belong to different lesions. “Missed duplicates” refer to image pairs that have differing lesion IDs according to the metadata, but their high visual similarity (measured by cosine similarity of their image embeddings) followed by manual review confirms that these are indeed images of the same lesion, and were therefore ‘missed’ by the metadata. Finally, “False duplicates” refer to pairs where images share the same lesion IDs but do not belong to the same lesion. In our analysis, we did not find any instances of “False duplicates” in HAM10000. For all these sample images, the image IDs and the lesion IDs are along the horizontal and the vertical axis, respectively. Images from HAM10000 are licensed under CC BY-NC 4.015.
The image pairs that lie along this confusion matrix’s diagonal, i.e., “confirmed duplicates” and “true non-duplicates”, are those where the metadata agrees with our analysis, and the possible errors in metadata arise out of the other two scenarios.
For detecting errors of the first kind, i.e., “missed duplicates”, we analyze the top 1,000 most similar image pairs, measured by similarity of the image embeddings. We look at these 1,000 pairs in intervals of 100 pairs, ordered by decreasing inter-image similarity. For the 100 image pairs in each interval, we look up the HAM10000 metadata for both images in a pair to exclude “confirmed duplicates”, since these duplicates are already accounted for in the metadata. For the remaining pairs, we manually review them to detect which, if any, of these are “missed duplicates” and “true non-duplicates”. We visualize these counts in Fig. 4. Of the 1,000 most similar image pairs in HAM10000, we discover 18 “missed duplicates” image pairs that were not accounted for in the metadata, which are visualized in Fig. 5. Moreover, the fraction of “true non-duplicates” keeps monotonically increasing as we look at intervals of 100 image pairs, going from 0% (0 “true non-duplicates” out of 2 duplicate pairs) in the top 100 most similar pairs to 100% (64 “true non-duplicates” out of 64 duplicate pairs) in the 501st to the 600th most similar pairs. This may be explained by the narrow field of view in dermoscopic images (HAM10000 contains dermoscopic images), allowing for fewer visual cues for accurate detection of duplicates. Additionally, the lack of any “missed duplicates” after the top 500 most similar pairs (Fig. 4) means it is highly unlikely that, apart from the 18 duplicate pairs discovered (Fig. 5), there are other undetected duplicate image pairs in HAM10000.
Analysis of the top 1,000 most similar pairs in HAM10000 detected by fastdup: in intervals of 100 images, we calculate how many of these 100 purported duplicate image pairs are not already present in the HAM10000 metadata, and manually review those to detect which of these are “Missed duplicates” (i.e., pairs where the two images have different lesion IDs, but are actually images of the same lesion; Fig. 3) and those that are “True non-duplicates” (i.e., pairs where the two images have different lesion IDs but are indeed images of different lesions; Fig. 3). For example, looking at the 301–400 range, we find that from the 301st to the 400th most similar image pairs detected by fastdup, 44 pairs contained images that did not belong to the same lesion ID according to the HAM10000 metadata. Of these 44 pairs, manual inspection revealed 3 pairs to be newly discovered “Confirmed Duplicates”, whereas the remaining 41 pairs were images of different lesion and were therefore “False Positives”. There were 18 confirmed duplicate image pairs detected in HAM10000 and they have been visualized in Fig. 5.
Visualizing the 18 “Missed duplicates” (Fig. 3) in HAM10000 obtained through the analysis of the top 1,000 most similar image pairs (Fig. 4). These 18 pairs of images (image IDs along the horizontal axis) should belong to different lesions (lesion IDs along the vertical axis) according to the metadata, but manual review shows that both images in these pairs belong to the same lesions, and are thus, duplicate image pairs. Images from HAM10000 are licensed under CC BY-NC 4.015.
Figure 6 shows an interesting discovery in the manual review of the highly similar image pairs. We found two instances of image pairs with high similarity scores, but manual inspection confirmed that the lesions had minor morphological differences. HAM10000 contains images acquired during follow-up clinical visits15, and it is possible that these highly similar image pairs are images of the same lesion acquired at different time durations.
Two image pairs (image IDs along the horizontal axis) that have a high visual similarity but belong to different lesion IDs (vertical axis) according to the HAM10000 metadata. Upon closer inspection, the images are near duplicates but exhibit inconspicuous differences, and are possibly images of the same lesion acquired at different times. Images from HAM10000 are licensed under CC BY-NC 4.015.
Next, we check of errors of the second kind, i.e., “false duplicates”. For all the lesion IDs that have more than 1 image per lesion, we measure image similarity between images that belong to the same lesion and manually review the 5 least similar image pairs, as visualized in Fig. 7. We find that there are no errors, meaning that all image pairs, despite their low similarities, indeed belong to the same lesion, and that visual dissimilarities can be attributed to one or more of: zoom and crop levels, rotation, flipping, and artifacts such as gel bubbles and rulers.
Visualizing the least similar image pairs that belong to the same lesion in HAM10000 to check for “False duplicates” (Fig. 3). For each lesion ID with {2, 3, 4, 5, 6} images per lesion (Fig. 1(b)), we look at pairs of images (image IDs along the horizontal axis) that belong to the same lesion (lesion ID along the vertical axis) but have the lowest similarity scores calculated using fastdup. We do this to detect whether images with mislabeled lesion IDs, since images of two different lesions that have been assigned the same lesion ID will be dissimilar and therefore have a low similarity score. In each row, we visualize the 5 least similar image pairs that share a lesion ID, and do so for lesions that have {2, 3, 4, 5, 6} images (Fig. 1(b)). We observe that all the image pairs indeed belong to the same lesion, and the low similarity score values can be easily explained by different zoom levels and/or geometric transformations (e.g., rotation and flipping). Images from HAM10000 are licensed under CC BY-NC 4.015.
Of these 18 newly discovered duplicate image pairs in HAM10000, 7 pairs leak across partitions: train-test: 5 images (5 lesions) and train-valid: 2 images (2 lesions). We correct this data leakage by moving both images in each of these 7 pairs to the train partition.
We name this “corrected” dataset version as DermaMNIST-C. Furthermore, the relative diagnosis-wise distribution of DermaMNIST-C across partitions is quite similar to that of the original DermaMNIST (Fig. 1(c)). We benchmark DermaMNIST-C using DermaMNIST’s publicly available disease classification codebase, repeating all experiments 3 times for robustness, and the results are presented in Table 1.
Results on 224 × 224 resolution
DermaMNIST is created by resizing images from HAM10000’s original 600 × 450 spatial resolution to MNIST-like 28 × 28 resolution using (bi)cubic spline interpolation. However, for their classification benchmark experiments on the 224 × 224 resolution, instead of resizing the original images to 224 × 224, the authors38,39 upsample the low-resolution 28 × 28 to obtain the 224 × 224 using nearest neighbor interpolation (“224 (resized from 28)”; verifiable through their source code70). This, unsurprisingly, leads to quite blurry images, given the unrecoverable information lost when downsampling from the original 600 × 450 to 28 × 28 and leads to significant loss of detail (e.g., dermoscopic structures and artifacts) in the images used to train models on 224 × 224 images (Fig. 2). Our approach of directly downsampling from the original high-resolution to 224 × 224 to create DermaMNIST-C and DermaMNIST-E, used when reporting results in Table 1, yields conspicuously more detailed outputs (Fig. 2).
Extending DermaMNIST
Finally, although DermaMNIST-C is a good light-weight dataset of choice for evaluating machine learning models on dermatological tasks and for educational purposes, as MedMNIST intended to be, the quantitative results of DermaMNIST-C (and DermaMNIST for that matter) perhaps paint a deceptively optimistic picture of the state of automated dermatological diagnosis models. We propose a more challenging extension of DermaMNIST named DermaMNIST-E. The original DermaMNIST and its corrected version DermaMNIST-C are based on HAM10000, which was used as the training partition for the ISIC Challenge 2018. However, the ISIC Challenge 2018 had, apart from the 10,015 training images from HAM10000, separate validation and testing partitions containing 193 and 1,512 images, respectively. Therefore, we create DermaMNIST-E with DermaMNIST as training set and ISIC 2018 validation and test partitions as validation and testing sets, respectively. Although the official testing partition of ISIC 2018 contained 1,512 images, we remove one image known as the “easter egg” (ISIC_0035068)68, resulting in a total of 1,511 images. While the resulting diagnosis distribution across partitions for DermaMNIST-E is similar to that of DermaMNIST-C and DermaMNIST (Fig. 1(c)), our benchmark results on DermaMNIST-E (Table 1) show that it is indeed a more challenging dataset and is guaranteed to be void of any data leakage. It should be noted that the DermaMNIST-E dataset is almost the same as the official partitions of the ISIC 2018 Challenge data with 2 distinctions: the images in DermaMNIST-E are resized (28 × 28 or 224 × 224) and the “easter egg” image has been removed from the testing partition.
A summary of the three datasets: DermaMNIST, DermaMNIST-C, and DermaMNIST-E is presented in Table 4, listing a brief description and statistics of the datasets.
Fitzpatrick17k
Released in 2021, Fitzpatrick17k is one of the largest publicly available datasets of clinical skin disease images. The large number of skin diseases covered (114), the in-the-wild nature of the images therein, and the availability of associated and diverse Fitzpatrick skin tone (FST) labels71 make it an immensely valuable dataset for skin image analysis research. However, unlike DermaMNIST, which is collected from clinical visits and whose labels were confirmed, Fitzpatrick17k was curated from 2 publicly available online dermatological atlases: DermaAmin72 (12,672 images) and Atlas Dermatologico73 (3,905 images). As such, the diagnosis labels of these images are not confirmed, through histopathology or otherwise. The authors conducted a small-scale study on only 3.04% of the entire dataset (504 out of 16,577 images) where 2 board-certified dermatologists assessed the diagnoses of the images, and the consensus was that only 69.0% of the images were clearly diagnostic of the disease label and, more importantly, 3.4% of the images were mislabeled. This is problematic since Fitzpatrick17k has been used to train models for high-stakes applications such as model explainability74,75, trustworthiness76, skin tone detection77, model calibration78 and fairness79,80,81,82,83,84. Fitzpatrick17k has also been used for for training and evaluating large vision-language models85,86,87,88,89, visual question answering90,91, clinical decision support for differential diagnosis92, generative modeling93,94,95, federated learning96, and for creating a derivative dataset: SkinCon97. Pakzad et al.80 previously highlighted the existence of erroneous and wrongly labeled images in Fitzpatrick17k, and for these reasons, we investigate the extent of labeling inaccuracy in this dataset.
