Abstract
Multiple Myeloma (MM) is a cytogenetically heterogeneous clonal plasma cell proliferative disease whose diagnosis is supported by analyses on histological slides of bone marrow aspirate. In summary, experts use a labor-intensive methodology to compute the ratio between plasma cells and non-plasma cells. Therefore, the key aspect of the methodology is identifying these cells, which relies on the experts’ attention and experience. In this work, we present a valuable dataset comprising more than 5,000 plasma and non-plasma cells, labeled by experts, along with some patient diagnostics. We also share a Deep Neural Network model, as a benchmark, trained to identify and count plasma and non-plasma cells automatically. The contributions of this work are two-fold: (i) the labeled cells can be used to train new practitioners and support continuing medical education; and (ii) the design of new methods to identify such cells, improving the results presented by our benchmark. We emphasize that our work supports the diagnosis of MM in practical scenarios and paves new ways to advance the state-of-the-art.
Similar content being viewed by others
Background & Summary
Multiple Myeloma (MM) is a hematological malignancy that originates from a clone of malignant plasma cells (mPCs) infiltrated in the bone marrow (BM)1,2. The MM diagnosis is confirmed by the presence of ≥10% of clonal plasma cells (PCs) in the bone marrow (or plasmacytoma confirmed by biopsy), and by the occurrence of end-organ damage such as hypercalcemia (serum calcium > 0.25 mmol/L, > 1 mg/dL, higher than the upper limit of normal or > 2.75 mmol/L, > 11 mg/dL), renal insufficiency (creatinine clearance < 40 mL per min† or serum creatinine > 177 μmol/L, > 2 mg/dL), anemia (haemoglobin value of > 20 g/L below the lower limit of normal, or a haemoglobin value < 100 g/L), and bone lesions (one or more osteolytic lesions on skeletal radiography, CT, or PET-CT)3.
Regarding the assessment of the 10% ratio between plasma and non-plasma cells, experts individually analyze hematological slides to identify and count these cells4, encountering several challenges even when using advanced equipment to compute them, such as: 1) manual identification of plasma cells on histological slides requires significant time and resources, potentially delaying diagnosis and treatment; 2) the identification process strongly depends on the expert’s attention and experience, leading to inconsistencies in result interpretation, thereby compromising the sensitivity and overall diagnostic accuracy; and 3) in resource-constrained communities, there is often a lack of access to trained professionals, significantly barring effective disease screening and diagnostic practices.
Therefore, considering the challenges associated with manually identifying plasma cells, we have employed a low-cost setup consisting of a microscope and a smartphone camera to create a comprehensive dataset of over 5,000 plasma and non-plasma cells manually labeled by experts.
Our dataset, PCMMD (Plasma Cells for Multiple Myeloma Diagnosis)5, is an important resource for training and validating Artificial Intelligence (AI) approaches to automate cell identification, supporting studies on MM in developing countries, and discovering approaches regarding other diseases affecting various cell types in diverse population conditions. In previous work6, plasma cells were labeled as part of a preliminary study. However, this dataset had several limitations, including a small number of cells, lack of information on other non-plasma cells, absence of patient diagnoses, and insufficient details about the cells’ nuclei, shapes, and membrane structures. In this manuscript, we address all these limitations. Moreover, the computational findings presented here serve as a benchmark, demonstrating the feasibility and effectiveness of AI-assisted diagnosis in hematological studies, with distinct contributions to the field:
-
The labeled-cell dataset assists with training new practitioners and continuing medical education, ensuring that future experts can accurately identify plasma cells;
-
Using a Deep Neural Network (DNN) model, our Machine Learning approach provides a benchmark that encourages the development of new AI methods and technologies that surpass the current state-of-the-art methods in cell identification;
-
Combining microscopes and smartphone cameras, our AI model offers a low-cost, accessible solution for diagnosing MM, making advanced diagnostic techniques available in resource-constrained settings;
-
The availability of our dataset and benchmark model support ongoing research and development in the field, promoting continuous improvement in the accuracy and efficiency of MM diagnostics.