Data duplication and leakage
To investigate the presence of duplicates in Fitzpatrick17k, we use fastdup to calculate inter-image embedding similarity scores \({\mathscr{S}}({x}_{i},{x}_{j})\) for all \(\left(\begin{array}{c}16,577\\ 2\end{array}\right)\) pairs of images (xi, xj). These are shown as a 16,577 × 16,577 similarity matrix in Fig. 8, and we can clearly see that there are several pairs with high similarities, denoted by darker shades, spread throughout the dataset. For subsequent analyses, we restrict ourselves to pairs with a high similarity by setting thresholds to \({\mathscr{S}}\ge \tau ;\,\tau \in \{0.90,0.95\}\). The distributions of image embedding pairs with these similarity thresholds are shown in Fig. 9(a,b), respectively. Note that there are 6,622 and 1,425 image embedding pairs with similarity scores greater than 0.90 and 0.95, respectively. Manual verification by a human reviewer of these 1,425 image pairs, whose embeddings had similarities greater than 0.95, revealed that 98.39% of these images (1,402 images) were indeed duplicates, with 16 pairs (1.12%) being false positive and 7 pairs (0.49%) being ambiguous. A second reviewer agreed with 1,419 labels of the first reviewer (99.58% match), exhibiting a near-perfect agreement98,99 (Cohen’s kappa κ = 0.87). Due to the publisher’s human data policy, we are unable to visualize these samples in the published manuscript, and instead direct the readers to our arXiv pre-print100 for additional visualizations and accompanying descriptions, where we visualize some of these pairs, categorizing them according to traits exhibited by the pairs. Since the filenames of the images in Fitzpatrick17k are of the format {MD5hash}.jpg, for each image, we also display their diagnosis abbreviation, their FST label (set to ‘N/A’ when the FST label is missing), and a truncated MD5 hash to uniquely identify the images. Notice that duplicate image pairs exist because of:
-
different crop/zoom levels,
-
different illumination setups,
-
different image resolutions, and
-
simple geometrical transformations (e.g., mirroring).
Inter-image similarity matrix (16, 577 × 16, 577) computed and visualized for all pairs of images in the Fitzpatrick17k dataset, where image pairs with higher similarity are represented by darker colors. Note how there are several regions of dark-colored pairs, indicating the presence of potential duplicates in the dataset.
Visualizing the distributions of duplicates in Fitzpatrick17k, filtered by different combinations of criteria. Total counts are inset in each plot. \({{\mathscr{S}}}_{0.90}\) and \({{\mathscr{S}}}_{0.95}\) denotes pairs with similarity scores of at least 0.90 and 0.95 respectively. \(\widehat{{\mathscr{D}}}\) denotes pairs that differ in their diagnoses labels. \({\widehat{{\mathscr{F}}}}^{\ge 1}\) and \({\widehat{{\mathscr{F}}}}^{ > 1}\) denotes pairs that ‘differ in their FST labels by 1’ versus ‘by more than 1’, respectively. Best viewed online.
Worryingly, duplicate image pairs containing multiple disjoint objects of interest or multiple people also exist, making it difficult to determine to which of these the diagnosis and the FST labels apply. Finally, several duplicate pairs with more than one of these issues were also detected.
For a more detailed duplicate detection, we employ another Python library, cleanvision101, to further assess the dataset. Aside from the duplicate image pairs found by fastdup based on our similarity threshold of \({\mathscr{S}}\ge 0.90\), the cleanvision analysis found 19 more duplicate pairs that have slightly lower inter-image similarity (\(0.85\le {\mathscr{S}} < 0.90\)), primarily because of the large difference in spatial resolutions between duplicate pairs.
Unfortunately, data duplication in Fitzpatrick17k is not limited to image pairs only. We use fastdup to cluster images whose intra-cluster image similarity is greater than 0.90, i.e., clusters of images where the mean similarity for all the image pairs is greater than 0.90. We consider clusters of at least 3 images, since those with 2 images (i.e., duplicate images pairs) are already covered in our analysis of duplicate pairs. Mathematically, we find all image clusters {x1, x2, …, xN}; N ≥ 3 such that \(\frac{1}{\left(\begin{array}{c}N\\ 2\end{array}\right)}{\sum }_{i=1}^{N}{\sum }_{j=i+1}^{N}{\mathscr{S}}({x}_{i},{x}_{j}) > 0.90\). Manual verification of the clustering outputs yielded 139 image clusters, with 3.71 ± 1.11 images per cluster on average. Visual inspection of these image clusters (we direct the readers to our arXiv pre-print100 for these visualizations since they could not be put in the published manuscript), shows that these image clusters exhibit similar traits as the duplicate pairs, i.e., the images in each cluster are one or more of: exact matches, zoomed-in or cropped-out duplicates, duplicates with different illumination setups (captured with and without camera flash), or acquired at slightly different viewing angles. Finally, we merge the results of duplicate pairs with duplicate clusters, forming larger clusters as they are discovered. This results in some large duplicate image clusters with as many as 10 images in a cluster, and a total of 2,297 clusters with 2.18 ± 0.66 images per cluster on average.
Mislabeled diagnosis and FST labels
In addition to the presence of duplicates, Fitzpatrick17k contains images with mislabeled diagnoses and FSTs. For a more concrete estimation of the extent of mislabeling, we use similarity thresholds of 0.90 (denoted by \({{\mathscr{S}}}_{0.90}\)) and 0.95 (denoted by \({{\mathscr{S}}}_{0.95}\)). We report the number of image pairs that have a similarity higher than {0.90, 0.95} but differ in their labels. Further, given the subjectivity of FST labels, Groh et al.20 evaluated the accuracy of human annotations (HA) to the gold standard (GT) subset using two metrics: accuracy and “off-by-one” accuracy, where the latter considers an annotation to be correct if \(| {{\mathscr{F}}}_{{\rm{HA}}}-{{\mathscr{F}}}_{{\rm{GT}}}| \le 1\). Similar to previous works that accounted for this “off-by-one” margin77,80, we count similar images that differ by at least 1 (\({\widehat{{\mathscr{F}}}}^{\ge 1}\)) and those that differ by strictly more than 1 FST (\({\widehat{{\mathscr{F}}}}^{ > 1}\)). Visualizations in our arXiv pre-print100 show sample pairs with very high inter-image similarity (\({\mathscr{S}} > 0.95\)) that differ in diagnoses and FST by 1 and more than 1, respectively. Fig. 9 shows the distributions of duplicate image pairs filtered by one or more of: their similarity scores, whether their diagnoses are different, and whether and how much their FST scores differ by. For image pair similarity thresholds of [0.90; 0.95], there are [2498; 93] image pairs that differ in their diagnoses (\(\widehat{{\mathscr{D}}}\)). [4030; 803] image pairs differ in their FST labels (\({\widehat{{\mathscr{F}}}}^{\ge 1}\)), while [1236; 199] pairs strictly differ in their FST labels (\({\widehat{{\mathscr{F}}}}^{ > 1}\)). [4947; 841] images pairs differ in either their diagnosis or in their FST label (\(\{\widehat{{\mathscr{D}}}\cup {\widehat{{\mathscr{F}}}}^{\ge 1}\}\)), and [3172; 277] differ in their diagnosis or strictly differ in the FST label (\(\{\widehat{{\mathscr{D}}}\cup {\widehat{{\mathscr{F}}}}^{ > 1}\}\)). Finally, there are [1581; 55] and [562; 15] image pairs for the \(\{\widehat{{\mathscr{D}}}\cap {\widehat{{\mathscr{F}}}}^{\ge 1}\}\) and \(\{\widehat{{\mathscr{D}}}\cap {\widehat{{\mathscr{F}}}}^{ > 1}\}\) categories, respectively.
Erroneous images
In a recent study, Pakzad et al.80 reported the presence of erroneous or outlier non-skin images in Fitzpatrick17k. Using an outlier detection approach based on distance to the nearest neighbors in the embedding space, we rank images in the dataset based on their probability of being an outlier. Sample outliers include non-dermatological imaging modalities (e.g., histopathology, radiology, microscopy, fundus), images of plants (leaves, trees) and animals (e.g., rodents, bugs, poultry), etc. Worryingly, Fitzpatrick17k does not contain information regarding which images are non-dermatological, which consequently impacts the training and evaluation of models.
Non-standardized data partitioning
The Fitzpatrick17k benchmarks by Groh et al.20, as well as several works that followed75,79,82,84, also suffer from another major problem: the lack of a strictly held-out test partition. For all their skin condition prediction experiments, the authors only used a training and a validation set, and used the terms “validation” and “testing” interchangeably in the paper. This can also be verified in their accompanying code implementation, where the data partitions used to select the best epoch during training102 (“the epoch with the lowest loss on the validation set”) and to report the final results103 are the same. This violates the fundamental rules of machine learning model training and evaluation, where the validation and the testing partitions must be separate disjoint sets, and the former is used for choosing the best performing model during training and hyperparameter selection, while the latter is reserved only for the final model evaluation and is never used during training.
Correcting Fitzpatrick17k
In light of the numerous aforementioned issues with Fitzpatrick17k: data duplication, conflicting labels, the presence of erroneous images, and the absence of a well-defined test partition, we attempt to clean up Fitzpatrick17k and present a smaller, yet more reliable dataset. Specifically, we remove clusters of duplicates (this includes duplicate pairs), keeping one image from each cluster if there are no conflicting diagnosis or FST labels within the cluster, i.e., a “homogenous cluster”. Next, we remove the erroneous images from the dataset and refer to this “cleaned” version of Fitzpatrick17k as Fitzpatrick17k-C.
In the absence of standardized dataset partitions, researchers who used Fitzpatrick17k for their models had to resort to generating their own splits74,79,80, making it very hard for models across papers to be compared. To resolve this, we present standardized training, validation, and testing partitions for Fitzpatrick17k-C for the skin image analysis community to use, obtained by splitting Fitzpatrick17k-C in the ratio of 70:10:20, stratified on the diagnosis labels. Table 4 summarizes the two datasets: Fitzpatrick17k and Fitzpatrick17k-C, listing the number of images in their respective partitions.
Finally, we also provide benchmarks for Fitzpatrick17k-C using all the different experimental settings proposed by Groh et al.20 in Table 2. We perform a hyperparameter search for each experimental setting over the space of optimizers ({Adam, SGD}), learning rate ({1e − 2, 1e − 3, 1e − 4}), and number of training epochs ({20, 50, 100, 200}), and list the number of images in the training, validation, and testing partitions. For added robustness, we repeat each experiment using 3 random seeds. We also observed that using one setting’s optimal hyperparameter choices to evaluate another setting’s test partition does not considerably degrade the classification performance (Table 3).