In summary, our work improves the practical diagnosis of MM and opens new avenues for advancing the field through improved training, innovative method development, and enhanced accessibility.
Methods
The dataset was created in accordance with the relevant ethical regulations established by the Institutional Research Committee - CEP ICS (CAAE - 70193723.5.0000.5662) from the Federal University of Bahia. The research ethics committee have waived the necessity for researchers to obtain informed consent from patients.
The methodology considered in this work adapted the CRISP-DM (CRoss-Industry Standard Process for Data Mining) process, which is a standard widely adopted in Machine Learning projects7. In the first step, we defined the problem by focusing on using images from histological slides to support experts in diagnosing MM, as discussed in the previous section. The second step comprises the tasks performed to collect, preprocess, and analyze such images. In the third step, we trained a DNN model to automatically identify and count plasma and non-plasma cells. After validating the results, in the fourth step, we deployed the model to assess its capability of suggesting diagnoses to patients with known positive and negative conditions, according to experts. In the following section, we provide more details about these steps.
Collection and Preparation
This section details the second step of CRISP-DM, which focuses on collecting images extracted from bone marrow (BM) histological slides and organizing the MM dataset. In this study, a team of hematology experts, including two hematologists, two biomedical hematologists and immunologists, and one biotechnologist immunologist, selected patients with confirmed oncohematological diseases, including MM.
All these patients were diagnosed and treated by the Brazilian Public Health System (Sistema Único de Saúde - SUS)8,9,10,11. The diagnosis by immunophenotyping was carried out in the Oncohematology and Immunophenotyping sector of the Laboratory of Immunology and Molecular Biology at the Federal University of Bahia (UFBA), which provided the samples for the present study. We emphasize that the slides were coded, respecting the users’ privacy, and categorized into groups of patients with and without MM.
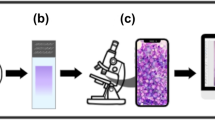
In Fig. 1, we illustrate the entire collection and preparation process. In the first stage, groups of patients with (Stage a2) and without (Stage a1) MM, who were submitted to a bone marrow aspirate procedure, thus resulting in a set of histological slides (Stage b). The BM slides were examined using a Nikon ECLIPSE CI visible light optical microscope with immersion oil, employing a 100x objective lens and a 10x ocular lens (Stage c). Observations were conducted at the smear’s feathered edge. Nucleated cells were photographed using a smartphone camera mounted on the microscope with a universal holder. Notably, our dataset comprises both mono- and multinucleated cells. In the last stage, cell images from patients with (Stage d2) and without MM (Stage d1) were individually analyzed and labeled, as depicted in the next section, to be later modeled by our DNN architectures.
(a1) Bone marrow aspirate procedure of patients without MM; (a2) Bone marrow aspirate procedure of patients with MM; (b) Wright-Giemsa (SIGMA-ALDRICH, MERCK) stained bone marrow aspirate smear slides, analyzed by the oncohematology and immunophenotyping service of the Labimuno; (c) Observation of stained slides in visible light optical microscopes and image capture by smartphone device; (d1 and d2) Identification, labeling and validation of detected non-plasma and plasma cells from patients with and without MM; and (e) Dataset organization to be later trained by DNN models.
Cell Labeling
After analyzing and validating the collected images, our expert team was divided into two groups to label all cells as plasma and non-plasma. This process was carried out using the LabelImg tool12. Each group manually labeled a separate set of images and validated the labels set by the other group.
The labeling process was performed based on cytomorphological characteristics and Wright-Giemsa staining to determine each identifiable cell type. Figure 2 shows 4 examples of images with labeled cells, highlighting plasma (in red) and non-plasma (in green) bounding boxes.