Discussion
In this paper, we examine the data quality of three popular and large skin image analysis datasets: DermaMNIST from the MedMNIST dataset and its source HAM10000 dataset (10,015 dermoscopic images of skin lesions) and Fitzpatrick17k (16,577 clinical images of skin diseases). For DermaMNIST, we investigate the extent of data leakage across its training, validation, and testing partitions, and propose corrected (DermaMNIST-C) and extended (DermaMNIST-E) versions. We conducted benchmark evaluations using multiple methods and compare the results to those of DermaMNIST across all datasets. Table 4 For Fitzpatrick17k, we perform a systematic analysis encompassing data duplication, mislabeling of diagnosis and Fitzpatrick skin tone labels, identification of erroneous images, as well as highlighting the use of non-standard data partitions. Finally, we propose a cleaned version of the dataset with standardized partitions called Fitzpatrick17k-C, and release the corresponding updated benchmarks.
DermaMNIST
Data leakage and benchmarks
The extent of data leakage in DermaMNIST emanating from improper data partitioning is quite severe, with 1,006 of the 7,470 unique lesions (~13.47%) in the dataset being present in more than 1 partition (Fig. 1(c)). Patient-level stratification, or in this case, lesion-level stratification, is crucial to ensure that the model does not “see” the lesions in the held-out test set while training. Consequently, the lack of such a stratification implies that the corresponding benchmark classification results are not truly reflective of the models’ generalization capability and, therefore, we strongly suspect that the models’ performance could be inflated. On the other hand, since most works use the entire HAM10000 dataset for training instead of splitting it like in the case of DermaMNIST, undetected duplicates in HAM10000 are arguably a less severe issue.
However, a word of caution on comparing the benchmark results of DermaMNIST and DermaMNIST-C (Table 1): the two datasets do not share the same test partitions. Specifically, DermaMNIST-C’s test set was obtained from DermaMNIST’s test set by removing all images whose lesion IDs were present in the test set, and therefore, the former is a subset of the other (i.e., DermaMNIST-C test set ⊂ DermaMNIST test set). Similarly, as shown in Fig. 1(c), the training set of DermaMNIST-C is larger than that of DermaMNIST. For these reasons, contrary to the assumption that fixing data leakage in DermaMNIST should decrease performance benchmarks, we emphasize that performance benchmarks for DermaMNIST and DermaMNIST-C should not be compared since these models have been trained and evaluated on dissimilar partitions.
A more challenging dataset
While HAM10000 is indeed a valuable dataset for skin image analysis research, models trained on HAM10000 do not necessarily perform well when evaluated on other skin lesion image datasets104. This was also observed by the organizers of the ISIC 2018 Challenge65 where they relied on external testing data for ranking the submissions ("multipartition test sets containing data not reflected in training dataset are an effective way to differentiate the ability of algorithms to generalize”). Therefore, following the ISIC 2018 Challenge, we use the images from the Challenge’s validation and testing partitions, resized to 28 × 28 and 224 × 224, to create the validation and testing partitions, respectively, of the newly proposed DermaMNIST-E dataset, thus allowing for a more robust assessment of the skin lesion diagnosis models trained on this dataset.
Incorrect scaling of images
Another issue with the DermaMNIST benchmarks was the use of 28 × 28 images to report the results on 224 × 224 resolutions. As we show in Fig. 2, the information lost when downsampling from 600 × 450 to 28 × 28 is quite significant, and it is impossible to recover this when upsampling from 28 × 28 to 224 × 224. This can also be observed in the quantitative results (Table 1): intuitively, given that we ensure that the models do not overfit, we would expect that a model with larger capacity (ResNet-50) trained on high- resolution images (224 × 224) should perform better than a lower capacity model (ResNet-18) trained on lower resolution images (28 × 28). However, this is not the case with DermaMNIST results, where ResNet-18/28 × 28 models perform [AUC; ACC] better than ResNet-50/224 × 224 models: [0.917; 0.735] versus [0.912; 0.731]. On the other hand, with DermaMNIST-C and DermaMNIST-E, ResNet-50/224 × 224 models do perform better than their ResNet-18/28 × 28 counterparts.
Fitzpatrick17k
Duplicate detection and guarantees
Because of the large scale of the Fitzpatrick17k dataset (16,577 images), manual review of images to verify duplicates is virtually impossible due to the huge combinatorial space: there are \(\left(\begin{array}{c}16,577\\ 2\end{array}\right)=137\) million pairs of images. Even worse, if we want to verify triplicates (i.e., 3 images that are copies of one another), we would have to review \(\left(\begin{array}{c}16,577\\ 3\end{array}\right)=759\) billion clusters of 3 images, and the number keeps growing as the size of the clusters being reviewed increases. Therefore, we rely on automated methods for duplicate detection followed by a manual review of duplicates above a reasonable similarity threshold, and our manual review results confirmed a near-perfect agreement with the algorithm’s results. Our experiments showed that using a second method of duplicate detection (cleanvision) was able to discover an additional 19 pairs of duplicates. These pairs were also detected by fastdup, but their similarity scores fell just short of the chosen thresholds of 0.90 and 0.95, and therefore were absent from our manual review of fastdup’s results. Our dataset cleaning pipeline, publicly available on GitHub105, is highly modular and configurable, allowing users to adjust similarity thresholds to exclude and use multiple duplicate lists (fastdup and cleanvision) for cleaning, decide whether they want to remove duplicate clusters altogether or retain one representative image from each cluster, decide whether they want to remove images with unknown FST labels, and decide which outliers to exclude based on a similarity threshold. While we can claim with a high degree of certainty that the new cleaned dataset Fitzpatrick17k-C will be devoid of any duplicates, the large scale of manual review required makes it nearly impossible to guarantee it.
Dealing with conflicting diagnosis and FST labels
Several duplicates in Fitzpatrick17k, despite being near identical copies, do not share the same diagnosis labels. This affects the training and evaluation of models, since a model can be incorrectly penalized for its prediction because of the conflicting labels. However, correcting these labels, so that copies of the same image have the same diagnosis label, requires a domain expert (i.e., a dermatologist) to go through all the images and confirm and correct their labels. Unfortunately, even if such an endeavor were to be undertaken, these diagnoses will not be histopathology-confirmed and the accuracy of diagnoses confirmed through images alone (a scenario similar to “store-and-forward” teledermatology) is expected to be lower than those confirmed through in-person patient visits106,107,108.
Another approach to resolving the diagnosis label conflicts could be through mapping the diagnoses in Fitzpatrick17k to the World Health Organization (WHO) International Classification of Diseases, Eleventh Revision (ICD-11)109, and if multiple diagnoses belong to the same “parent”, their label conflict can be resolved by assigning the “parent”’s label to both. However, when mapping the diagnoses in Fitzpatrick17k to using the ICD-11 Browser110 we ran into the following issues:
-
Several diagnosis labels did not yield any matches (e.g., “acquired autoimmune bullous diseaseherpes gestationis”, “nematode infection”, “neurotic excoriations”, “pediculosis lids”).
-
There were diagnosis labels where one of the labels has an entry in ICD-11, but other seemingly related labels do not (e.g., “basal cell carcinoma” exists in ICD-11, but possibly related “basal cell carcinoma morpheiform” and “solid cystic basal cell carcinoma” do not).
-
There were labels where we found near but not exact matches (e.g., the label “erythema annulare centrifigum” in Fitzpatrick17k does not have an exact match, but ICD-11 contains “Erythema annulare”; other partial matching [Fitzpatrick17k; ICD-11] labels include ["hidradenitis”; “Hidradenitis suppurativa”], ["fixed eruptions”, “Fixed drug eruption”], ["lupus subacute”, “Subacute cutaneous lupus erythematosus”], ["porokeratosis actinic”, “Disseminated superficial porokeratosis actinic”], etc.
Overall, 8 diagnosis labels had no matches in ICD-11. An additional 29 diagnoses yielded partial matches to entries in ICD-11, and therefore could not be reliably mapped to a single entry. For these diagnoses, i.e., those that had partial or no matches in ICD-11, we expanded our search to the ICD-11 Coding Tool Mortality and Morbidity Statistics (MMS)111 and the ICD-11 Classification of Dermatological Diseases112,113, but this did not resolve any issues.
Additionally, we also carried out a similar diagnosis lookup on the Systematized Medical Nomenclature of Medicine - Clinical Terminology (SNOMED-CT), a comprehensive clinical global healthcare terminology that may better represent medical vocabulary than ICD-11114. Although we were able to find more matches to SNOMED-CT Identifiers115 (SCTIDs) than to entries in ICD-11 and its derivatives, it was still not perfect: we ended up with 17 and 3 diagnosis labels with partial and no matches, respectively. We make the results of our Fitzpatrick17k diagnosis-to-{ICD-11 classification code, SCTID} mapping publicly available online.
As a recommendation for the future datasets containing clinical images of skin diseases, we believe that adding either ICD classification codes or SNOMED-CT Identifiers (SCTIDs) for the disease labels would be a helpful addition to the metadata, and would greatly enhance the usability of such datasets for hierarchical diagnosis methods7,8,9.
Similar conflicts were observed for FST labels of images, where images with very high similarity varied in their FST labels, sometimes by as much as 4 tones (Fig. 9). A potential solution to this would be to re-assess the images with conflicting FST labels. This would involve either obtaining manual and verified healthy skin segmentation masks or using an automated healthy skin segmentation method, followed by mapping the skin tone estimated using these healthy skin pixels to an FST label. However, as noted by Groh et al.20, collecting manual segmentation masks is expensive, and automated skin segmentation approaches suffer from their own set of challenges116,117, including but not limited to susceptibility to non-standardized illumination, the presence of multiple objects, and/or low quality images (we direct the readers to our arXiv pre-print100 which contains several more visualizations which could not make it to the publisher’s version of the paper). The presence of diffuse pathologies such as “acne”, “acne vulgaris”, and “disseminated actinic porokeratosis”, where the diseased regions of the skin are spread over a wide region and the exact boundaries between diseased and healthy regions are not well-defined, further compounds the task for any automated healthy skin detection approach.
Because of these intractable issues, we make the decision of removing duplicate images that have conflicting diagnosis or FST labels. While we realize that relabeling might be able to solve these label conflicts, we also acknowledge the enormous time, effort, and expenses such an endeavor would entail.
Updated benchmarks
As discussed above, Groh et al.20 used non-standardized dataset partitions for training and evaluating their models for benchmarking, the most notable issue being the absence of a well-defined and held-out test set, thus violating one of the most basic rules of machine learning. The use of the same dataset partition for validation, i.e., choosing the best performing models, and for testing ensures that the reported performance is the best possible, but it is a poor reflection of the models’ generalization capabilities, since the models have been overfit to the testing data. This issue, coupled with the presence of duplicates in Fitzpatrick17k, which would inevitably lead to data leakage across partitions, is the reason for the difference between Fitzpatrick17k model benchmarks by Groh et al. and our Fitzpatrick17k-C benchmarks (Table 2). For the “Verified” experiments (i.e., “testing on the subset of images labeled by a board-certified dermatologist as diagnostic of the labeled condition and training on the rest of the data”20), the number of training images for Fitzpatrick17k decreased from 16,229 to 10,060, meaning the models were trained on considerably fewer images. On the other hand, the number of testing images in Fitzpatrick17k-C decreased by ~ 38% (from 348 in Fitzpatrick17k to 215) after dataset cleaning, implying that 133 images were duplicates in Fitzpatrick17k’s testing partition, which explains why their benchmark results were inflated. Similar trends can be observed across all the benchmark experiments: the number of training images for “Source A”, “Source B”, and “Fitz 1-2 & 5-6” experiments nearly halved after dataset cleaning, and the number of both training and testing images for the “Fitz 1-4” experiments decreased by more than 50%.
We believe that the newly proposed Fitzpatrick17k-C along with the well-defined and disjoint training, validation, and testing partitions, the benchmarks, and the publicly available training and evaluation code will help other researchers better utilize the dataset and make comparison across methods easier and standardized.
Comparison to other works
One of the first works on analyzing skin image analysis datasets by Abhishek34 focused on ISIC Challenge datasets for the skin lesion segmentation task, specifically on the challenges from 2016, 2017, and 2018. They found these datasets to have considerable overlap across their training partitions, and this can be attributed to all of them being subsets of the ISIC Archive118. Their analysis, however, was limited to overlap detection simply based on the filenames, since all the filenames in the ISIC Archive follow the template ISIC_{ISIC identifier}.jpg. Specific to Fitzpatrick17k, Pakzad et al.’s work80 was the first to mention the presence of “erroneous and wrongly labeled images”, and how the images therein have non-standard illumination and camera perspectives, highlighting the need for cleaning Fitzpatrick17k. Groger et al.37 conducted a data quality analysis on the presence of “irrelevant samples”, near duplicates, and label errors in 6 datasets. They used a self-supervised DL-based method for generating rankings of images potentially containing data quality issues, following which, 3 experts including a board-certified dermatologist manually reviewed the images and answered a questionnaire. The authors also noted that non-expert annotations may be sufficient for confirming duplicate images. Vega et al.36 analyzed a popular and well-cited monkeypox skin image dataset, and found it to contain “medically irrelevant images”. They discovered that the images were extracted from online repositories through web-scraping and lacked medical validation. Finally, their experiments showed that the claims made by the dataset’s authors about the utility of the images might not have been true, since a model trained by Vega et al. on “blinded” images (i.e., images where the regions of interest related to the disease were covered by black rectangles) was also able to accurately classify the diseases.
The closest dataset analysis to our work, both in the scale of the analyses and the sizes of the datasets analyzed, is the work by Cassidy et al.35 on the ISIC Challenge datasets from 2016 through 2020, where the authors employed a multi-step duplicate removal strategy. First, similar to our previous work34, they removed duplicates across dataset partitions based on filenames. Next, they used the following tools and methods on “a random selection of training images for 72 hours”: (a) a Python library called ImageHash to measure similarity based on several hashing methods (average, perceptual, difference, and wavelet), (b) mean-squared error (MSE)-based image similarity detection, (c) structural similarity index measure (SSIM)-based image similarity detection, and (d) cosine similarity-based detection. Finally, they used a Python library called FSlint to detect duplicates based on MD5 and SHA-1 file checksum signatures.
The analyses of the three datasets in this paper: DermaMNIST, HAM10000, and Fitzpatrick17k arguably goes beyond those presented in these previous works. For DermaMNIST, since we were able to request the exact training-validation-testing splits’ filenames, we could simply cross-reference those to the publicly available HAM10000 metadata to detect and correct data leakage. Our analysis of Fitzpatrick17k is much more in-depth and involved: we start with finding similar images and erroneous images in the embeddings’ latent space, followed by a manual non-expert review to confirm the duplicates, and then coalesce duplicates to form larger clusters wherever applicable. The filenames in Fitzpatrick17k follow the template <MD5_hash>.jpg, meaning that all the images have a unique MD5 checksum, which in turn implies that file checksum-based duplicate detection would not be possible. The Python library cleanvision that we use for duplicate detection already relies upon ImageHash for hashing-based similarity measurement. Measuring similarity scores in the embedding space is arguably superior to doing so in the input space (e.g., MSE, SSIM-based duplication checks by Cassidy et al.35), since a latent representation goes beyond pixel-level information and is able to better capture semantic information about the image as a whole. Finally, because of the large scale of Fitzpatrick17k (16,577 images), manual review by experts become cost prohibitive.
Data leakage and reproducibility crisis
Kapoor et al.25 presented an in-depth review of instances of data leakage in machine learning (ML) applications across 17 scientific domains, and how such leakage can lead to a reproducibility crisis in ML-based science. They also proposed a hierarchical taxonomy of leakage types, which has 8 types of leakage across 2 levels. Our analysis of DermaMNIST shows that it suffers from “[L1.4]: Duplicates in datasets” and “[L3.2]: Non-independence between train and test samples”, where the latter is a direct consequence of the former. Concerningly, Fitzpatrick17k exhibited three types of data leakage: “[L1.1]: No test set”, since it did not have a disjoint held-out test set and had duplicate images, along with the aforementioned [L1.4] and [L3.2].
In conclusion, in this paper, we examined the data quality of three popular skin image analysis datasets: DermaMNIST from the MedMNIST collection, its source HAM10000, and Fitzpatrick17k. For DermaMNIST, we investigated the extent of data leakage across its partitions and proposed two new and improved versions: a corrected dataset that does not have any leakage (DermaMNIST-C) and an extended and arguably more challenging dataset (DermaMNIST-E) that is almost the same as the ISIC 2018 Challenge dataset except the images are resized to 28 × 28 and 224 × 224 and the “easter egg” image is removed from the test partition. We also investigated the presence of duplicates in HAM10000, the source dataset for DermaMNIST, and discovered 18 new duplicate image pairs that were unaccounted for in the metadata. For Fitzpatrick17k, we conducted a systematic analysis encompassing data duplication, mislabeling of diagnosis and skin tone labels, as well as the identification of outlier images, followed by cleaning the dataset to propose a cleaned version Fitzpatrick17k-Cwith standardized training, validation, and testing partitions. We also showed how Fitzpatrick17k contains labels that do not fully align, which we recommend they should, with internationally recognized standards such as ICD-11 and SNOMED-CT, limiting its utility for hierarchical and differential diagnosis-based approaches. For all the datasets, we conducted benchmark evaluations using multiple methods, repeated 3 times for robustness. The primary objective of this paper is to raise awareness about potential data quality issues that may arise in large datasets, and how these issues can go unnoticed even in popular datasets, casting doubts on the conclusions made about the robustness and the generalizability of the models trained on these datasets. We hope this can serve as a call to action for more stringent data quality assessments. To facilitate this, we plan to make our evaluation method and code publicly available so that they can be used and extended upon for evaluating more existing and new datasets.
Methods
DermaMNIST
Detecting and correcting data leakage
We used HAM10000’s publicly available metadata, which contains image ID to lesion ID mappings, and DermaMNIST partitions’ filenames, which contain image IDs, to perform an inner-join using the Python library pandas. By doing so, we were able to obtain the list of images belonging to the same lesion that were common across train-valid, train-test, valid-test, and train-valid-test partitions.
To correct the leakage, we follow this approach: if images of a lesion ID exist in the training partition, move all images belonging to that lesion ID from validation and testing partitions to the training. For instance, for the images visualized in Fig. 1(a), since images belonging to lesion ID HAM_0002364, i.e., ISIC_0024712, ISIC_0025446, and ISIC_0030348, are present in the training partition, we move the other images of the same lesion, i.e., ISIC_0029838 and ISIC_0032042 from testing and validation partitions, respectively to the training partition.
For detecting duplicates in HAM10000 based on image embedding similarity, we used fastdup to extract a 960-dimensional embedding for all the 10,015 images. The similarities between all the \(\left(\begin{array}{c}10,015\\ 2\end{array}\right)\) pairs of images, denoted by \({\mathscr{S}}\), were calculated using the cosine similarity, where \(0\le {\mathscr{S}}\le 1\) and a higher cosine similarity score corresponds to a higher similarity. It should be noted that using cosine similarity in a lower-dimensional embedding space yields more accurate matches, as compared to using it in the high-dimensional image space, where it results in “only false positive results”, as noted by Cassidy et al.35.
Image resizing
The 28 × 28 images in DermaMNIST-C and DermaMNIST-E were obtained by resizing the original high-resolution skin lesion images from HAM10000 and other sources using bicubic interpolation. For the 224 × 224 resolution, unlike DermaMNIST which first resized the images from the original resolution to 28 × 28 using bicubic interpolation and then resizing to 224 × 224 using nearest neighbor interpolation, the images in DermaMNIST-C and DermaMNIST-E were obtained by directly resizing the original resolution images to 224 × 224 using bicubic interpolation. All image resizing operations were performed using the Python library PIL.
Model training and evaluation
We used the official MedMNIST training and evaluation code119 by Yang et al.38,39. Both the model architectures: ResNet-18 and ResNet-50, are trained for 100 epochs with the cross-entropy loss and the Adam120 optimizer with a batch size of 128. An initial learning rate of 0.001 was used, and a learning rate scheduler reduced it by a factor of 10 after the 50th and the 75th epochs. Over the training epochs, the model with the best area under the ROC curve (AUC) on the validation partition was used for testing. The reported metrics for evaluation were the AUC and the overall classification accuracy (ACC). All models were trained and evaluated using PyTorch121.
Fitzpatrick17k
Duplicate detection and manual verification
The images in Fitzpatrick17k were downloaded using the image URLs in the dataset’s metadata. For all the 16,577 images in Fitzpatrick17k, a 960-dimensional embedding was extracted using fastdup. All subsequent analysis (duplicate pairs detection, duplicate clusters detection, outlier detection) used these embeddings. The similarities between all possible pairs of images (i.e., \(\left(\begin{array}{c}16,577\\ 2\end{array}\right)\) possible pairs), denoted by \({\mathscr{S}}\), were calculated using the cosine similarity.