In Fig. 2(a), we show 4 non-plasma and 2 plasma cells labeled by our experts. Moreover, one may also notice a set of red cells (not labeled) as background. In Figure 2(b) and (c), it is possible to realize two artifacts not labeled for being partially cut by the top-most and left-most image limits, respectively. In this situation, experts tend to disregard such artifacts to avoid misclassification. In Fig. 2(d), there are regions, also considered artifacts, affected by the staining process. In the next section, we present more details about the final dataset.
Dataset Description
The dataset is organized into two sets of cells. The first one contains 3,546 cells, of which 54% were labeled as non-plasma and 46% as plasma cells. This set was used to train our DNN model, as detailed in the following section. Each cell was individually segmented to remove background elements, enabling the analysis of key features such as the number of nuclei, cell shapes, and membrane structures. From this segmentation process, 1,613 plasma cells and 1,927 non-plasma cells were identified and shared as complementary data.
The second set contains 2,021 cells that we have used to test the performance of our model. Besides measuring the agreement between labels provided by experts and predicted by our model, we also separated the cells according to the patients’ diagnoses, as shown in Table 1. According to the literature, the MM diagnosis depends on counting 200-500 cells in myelogram slides of bone marrow (BM) aspirate. Columns Non-Plasma Cells, Plasma Cells, and Total Cells show the number of cells in each patient’s slide. If more than 10% are plasma cells (Percentage column), it indicates a positive diagnosis. In our investigation, we have considered at least 200 cells to create the dataset with patients diagnosed with (diseased) and without (healthy) MM, in which hematologist experts carefully curated all images. In the following section, we describe the experimental setup considered in this work.
Experimental Setup
We have designed two experimental setups to assess the relevance of our dataset. In the first one, we have used the labeled cells, as illustrated in Fig. 2, to train a DNN model capable of learning patterns from such cells. In our second experimental setup, we analyzed the possibility of automatically suggesting diagnoses to patients (Table 1) based on the ratio of plasma cells in myelogram slides.
Our model allows automatic analysis of images from slides of BM aspirate to detect and identify plasma and non-plasma cells, thus supporting hematologists during the diagnosis procedure. Details about our DNN model are discussed in the following section.
The evaluation methodology was based on conventional practices used in computer vision (CV) for object detection tasks. Our study assessed the bounding boxes surrounding cells defined by the experts (ground truth) and those predicted by our model. By considering the bounding boxes, it is possible to compute the Intersection Over Union (IOU) ratio as defined in Equation (1) to estimate the number of true positives (TP), false positives (FP), and false negatives (FN). This estimation is based on a threshold, which works as an interval for accepting or rejecting a predicted bounding box. In brief, if IOU ≥ τ1, there is enough overlapping between the predicted and ground truth bounding boxes, thus classifying the detected cell as true positive (TP). If the predicted bounding box does not have enough overlapped area with ground truth, a false positive (FP) is detected. Similarly, when a prediction bounding box does not overlap a ground truth area, it is considered a false negative (FN).
In Fig. 3, we show 4 images to visually illustrate the performance of our DNN, in which the bounding boxes contain the predicted labels and their respective probability. The ground truth for all cells presented in these images was previously shown in Fig. 2. Based on these figures, it is possible to exemplify different classification performances in terms of TP, FP, and FN estimated from IOU.
Example of stained cells labeled by our DNN model as plasma (red) and non-plasma (blue). The label probabilities were also plotted along with the bounding box captions. The ground truth bounding boxes for these cells were shown in Fig. 2.
For example, in Fig. 3(a), we can notice a TP prediction considering the top-most detected non-plasma cell. There is an overlapping between the prediction and expected (Fig. 2(a)) bounding boxes. These figures also show an example of FP prediction, by analyzing the cluster of three non-plasma cells in Fig. 2(d). In contrast, the DNN model, Fig. 3(a), predicted four non-plasma cells on the same region. In the center of Fig. 2(b), we can notice a cluster with one plasma and two non-plasma cells. In our prediction, Fig. 3(b), we noticed two examples of TP, but one non-plasma cell (the smaller one) is missing, exemplifying a FN. In Fig. 3(c), we illustrate a situation in which all expected cells were correctly detected in Fig. 2(c). However, a plasma cell was misclassified as non-plasma cell by our model (Fig. 3(c)). Finally, in Fig. 3(d), we illustrate a FN prediction, once our DNN was not capable of detecting a plasma cell in the right-most cluster presented in Fig. 2(d).