The original filenames in Fitzpatrick17k follow the format {MD5_hash}.jpg, which is arguably not helpful in interpreting either the diagnosis or the FST label without looking up the metadata. For an intuitive understanding of the images, we renamed them from their original filenames to a more interpretable format {diag. abbrv.}_f{FST label}_{image index}_{truncated MD5hash}.jpg. Images with missing FST labels are assigned FST 0. For example, a file originally called 0a94359e7eaacd7178e06b2823777789.jpg is renamed to ps_f1_0_0a94359e.jpg, which can be interpreted as: this is the first image (index 0) of “psoriasis” (ps) and has FST 1 (f1) and truncated MD5 hash 0a94359e. We chose to include a truncated MD5 hash of length 8 to make it easier to map new filenames to the original ones while still avoiding hash collision. A version of Fitzpatrick17k with renamed files and the metadata containing all the old and new filenames and the diagnosis labels’ abbreviations are available on Zenodo40.
To detect duplicates, all image pairs’ similarity scores were filtered based on the threshold(s), and this resulted in 6,622 unique image pairs with cosine similarity over 0.90 (i.e., \(| \{{{\mathscr{S}}}_{0.90}\}| \)) and 1,425 unique images pairs for with similarity over 0.95 (i.e., \(| \{{{\mathscr{S}}}_{0.90}\}| \)). For the manual review of duplicates, a GUI was created using the Python library tkinter that displayed the candidate duplicate image pairs detected by fastdup, and three clickable buttons: “Duplicate”, “Unclear”, and “Different”. Two annotators were tasked with independently reviewing the 1,425 duplicate images pairs each, and their responses were recorded in a CSV file.
For using cleanvision to detect duplicates, we filtered the image quality report for “near_duplicates” and “exact_duplicates”, which were 100 and 10 unique image pairs, respectively. Upon filtering these output pairs to those detected and verified using fastdup’s output, we found 19 unique duplicate pairs that were not detected using fastdup.
Interpreting fastdup duplicates visualization
Next, we describe how the table of duplicates in Fitzpatrick17k detected using fastdup, which is available on Zenodo40, can be interpreted. Consider the 21st row of the table (please see the arXiv pre-print100 version of our paper for this visualization and several others that have been omitted from this publisher’s version of the paper):
-
/rh_91/rh_f4_32_b6349594.jpg (Image 1): Image of “rhinophyma” (rh) with FST 4. This is image #33 of total 91 rhinophyma images.
-
/ro_102/ro_f2_90_0d586f26.jpg (Image 2): Image of “rosacea” (ro) with FST 2. This is image #91 of total 102 rosacea images.
-
“Blended” Image: Output of α-blending of the two images with α = 0.5.
-
Similarity: The cosine similarity between the embeddings of the two images is 0.991658.
The “distance = 0.99” watermark on Image 1 is a misnomer. Despite being called “distance” (a fastdup default), it is indeed reporting the cosine similarity value as expected.
Erroneous image detection
Using the image embeddings calculated for all the 16,577 images, we adopt the following approach for detecting erroneous images: for each image xi, we calculate its N nearest neighbors {xi1, …, xiN} in the embedding space, and their corresponding similarities \(\{{{\mathscr{S}}}_{i,i1},\cdots ,{{\mathscr{S}}}_{i,iN}\}\). An outlier would be dissimilar to other skin images in the dataset, and would therefore have low similarity scores with its nearest neighbors. To list all possible outliers in Fitzpatrick17k, we choose N = 5 and prepare tuples of \(\left({x}_{i},min\left(\{{{\mathscr{S}}}_{i,i1},\ldots ,{{\mathscr{S}}}_{i,i5}\}\right)\right)\), where \(min\left(\{{{\mathscr{S}}}_{i,i1},\ldots ,{{\mathscr{S}}}_{i,i5}\}\right)\), called the outlier score, is inversely proportional to the likelihood of an image being an erroneous image. These tuples are then sorted by the ascending order of their outlier score, i.e., the image with the lowest score and therefore the most likely to be an erroneous image is listed first, and are displayed on Zenodo40.
Correcting Fitzpatrick17k
The Fitzpatrick17k cleaning pipeline consists of the following steps:
-
1.
Similarity score-based filtering: We specify an image similarity threshold of 0.99, meaning any image whose maximum similarity score to any other image in the dataset is greater than this threshold is removed. This is done to remove the near-exact duplicates.
-
2.
Processing duplicates: We then process the duplicate pairs and clusters detected by fastdup and cleanvision to merge them into larger clusters if they exist, using the union-find algorithm122. For the final list of clusters, we check if the clusters are “homogenous”, meaning if all the duplicate images in a cluster share the same diagnosis and FST labels. If a cluster is not homogenous, we remove all the images in it from Fitzpatrick17k, but if it is, we retain the single largest image in the cluster by spatial resolution and remove the rest.
-
3.
Remove erroneous images: Finally, we remove the erroneous images detected in Fitzpatrick17k.
While we do not do this in Fitzpatrick17k-C, the data cleaning code also allows the users to remove images from Fitzpatrick17k that have missing FST labels.
Model training and evaluation
We use the official Fitzpatrick17k training and evaluation code123 by Groh et al.20 with some modifications. First, we use separate partitions for validation (i.e., picking the best performing model across training epochs based on the highest validation accuracy) and for testing (i.e., for reporting the final Fitzpatrick17k-C benchmarks in Table 2), and therefore the code is modified accordingly. Next, Next, the datasets for the seven experimental settings (i.e., “Verified”, “Random”, “Source A”, “Source B”, “FST 3–6”, “FST 1–2 & 5–6”, and “FST 1–4”) proposed by Groh et al.20 vary considerably in the number of images across training-validation-testing partitions (Table 2), we conduct a hyperparameter search for each setting. We vary the optimizer: {Adam, SGD}, the learning rate: {1e − 2, 1e − 3, 1e − 4}, and the number of training epochs: {20, 50, 100, 200}, and for each of the 7 experimental settings, we train 3 models with each of the above hyperparameter settings with different seed values, effectively training 2 × 3 × 4 × 7 × 3 = 504 models. For each experimental setting, the hyperparameter setting with the highest accuracy on the validation partition was used for the final testing, and the results are reported in Table 2. Finally, we used mixed precision training through Hugging Face Accelerate124 to speed up the training. For all the experiments, we use the same image transformations as those in Groh et al.’s work. The reported metrics for evaluation were the overall and the FST-wise classification accuracies. All models were trained and evaluated using PyTorch121 and Accelerate124.
To understand how sensitive these classification models are to the hyperparameter choices, we also evaluated models optimized on one experiment’s best hyperparameters on another experiment’s test set, and these results are presented in Table 3. The columns represent the optimal hyperparameters for each setting, and the rows represent the overall test set accuracies for all the settings when evaluated using those particular hyperparameters. We observe that varying the hyperparameters does not considerably affect the test accuracies. Additionally, the entries along the diagonal of Table 3 are the same as the overall accuracies in Table 2, since these are the test accuracies of models trained and evaluated on each particular setting’s optimal hyperparameters.
Hardware and Software Environments
All experiments were carried out on a workstation running Ubuntu 20.04 with AMD Ryzen 9 16-core 5950X CPU, 32 GB RAM, and NVIDIA RTX 3090 24 GB GPU. The following versions of software packages were used: Python 3.10, torch 1.11.0, torchvision 0.12.0, PIL 10.0.1, fastdup 1.71, cleanvision 0.3.4, and accelerate 0.9.0.
Source Datasets’ Licenses
All the source datasets are associated with the Creative Commons (CC) Licenses: DermaMNIST (CC BY-NC 4.0)125, Fitzpatrick17k (CC BY-NC-SA 3.0)123, and ISIC 2018 Challenge Datasets (CC BY-NC 4.0)126.
Data availability
All the datasets released with this work: DermaMNIST-C, DermaMNIST-E, and Fitzpatrick17k-C are publicly available on Zenodo40.
Code availability
The codebase for the analyses and the experiments in this paper is available at https://github.com/kakumarabhishek/Corrected-Skin-Image-Datasets. The results of analysis, including the visualizations, are available online on Zenodo40.
References
Schofield, J., Fleming, D., Grindlay, D. & Williams, H. Skin conditions are the commonest new reason people present to general practitioners in England and Wales. British Journal of Dermatology 165, 1044–1050, https://doi.org/10.1111/j.1365-2133.2011.10464.x (2011).
Hay, R. J. et al. The Global Burden of Skin Disease in 2010: An Analysis of the Prevalence and Impact of Skin Conditions. Journal of Investigative Dermatology 134, 1527–1534, https://doi.org/10.1038/jid.2013.446 (2014).
Flohr, C. & Hay, R. Putting the burden of skin diseases on the global map. British Journal of Dermatology 184, 189–190, https://doi.org/10.1111/bjd.19704 (2021).
Karimkhani, C. et al. Global Skin Disease Morbidity and Mortality: An Update From the Global Burden of Disease Study 2013. JAMA Dermatology 153, 406, https://doi.org/10.1001/jamadermatol.2016.5538 (2017).
Lim, H. W. et al. The burden of skin disease in the United States. Journal of the American Academy of Dermatology 76, 958–972.e2, https://doi.org/10.1016/j.jaad.2016.12.043 (2017).
World Health Organization. Cancer. https://www.who.int/news-room/fact-sheets/detail/cancer/. [Online. Accessed June 19, 2023].
Barata, C., Marques, J. S. & Celebi, M. E. Deep Attention Model for the Hierarchical Diagnosis of Skin Lesions. In 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), 2757–2765, https://doi.org/10.1109/CVPRW.2019.00334 (IEEE, Long Beach, CA, USA, 2019).
Benyahia, S., Meftah, B. & Lézoray, O. Hierarchical Approach for the Classification of Multi-class Skin Lesions Based on Deep Convolutional Neural Networks. In El Yacoubi, M., Granger, E., Yuen, P. C., Pal, U. & Vincent, N. (eds.) Pattern Recognition and Artificial Intelligence, vol. 13364, 139–149, https://doi.org/10.1007/978-3-031-09282-4_12 (Springer International Publishing, Cham, 2022).