Based on IOU, we also assess the model confidence (C) by measuring the performance of predicting a cell in a bounding box. In summary, this confidence is calculated by the product between the probability f existing a cell and IoU, such as C = Pr(Object) * IOU. Considering the confidence, the model accepts the predicted bounding box when C ≥ τ2. The thresholds τ1 and τ2 are hyperparameters (not trainable), which can be varied to find objects in images better. Once there is no well-defined approach to set their values, we used a grid search in our experiments and selected their best combination based on the performance measured with F1-score, which is detailed next.
After calculating IOU, we compute three metrics widely used in CV tasks to assess the performance of our model: Precision, Recall, and F1-score. In summary, Precision (Equation (2)) computes the rate of correct classifications for the positive label over the number of outcomes classified as positives. On the other hand, Recall (Equation (3)) measures the models’ completeness, which is calculated by the rate of correct classifications for the positive label over the number of elements expected to be under the positive label. F1-score works as a harmonic mean of precision and recall, as shown in Equation (4).
Deep learning architecture
To automatically detect plasma cells in our new dataset, we have used a DNN architecture based on YOLO (You Only Look Once), a popular real-time object detection technique. YOLO consists of three main components: the backbone, the neck, and the head. The backbone extracts features from images at varying levels of detail. These features are then passed through the neck, which improves processing efficiency by combining different scales and reducing dimensionality before reaching the head component for final object detection.
We have used the version Ultralytics YOLOv813, which is well-suited for real-time object detection tasks across various application areas by maintaining an optimal balance between accuracy and speed. This version incorporates state-of-the-art backbone and neck architectures, enhancing feature extraction and object detection performance. YOLOv8 also utilizes an anchor-free split Ultralytics head, which improves accuracy and efficiency compared to anchor-based methods. The head component detects objects, draws bounding boxes, and infers the estimated classes (labels), by using a self-attention mechanism, enabling the model to focus on various parts of the image and adjust the importance of different features based on their relevance to the task.
Over time, the backbone component of the YOLO series has evolved through several architectures: Darknet in the first version14, Cross-Stage Partial Network (CSP) in version 515, Extended Efficient Layer Aggregation Network (E-ELAN) in version 716, and CSPDarknet53 in version 813. CSPDarknet53 uses a split and merge strategy to improve the flow of gradients through the network, helping to improve the system’s accuracy by allowing it to learn more complex representations of the input data.
Another significant contribution of YOLOv8 is the model scaling, which allows the network to be adjusted on different devices and applications. Considering this functionality, we configured and personalized a DNN to distinguish between plasma and non-plasma cells in stained bone marrow aspirate smear slides. This configuration process involved the traditional construction of a DNN, including adding and removing layers, adjusting image resolution, modifying channels and filters, and tuning various parameters.
We also implemented transfer learning by initializing our models with pre-trained weights and then training them on our custom dataset. These weights are derived from a model trained on the COCO (Common Objects in Context)17 dataset, a large-scale dataset commonly used for object detection, segmentation, and captioning tasks in computer vision. By reusing the knowledge learned from previous tasks, we enhanced the performance of detecting plasma cells. Finally, we conducted a fine-tuning process on our images, which significantly improved our results in detecting bounding box sizes, probabilities, and classifications of plasma cells.
The final configuration of our training process was obtained after using a batch with a size equal to 32, 300 epochs, 12 GB of GPU memory, a learning rate equal to 0.01, and an Adam optimizer. After using our images during the training phase, the resultant model presented 225 layers with 3,157,200 parameters.
Data Records
The dataset is available at Mendeley Data5. The data folder is organized into two subfolders: “detection” and “segmentation”. The detection one contains randomized stained slide images and patient-specific datasets, which include the slides utilized for diagnostic analysis. In addition to the labeled images, this folder also provides the configuration files used for training the DNN models, following a cross-validation strategy.
The primary objective of this work is to differentiate plasma and non-plasma cells without using additional cell features such as the number of nuclei, shapes, and membrane structures. A key direction for future work involves developing models capable of learning from these features, as they hold the potential to provide deeper insights into the disease dynamics and uncover critical properties related to MM. To achieve this objective, the proposed dataset includes an subfold called “segmentation” containing numerous plasma and non-plasma cells segmented from the original stained slides, as illustrated in Fig. 4.
Beyond being utilized to train models capable of learning cellular features, this dataset also enables the development of generative AI models that can create synthetic cells resembling real ones. These synthetic cells can enhance the quality of DNNs, provide a richer variety of examples for model training, and support the education of new practitioners. This segmentation process was carried out using the AnyLabeling (https://anylabeling.nrl.ai/). Our dataset consists of the following files: image data in JPG format, label information in TXT format, segmentation coordinates in JSON format, cross-validation configurations in TXT and YAML files, and diagnosis data in CSV format.
Technical Validation
After creating the dataset, we conducted a thorough validation process from two perspectives. First, we performed a visual analysis of the cell distribution across all images from the stained slides, as illustrated in Fig. 5. Figure 5(a) displays the number of cells per label, while Fig. 5(b) summarizes the distribution of cells across the slide images. As expected, the slides are positioned on the microscope to centralize the clusters of cells. These figures are crucial for visualizing the overall placement of plasma and non-plasma cells in our dataset. Lastly, Fig. 5(c) illustrates the shape (width × height) of the bounding boxes, emphasizing the regions surrounding the labeled cells.
From the second perspective, we trained a DNN model to demonstrate the feasibility of extracting patterns and learning from our images. This process highlights the technical quality of our dataset and illustrates the potential for using both stained slides and segmented cells to enhance diagnostic processes and develop new AI-based methods to support specialists.
Initially, we focused our attention on the first experimental setup, which aims to detect and identify plasma and non-plasma cells. Using the DNN architecture previously defined, we started the training process using the 5-fold cross-validation strategy to find the optimal configuration for our DNN and to avoid obtaining results by chance. In Table 2, we present the cell distributions by labels and folds. In each fold, different cell configurations are used to train (train) the model and validate (val) its performance. As one may notice, we also used a nearly balanced amount of data to avoid inducing better results for a specific class.
Considering the train and validation cells, the obtained mean F1-score for all five folds was 78%. This result suggests our model was learning from the training data. The individual results obtained for each training and validation fold are available in the supplementary material.
In the next step, we used the entire dataset to find the optimal configuration to classify cells in our second experimental setup, i.e., analyzing the Diagnostic dataset presented in Table 1. In this sense, we have executed a grid search to better combine the thresholds and confidences. In our experiment, we noticed similar results were produced by varying τ1; hence, we set it as τ1 = 0.7. However, the relationship between τ2 and C significantly affects the final classification, as illustrated in Fig. 6(a). In our experiments, the best performance to detect and identify plasma and non-plasma cells achieved F1-score = 84% with τ2 = 0.532.
This F1 score indicates that our DNN model demonstrates significant overall performance in classifying cells. Next, we assessed its performance on each individual class. In Fig. 7, we present the relation between expected (true or ground truth) and predicted bounding boxes as a confusion (a.k.a. contingency) matrix.
Confusion (contingency) matrix showing the classification performance of the DNN model to predict the three classes: plasma_cell, non_plasma_cell, and background. Correct classifications are represented along the diagonal, while misclassifications are shown in the off-diagonal elements. The color gradient indicates the percentage of predictions, with darker shades corresponding to higher performances.