Yu, Z. et al. Skin Lesion Recognition with Class-Hierarchy Regularized Hyperbolic Embeddings. In Wang, L., Dou, Q., Fletcher, P. T., Speidel, S. & Li, S. (eds.) Medical Image Computing and Computer Assisted Intervention - MICCAI 2022, vol. 13433, 594–603, https://doi.org/10.1007/978-3-031-16437-8_57 (Springer Nature Switzerland, Cham, 2022).
Liu, Y. et al. A deep learning system for differential diagnosis of skin diseases. Nature Medicine 26, 900–908, https://doi.org/10.1038/s41591-020-0842-3 (2020).
Asgari Taghanaki, S., Abhishek, K., Cohen, J. P., Cohen-Adad, J. & Hamarneh, G. Deep semantic segmentation of natural and medical images: a review. Artificial Intelligence Review 54, 137–178, https://doi.org/10.1007/s10462-020-09854-1 (2021).
Kiryati, N. & Landau, Y. Dataset Growth in Medical Image Analysis Research. Journal of Imaging 7, 155, https://doi.org/10.3390/jimaging7080155 (2021).
Wen, D. et al. Characteristics of publicly available skin cancer image datasets: a systematic review. The Lancet Digital Health 4, e64–e74, https://doi.org/10.1016/S2589-7500(21)00252-1 (2022).
Mirikharaji, Z. et al. A survey on deep learning for skin lesion segmentation. Medical Image Analysis 88, 102863, https://doi.org/10.1016/j.media.2023.102863 (2023).
Tschandl, P., Rosendahl, C. & Kittler, H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Scientific Data 5, 180161, https://doi.org/10.1038/sdata.2018.161 (2018).
Combalia, M. et al. BCN20000: Dermoscopic Lesions in the Wild. arXiv https://doi.org/10.48550/ARXIV.1908.02288 (2019).
Sun, X., Yang, J., Sun, M. & Wang, K. A Benchmark for Automatic Visual Classification of Clinical Skin Disease Images. In Leibe, B., Matas, J., Sebe, N. & Welling, M. (eds.) Computer Vision - ECCV 2016, vol. 9910, 206–222, https://doi.org/10.1007/978-3-319-46466-4_13 (Springer International Publishing, Cham, 2016). Series Title: Lecture Notes in Computer Science.
Yang, J. et al. Self-Paced Balance Learning for Clinical Skin Disease Recognition. IEEE transactions on neural networks and learning systems 31, 2832–2846, https://doi.org/10.1109/TNNLS.2019.2917524 (2019).
Kawahara, J., Daneshvar, S., Argenziano, G. & Hamarneh, G. 7-Point Checklist and Skin Lesion Classification using Multi-Task Multi-Modal Neural Nets. IEEE journal of biomedical and health informatics https://doi.org/10.1109/JBHI.2018.2824327 (2018).
Groh, M. et al. Evaluating Deep Neural Networks Trained on Clinical Images in Dermatology with the Fitzpatrick 17k Dataset. In 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), 1820–1828, https://doi.org/10.1109/CVPRW53098.2021.00201 (IEEE, Nashville, TN, USA, 2021).
Norori, N., Hu, Q., Aellen, F. M., Faraci, F. D. & Tzovara, A. Addressing bias in big data and AI for health care: A call for open science. Patterns 2, 100347, https://doi.org/10.1016/j.patter.2021.100347 (2021).
Sambasivan, N. et al. “Everyone wants to do the model work, not the data work”: Data Cascades in High-Stakes AI. In Proceedings of the 2021 CHI Conference on Human Factors in Computing Systems, 1–15, https://doi.org/10.1145/3411764.3445518 (ACM, Yokohama Japan, 2021).
Munappy, A. R., Bosch, J., Olsson, H. H., Arpteg, A. & Brinne, B. Data management for production quality deep learning models: Challenges and solutions. Journal of Systems and Software 191, 111359, https://doi.org/10.1016/j.jss.2022.111359 (2022).
Budach, L. et al. The Effects of Data Quality on Machine Learning Performance. arXiv https://doi.org/10.48550/ARXIV.2207.14529 (2022).
Kapoor, S. & Narayanan, A. Leakage and the reproducibility crisis in machine-learning-based science. Patterns 4, 100804, https://doi.org/10.1016/j.patter.2023.100804 (2023).
Northcutt, C. G., Athalye, A. & Mueller, J. Pervasive label errors in test sets destabilize machine learning benchmarks. In Thirty-fifth Conference on Neural Information Processing Systems Datasets and Benchmarks Track (Round 1) (2021).
Oner, M. U., Cheng, Y.-C., Lee, H. K. & Sung, W.-K. Training machine learning models on patient level data segregation is crucial in practical clinical applications. medRxiv https://doi.org/10.1101/2020.04.23.20076406 (2020).
Bussola, N., Marcolini, A., Maggio, V., Jurman, G. & Furlanello, C. AI Slipping on Tiles: Data Leakage in Digital Pathology. In Del Bimbo, A. et al. (eds.) Pattern Recognition. ICPR International Workshops and Challenges, vol. 12661, 167–182, https://doi.org/10.1007/978-3-030-68763-2_13 (Springer International Publishing, Cham, 2021).
Samala, R. K., Chan, H.-P., Hadjiiski, L. & Koneru, S. Hazards of data leakage in machine learning: a study on classification of breast cancer using deep neural networks. In Hahn, H. K. & Mazurowski, M. A. (eds.) Medical Imaging 2020: Computer-Aided Diagnosis, 39, https://doi.org/10.1117/12.2549313 (SPIE, Houston, United States, 2020).
Samala, R. K., Chan, H., Hadjiiski, L. & Helvie, M. A. Risks of feature leakage and sample size dependencies in deep feature extraction for breast mass classification. Medical Physics 48, 2827–2837, https://doi.org/10.1002/mp.14678 (2021).
Tampu, I. E., Eklund, A. & Haj-Hosseini, N. Inflation of test accuracy due to data leakage in deep learning-based classification of OCT images. Scientific Data 9, 580, https://doi.org/10.1038/s41597-022-01618-6 (2022).
Yagis, E. et al. Effect of data leakage in brain MRI classification using 2D convolutional neural networks. Scientific Reports 11, 22544, https://doi.org/10.1038/s41598-021-01681-w (2021).
Rumala, D. J. How You Split Matters: Data Leakage and Subject Characteristics Studies in Longitudinal Brain MRI Analysis. In Wesarg, S. et al. (eds.) Clinical Image-Based Procedures, Fairness of AI in Medical Imaging, and Ethical and Philosophical Issues in Medical Imaging, vol. 14242, 235–245, https://doi.org/10.1007/978-3-031-45249-9_23 (Springer Nature Switzerland, Cham, 2023).
Abhishek, K.Input Space Augmentation for Skin Lesion Segmentation in Dermoscopic Images. Master’s thesis, Simon Fraser University, Burnaby, Canada (2020).
Cassidy, B., Kendrick, C., Brodzicki, A., Jaworek-Korjakowska, J. & Yap, M. H. Analysis of the ISIC image datasets: Usage, benchmarks and recommendations. Medical Image Analysis 75, 102305, https://doi.org/10.1016/j.media.2021.102305 (2022).
Vega, C., Schneider, R. & Satagopam, V. Analysis: Flawed Datasets of Monkeypox Skin Images. Journal of Medical Systems 47, 1–16, https://doi.org/10.1007/s10916-023-01928-1 (2023).
Groger, F. et al. Towards Reliable Dermatology Evaluation Benchmarks. In Proceedings of the 3rd Machine Learning for Health Symposium, vol. 225 of Proceedings of Machine Learning Research, 101–128 (PMLR, 2023).
Yang, J., Shi, R. & Ni, B. MedMNIST Classification Decathlon: A Lightweight AutoML Benchmark for Medical Image Analysis. In 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI), 191–195, https://doi.org/10.1109/ISBI48211.2021.9434062 (IEEE, Nice, France, 2021).
Yang, J. et al. MedMNIST v2 - A large-scale lightweight benchmark for 2D and 3D biomedical image classification. Scientific Data 10, 41, https://doi.org/10.1038/s41597-022-01721-8 (2023).
Abhishek, K., Jain, A. & Hamarneh, G. Investigating the quality of DermaMNIST and Fitzpatrick17k dermatological image datasets, https://doi.org/10.5281/ZENODO.11101337 (2024).
Sportisse, A., Schmutz, H., Humbert, O., Bouveyron, C. & Mattei, P.-A. Are labels informative in semi-supervised learning? Estimating and leveraging the missing-data mechanism. In Krause, A. et al. (eds.) Proceedings of the 40th international conference on machine learning, vol. 202 of Proceedings of machine learning research, 32521–32539 (PMLR, 2023).
Kang, M. et al. Label-Assemble: Leveraging Multiple Datasets with Partial Labels. In 2023 IEEE 20th International Symposium on Biomedical Imaging (ISBI), 1–5, https://doi.org/10.1109/ISBI53787.2023.10230766 (IEEE, Cartagena, Colombia, 2023).
Chen, W., Li, C., Chen, D. & Luo, X. A knowledge-based learning framework for self-supervised pre-training towards enhanced recognition of biomedical microscopy images. Neural Networks 167, 810–826, https://doi.org/10.1016/j.neunet.2023.09.001 (2023).
Lu, C., Yu, Y., Karimireddy, S. P., Jordan, M. & Raskar, R. Federated conformal predictors for distributed uncertainty quantification. In Krause, A. et al. (eds.) Proceedings of the 40th international conference on machine learning, vol. 202 of Proceedings of machine learning research, 22942–22964 (PMLR, 2023).
Zhu, W. & Luo, J. Federated Medical Image Analysis with Virtual Sample Synthesis. In Wang, L., Dou, Q., Fletcher, P. T., Speidel, S. & Li, S. (eds.) Medical Image Computing and Computer Assisted Intervention - MICCAI 2022, vol. 13433, 728–738, https://doi.org/10.1007/978-3-031-16437-8_70 (Springer Nature Switzerland, Cham, 2022).
Kim, S., Bae, S., Song, H. & Yun, S.-Y. Re-Thinking Federated Active Learning Based on Inter-Class Diversity. In 2023 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 3944–3953, https://doi.org/10.1109/CVPR52729.2023.00384 (IEEE, Vancouver, BC, Canada, 2023).
Tang, X., Panda, A., Sehwag, V. & Mittal, P. Differentially private image classification by learning priors from random processes. In Oh, A. et al. (eds.) Advances in neural information processing systems, vol. 36, 35855–35877 (Curran Associates, Inc., 2023).
Lee, S., Lee, G., Kim, J. W., Shin, J. & Lee, M.-K. HETAL: Efficient privacy-preserving transfer learning with homomorphic encryption. In Krause, A. et al. (eds.) Proceedings of the 40th international conference on machine learning, vol. 202 of Proceedings of machine learning research, 19010–19035 (PMLR, 2023).
Xu, H., Wang, D., Sowmya, A. & Katz, I. Detection of Basal Cell Carcinoma in Whole Slide Images. In Greenspan, H. et al. (eds.) Medical Image Computing and Computer Assisted Intervention - MICCAI 2023, vol. 14225, 263–272, https://doi.org/10.1007/978-3-031-43987-2_26 (Springer Nature Switzerland, Cham, 2023).
Wang, Y. et al. MedNAS: Multi-Scale Training-Free Neural Architecture Search for Medical Image Analysis. IEEE Transactions on Evolutionary Computation 1–1, https://doi.org/10.1109/TEVC.2024.3352641 (2024).
Xu, M., Zhang, T. & Zhang, D. MedRDF: A Robust and Retrain-Less Diagnostic Framework for Medical Pretrained Models Against Adversarial Attack. IEEE Transactions on Medical Imaging 41, 2130–2143, https://doi.org/10.1109/TMI.2022.3156268 (2022).
Ahmed, S., Dera, D., Hassan, S. U., Bouaynaya, N. & Rasool, G. Failure Detection in Deep Neural Networks for Medical Imaging. Frontiers in Medical Technology 4, 919046, https://doi.org/10.3389/fmedt.2022.919046 (2022).
Yang, K. et al. Adversarial auto-augment with label preservation: A representation learning principle guided approach. In Koyejo, S. et al. (eds.) Advances in neural information processing systems, vol. 35, 22035–22048 (Curran Associates, Inc., 2022).
Abhishek, K., Brown, C. J. & Hamarneh, G. Multi-sample ζ-mixup: richer, more realistic synthetic samples from a p-series interpolant. Journal of Big Data 11, 43, https://doi.org/10.1186/s40537-024-00898-6 (2024).
Wang, J. et al. Autoencoder-based conditional optimal transport generative adversarial network for medical image generation. Visual Informatics 8, 15–25, https://doi.org/10.1016/j.visinf.2023.11.001 (2024).
Nguyen, A.-P., Moreno, D. L., Le-Bel, N., Rodríguez Martínez, M., Nguyen, A.-P., Moreno, D. L., Le-Bel, N. & Rodríguez Martínez, M. MonoNet: enhancing interpretability in neural networks via monotonic features. Bioinformatics Advances 3, vbad016, https://doi.org/10.1093/bioadv/vbad016 (2023).
Kumar, R., Romano, J., Ritchie, M. & Moore, J. Extending Tree-Based Automated Machine Learning to Biomedical Image and Text Data Using Custom Feature Extractors. In Proceedings of the Companion Conference on Genetic and Evolutionary Computation, 599–602, https://doi.org/10.1145/3583133.3590584 (ACM, Lisbon Portugal, 2023).
Das, A. M. et al. Accelerating batch active learning using continual learning techniques. Transactions on Machine Learning Research (2023).
Cherrat, E. A. et al. Quantum Vision Transformers. Quantum 8, 1265, https://doi.org/10.22331/q-2024-02-22-1265 (2024).
Lin, W. et al. PMC-CLIP: Contrastive Language-Image Pre-training Using Biomedical Documents. In Greenspan, H. et al. (eds.) Medical Image Computing and Computer Assisted Intervention - MICCAI 2023, vol. 14227, 525–536, https://doi.org/10.1007/978-3-031-43993-3_51 (Springer Nature Switzerland, Cham, 2023).
Zhang, K. et al. A generalist vision-language foundation model for diverse biomedical tasks. Nature Medicine 30, 3129–3141, https://doi.org/10.1038/s41591-024-03185-2 (2024).
Zhu, X., Hu, Y., Mo, F., Li, M. & Wu, J. Uni-med: a unified medical generalist foundation model for multi-task learning via connector-MoE. In The thirty-eighth annual conference on neural information processing systems (2024).
Di Salvo, F., Doerrich, S. & Ledig, C. MedMNIST-C: Comprehensive benchmark and improved classifier robustness by simulating realistic image corruptions, https://doi.org/10.48550/ARXIV.2406.17536 (2024).
Doerrich, S., Di Salvo, F., Brockmann, J. & Ledig, C. Rethinking Model Prototyping through the MedMNIST+ Dataset Collection, https://doi.org/10.48550/ARXIV.2404.15786 (2024).
Codella, N. et al. Skin Lesion Analysis Toward Melanoma Detection 2018: A Challenge Hosted by the International Skin Imaging Collaboration (ISIC). arXiv https://doi.org/10.48550/ARXIV.1902.03368 (2019).
ISIC. International Skin Imaging Collaboration (ISIC) 2019 Challenge (2019).
Tschandl, P. et al. Human-computer collaboration for skin cancer recognition. Nature Medicine 26, 1229-1234, https://doi.org/10.1038/s41591-020-0942-0 (2020).
Barata, C. et al. A reinforcement learning model for AI-based decision support in skin cancer. Nature Medicine 29, 1941–1946, https://doi.org/10.1038/s41591-023-02475-5 (2023).
visual layer. Github – fastdup. https://github.com/visual-layer/fastdup. [Online. Accessed 7-Jan-2024].
MedMNIST. Github – experiments/medmnist2d/train_and_eval_pytorch.py at 8b0f553. https://github.com/MedMNIST/experiments/blob/8b0f553f95ea6b5f5517e49c539952cb21c79d89/MedMNIST2D/train_and_eval_pytorch.py#L54. [Online. Accessed 7-Jan-2024].
Alipour, N., Burke, T. & Courtney, J. Skin Type Diversity: a Case Study in Skin Lesion Datasets. Current Dermatology Reports 13, 198–210, https://doi.org/10.1007/s13671-024-00440-0 (2024).
Amin AlKattash, J. DermaAmin. https://www.dermaamin.com/site/. [Online. Accessed 19-Jun-2023].
Freire da Silva, S. Atlas Dermatologico. https://www.atlasdermatologico.com.br/. [Online. Accessed 19-Jun-2023].
Abid, A., Yuksekgonul, M. & Zou, J. Meaningfully debugging model mistakes using conceptual counterfactual explanations. In Chaudhuri, K. et al. (eds.) Proceedings of the 39th international conference on machine learning, vol. 162 of Proceedings of machine learning research, 66–88 (PMLR, 2022).
Ghadiri, A., Pagnucco, M. & Song, Y. XTranPrune: eXplainability-Aware Transformer Pruning for Bias Mitigation in Dermatological Disease Classification. In Linguraru, M. G. et al. (eds.) Medical Image Computing and Computer Assisted Intervention - MICCAI 2024, vol. 15010, 749–758, https://doi.org/10.1007/978-3-031-72117-5_70 (Springer Nature Switzerland, Cham, 2024).
Yan, S. et al. Towards Trustable Skin Cancer Diagnosis via Rewriting Model’s Decision. In 2023 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 11568–11577, https://doi.org/10.1109/CVPR52729.2023.01113 (IEEE, Vancouver, BC, Canada, 2023).
Bevan, P. J. & Atapour-Abarghouei, A. Detecting Melanoma Fairly: Skin Tone Detection and Debiasing for Skin Lesion Classification. In Kamnitsas, K. et al. (eds.) Domain Adaptation and Representation Transfer, vol. 13542, 1–11, https://doi.org/10.1007/978-3-031-16852-9_1 (Springer Nature Switzerland, Cham, 2022).
Yuksekgonul, M., Zhang, L., Zou, J. & Guestrin, C. Beyond confidence: Reliable models should also consider atypicality. In Thirty-seventh conference on neural information processing systems (2023).
Du, S., Hers, B., Bayasi, N., Hamarneh, G. & Garbi, R. FairDisCo: Fairer AI in Dermatology via Disentanglement Contrastive Learning. In Karlinsky, L., Michaeli, T. & Nishino, K. (eds.) Computer Vision - ECCV 2022 Workshops, vol. 13804, 185–202, https://doi.org/10.1007/978-3-031-25069-9_13 (Springer Nature Switzerland, Cham, 2022).
Pakzad, A., Abhishek, K. & Hamarneh, G. CIRCLe: Color Invariant Representation Learning for Unbiased Classification of Skin Lesions. In Karlinsky, L., Michaeli, T. & Nishino, K. (eds.) Computer Vision - ECCV 2022 Workshops, vol. 13804, 203–219, https://doi.org/10.1007/978-3-031-25069-9_14 (Springer Nature Switzerland, Cham, 2022).
Seth, P. & Pai, A. K. Does the Fairness of Your Pre-Training Hold Up? Examining the Influence of Pre-Training Techniques on Skin Tone Bias in Skin Lesion Classification. In 2024 IEEE/CVF Winter Conference on Applications of Computer Vision Workshops (WACVW), 580–587, https://doi.org/10.1109/WACVW60836.2024.00067 (IEEE, Waikoloa, HI, USA, 2024).
Wang, J., Zhang, Y., Ding, Z. & Hamm, J. Achieving Reliable and Fair Skin Lesion Diagnosis via Unsupervised Domain Adaptation. In 2024 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), 5157–5166, https://doi.org/10.1109/CVPRW63382.2024.00523 (IEEE, Seattle, WA, USA, 2024).
Chiu, C.-H., Chen, Y.-J., Wu, Y., Shi, Y. & Ho, T.-Y. Achieve fairness without demographics for dermatological disease diagnosis. Medical Image Analysis 95, 103188, https://doi.org/10.1016/j.media.2024.103188 (2024).
Aayushman, Gaddey, H., Mittal, V., Chawla, M. & Gupta, G. R. Fair and Accurate Skin Disease Image Classification by Alignment with Clinical Labels. In Linguraru, M. G. et al. (eds.) Medical Image Computing and Computer Assisted Intervention - MICCAI 2024, vol. 15001, 394–404, https://doi.org/10.1007/978-3-031-72378-0_37 (Springer Nature Switzerland, Cham, 2024).
Kim, C. et al. Transparent medical image AI via an image-text foundation model grounded in medical literature. Nature Medicine 30, 1154–1165, https://doi.org/10.1038/s41591-024-02887-x (2024).
Akuffo-Addo, E. et al. Assessing GPT-4’s diagnostic accuracy with darker skin tones: underperformance and implications. Clinical and Experimental Dermatology 49, 1244–1245, https://doi.org/10.1093/ced/llae158 (2024).
Chen, P. et al. GMAI-MMBench: A Comprehensive Multimodal Evaluation Benchmark Towards General Medical AI, https://doi.org/10.48550/arXiv.2408.03361 (2024).
Yan, S. et al. A General-Purpose Multimodal Foundation Model for Dermatology, https://doi.org/10.48550/arXiv.2410.15038 (2024).
Zhou, J. et al. Pre-trained multimodal large language model enhances dermatological diagnosis using SkinGPT-4. Nature Communications 15, 5649, https://doi.org/10.1038/s41467-024-50043-3 (2024).
Hu, Y. et al. OmniMedVQA: A New Large-Scale Comprehensive Evaluation Benchmark for Medical LVLM. In 2024 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 22170–22183, https://doi.org/10.1109/CVPR52733.2024.02093 (IEEE, Seattle, WA, USA, 2024).
Yim, W.-w. et al. DermaVQA: A Multilingual Visual Question Answering Dataset for Dermatology. In Linguraru, M. G. et al. (eds.) Medical Image Computing and Computer Assisted Intervention - MICCAI 2024, vol. 15005, 209–219, https://doi.org/10.1007/978-3-031-72086-4_20 (Springer Nature Switzerland, Cham, 2024).
Groh, M. et al. Deep learning-aided decision support for diagnosis of skin disease across skin tones. Nature Medicine 30, 573–583, https://doi.org/10.1038/s41591-023-02728-3 (2024).
Sagers, L. W. et al. Improving dermatology classifiers across populations using images generated by large diffusion models. In NeurIPS 2022 workshop on synthetic data for empowering ML research (2022).
Lin, B. et al. SkinGEN: an Explainable Dermatology Diagnosis-to-Generation Framework with Interactive Vision-Language Models, https://doi.org/10.48550/ARXIV.2404.14755 (2024).
Mekala, R. R. et al. Synthetic Generation of Dermatoscopic Images with GAN and Closed-Form Factorization, https://doi.org/10.48550/ARXIV.2410.05114 (2024).
Alhamoud, K. et al. FedMedICL: Towards Holistic Evaluation of Distribution Shifts in Federated Medical Imaging. In Linguraru, M. G. et al. (eds.) Medical Image Computing and Computer Assisted Intervention - MICCAI 2024, vol. 15010, 383–393, https://doi.org/10.1007/978-3-031-72117-5_36 (Springer Nature Switzerland, Cham, 2024).
Daneshjou, R., Yuksekgonul, M., Cai, Z. R., Novoa, R. A. & Zou, J. SkinCon: A skin disease dataset densely annotated by domain experts for fine-grained debugging and analysis. In Thirty-sixth conference on Neural Information Processing Systems Datasets and Benchmarks Track (2022).
Cohen, J. A Coefficient of Agreement for Nominal Scales. Educational and Psychological Measurement 20, 37-46, https://doi.org/10.1177/001316446002000104 (1960).
Landis, J. R. & Koch, G. G. The Measurement of Observer Agreement for Categorical Data. Biometrics 33, 159, https://doi.org/10.2307/2529310 (1977).
Abhishek, K., Jain, A. & Hamarneh, G. Investigating the Quality of DermaMNIST and Fitzpatrick17k Dermatological Image Datasets, https://doi.org/10.48550/arXiv.2401.14497 (2024).
cleanlab. Github – cleanvision. https://github.com/cleanlab/cleanvision. [Online. Accessed 7-Jan-2024].
Groh, M. Github – fitzpatrick17k/train.py at 26d5074. https://github.com/mattgroh/fitzpatrick17k/blob/26d50745348f82a76f872ed7924361d1dccd629e/train.py#L82. [Online. Accessed 7-Jan-2024].
Groh, M. Github – fitzpatrick17k/train.py at 26d5074. https://github.com/mattgroh/fitzpatrick17k/blob/26d50745348f82a76f872ed7924361d1dccd629e/train.py#L342. [Online. Accessed 7-Jan-2024].
Yoon, C., Hamarneh, G. & Garbi, R. Generalizable Feature Learning in the Presence of Data Bias and Domain Class Imbalance with Application to Skin Lesion Classification. In Shen, D. et al. (eds.) Medical Image Computing and Computer Assisted Intervention - MICCAI 2019, vol. 11767, 365–373, https://doi.org/10.1007/978-3-030-32251-9_40 (Springer International Publishing, Cham, 2019).
Abhishek, K. Github – corrected-skin-image-datasets. https://github.com/kakumarabhishek/Corrected-Skin-Image-Datasets. [Online. Accessed 12-May-2024].
S. Levin, Y. & Warshaw, E. M. Teledermatology: A Review of Reliability and Accuracy of Diagnosis and Management. Dermatologic Clinics 27, 163–176, https://doi.org/10.1016/j.det.2008.11.012 (2009).
Gerhardt, C. A., Foels, R., Grewe, S. & Baldwin, B. T. Assessing the Diagnostic Accuracy of Teledermatology Consultations at a Local Veterans Affairs Dermatology Clinic. Cureushttps://doi.org/10.7759/cureus.15406 (2021).
Gunn, H. & Argáez, C. Asynchronous Teledermatology Consultations Using Store-and-Forward Technology: Diagnostic Accuracy, Clinical Utility, and Cost-Effectiveness. Tech. Rep. RB1523-000, Canadian Agency for Drugs and Technologies in Health (CADTH), Canada (2020).
WHO. International Classification of Diseases, Eleventh Revision (ICD-11) (2019).
WHO. ICD-11 for Mortality and Morbidity Statistics — icd.who.int. https://icd.who.int/browse11. [Online. Accessed 23-Mar-2024].
WHO. ICD-11 Coding Tool Mortality and Morbidity Statistics (MMS) — icd.who.int. https://icd.who.int/ct/icd11_mms/en/release. [Online. Accessed 23-Mar-2024].
White, J., Lui, H., Chute, C., Jakob, R. & Chalmers, R. The WHO ICD-11 Classification of Dermatological Diseases: a new comprehensive online skin disease taxonomy designed by and for dermatologists. British Journal of Dermatology 186, 178–179, https://doi.org/10.1111/bjd.20656 (2022).
WHO. ICD-11 Classification of Dermatological Diseases — icd.who.int. https://icd.who.int/dev11/l-derma/en. [Online. Accessed 23-Mar-2024].
Thun, S. & Dewenter, H. ICD-11, ICHI und SNOMED CT - was bedeuten die Systematiken für E-Health-Anwendungen? Bundesgesundheitsblatt - Gesundheitsforschung - Gesundheitsschutz 61, 812–820, https://doi.org/10.1007/s00103-018-2759-2 (2018).
International, S. SNOMED International Browser — browser.ihtsdotools.org. https://browser.ihtsdotools.org/?perspective=full&conceptId1=404684003&edition=MAIN/2024-05-01&release=&languages=en [Online. Accessed 23-Mar-2024].
Naji, S., Jalab, H. A. & Kareem, S. A. A survey on skin detection in colored images. Artificial Intelligence Review 52, 1041–1087, https://doi.org/10.1007/s10462-018-9664-9 (2019).
Nanni, L., Loreggia, A., Lumini, A. & Dorizza, A. A Standardized Approach for Skin Detection: Analysis of the Literature and Case Studies. Journal of Imaging 9, 35, https://doi.org/10.3390/jimaging9020035 (2023).
ISIC. International Skin Imaging Collaboration: Melanoma Project (2023).
MedMNIST. Github – experiments at 8b0f553. https://github.com/MedMNIST/experiments/tree/8b0f553f95ea6b5f5517e49c539952cb21c79d89. [Online. Accessed 7-Jan-2024].
Kingma, D. P. & Ba, J. Adam: A Method for Stochastic Optimization. arXivhttps://doi.org/10.48550/ARXIV.1412.6980 (2014).
Paszke, A. et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. In Proceedings of the 33rd International Conference on Neural Information Processing Systems, 8026–8037 (Curran Associates Inc., Red Hook, NY, USA, 2019).
Wikipedia contributors. Disjoint-set data structure — Wikipedia, the free encyclopedia [Online. Accessed 7-Jan-2024] (2023).
Groh, M. Github – fitzpatrick17k at 26d5074. https://github.com/mattgroh/fitzpatrick17k/tree/26d50745348f82a76f872ed7924361d1dccd629e. [Online. Accessed 7-Jan-2024].
Gugger, S. et al. Accelerate: Training and inference at scale made simple, efficient and adaptable. https://github.com/huggingface/accelerate [Online. Accessed 19-Jun-2023] (2022)
MedMNIST. MedMNIST – 18x standardized datasets for 2D and 3D biomedical image classification. https://www.medmnist.com [Online. Accessed 19-Jun-2023].
ISIC. ISIC Challenge Datasets – 2018. https://challenge.isic-archive.com/data/#2018 [Online. Accessed 19-Jun-2023].
Acknowledgements
K.A. is funded by Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery grant RGPIN 06795 and Simon Fraser University. The authors would like to thank Rui Shi for sharing the image IDs of the training, validation, and testing partitions of the DermaMNIST dataset. The authors are grateful to Prof. Rafeef Garbi for suggesting the idea of documenting and publicly disseminating these important data quality issues in a paper. The authors are also thankful to Hanene Ben Yedder and Arezou Pakzad of the Medical Image Analysis Lab for the helpful discussions, and to the NVIDIA Corporation and the Digital Research Alliance of Canada for providing the computational resources.
Author information
Authors and Affiliations
Contributions
K.A. worked on writing the code, performing the formal analysis and the experiments, and preparing the figures, with support from A.J. and G.H. K.A. and A.J. worked on data curation. K.A. worked on writing the initial draft. G.H. supervised the project and provided the resources and funding for the project. All authors contributed to the design and the evaluation of the algorithms. All authors contributed to writing, reviewing, and editing the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Abhishek, K., Jain, A. & Hamarneh, G. Investigating the Quality of DermaMNIST and Fitzpatrick17k Dermatological Image Datasets. Sci Data 12, 196 (2025). https://doi.org/10.1038/s41597-025-04382-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41597-025-04382-5
This article is cited by
-
MedNet: a lightweight attention-augmented CNN for medical image classification
Scientific Reports (2025)
-
MTAKD: multi-teacher agreement knowledge distillation for edge AI skin disease diagnosis
Scientific Reports (2025)
-
Evaluating skin tone scales for dermatologic dataset labeling: a prospective-comparative study
npj Digital Medicine (2025)
-
Mask of Truth: Model Sensitivity to Unexpected Regions of Medical Images
Journal of Imaging Informatics in Medicine (2025)