The diagonal results for plasma and non-plasma cells emphasize that most bounding boxes were correctly predicted (true positives, TP). False positives (FP) and false negatives (FN) are represented by the other values in this matrix. According to this matrix, 381 plasma cells were correctly identified by our DNN model, with only 37 non-plasma cells misclassified as plasma cells. Regarding non-plasma cells, 1,378 were correctly classified, while only 7 were misclassified as plasma cells.
In relation to the background, 71 areas were incorrectly marked as plasma cells and 152 as non-plasma cells. These numbers are considerably low, as the protocol followed by specialists avoids drawing bounding boxes around cells that are not fully visible. Nevertheless, even when partially omitted or not fully visible, our DNN model can still identify cells. Although this may increase classification errors, it does not pose a significant issue, as such cases are typically disregarded during the diagnostic process.
In Fig. 6(b), we show the relevant performance of our DNN model, presenting a mAP50 result superior to 90%. Considering all analyses summarized in these figures, we are confident that our dataset contains valuable patterns to identify plasma and non-plasma cells, providing an important and low-cost setup to support hematologists. Moreover, our dataset can also be used to train new hematologists, providing different cell patterns.
The final challenge presented in this work lies in verifying the performance of our approach in effectively aiding specialists in diagnosing MM. To reach this goal, we analyzed slides for each patient in Table 3. In summary, we have used our DNN model to detect, identify, and count plasma and non-plasma cells. Next, we calculate the ratio between them. As aforementioned, if the ratio is greater than 10%, the MM diagnosis is positive. As one may notice, there is narrow agreement between the expert and predicted diagnoses. The only different was found in Patient 07, in which the ratio was 8.8% and the ratio predicted by our model was 11.1%. However, once were are using the DNN model to support experts, we consider it a less serious mistake compared to incorrectly labeling a patient’s exam with MM as healthy (negative for MM). In this case, the expert is warned to recount the cells.
To better understand the performance of the DNN model in every patient, Table 4 shows the precision, recall, and F1 scores. These metrics allow for an individual assessment of the model’s effectiveness, highlighting its strong performance on cell slides collected from both healthy and diseased patients.
Our contribution is distinct from usual approaches due to focusing on modeling images captured using standard smartphone cameras. This feature is particularly pertinent as it facilitates the decision-making process in hospitals with resource-constrained settings. Moreover, we also provide a fine-tuned DNN model capable of detecting and identifying plasma and non-plasma cells. This model is especially important to support hematologists and scientists devoted to using it as a benchmark to improve the state-of-the-art in DNN to detect cells. Further results using the DNN architecture are provided in the supplementary material.
Usage Notes
The dataset is under license Creative Commons (CC) BY 4.0. We encourage all interested researchers to download and utilize this dataset for detecting and identifying plasma cells, as well as investigating new machine learning methods for classifying patients with MM.
Code availability
The source code, models, and datasets used in the study are freely available at https://github.com/LabIA-UFBA/MMDB. The repository is structured into two primary folders: “src”, which contains all source code and the DNN model utilized in this study, and “data”, which includes all cell data shared in this research. Besides this dataset, we also created a repository with all codes used to preprocess the cells, organize the experiments, and train our DNN model. Such codes are available at https://github.com/LabIA-UFBA/MMDB. Aiming to assure the reproducibility of our results, the reader can find in our repositories all configurations to split the dataset into train, validate, and test folds. Notebooks with codes illustrating every step of our investigation are also available to easily support researchers interested in deeply understanding our work. Finally, in supplementary material, we present a datasheet that provides general information on our dataset.
References
Rajkumar, S. V. et al. International myeloma working group updated criteria for the diagnosis of multiple myeloma. The lancet oncology 15, e538–e548 (2014).
Horenstein, A., Morandi, F., Bracci, C., Pistoia, V. & Malavasi, F. Functional insights into nucleotide-metabolizing ectoenzymes expressed by bone marrow-resident cells in patients with multiple myeloma. Immunology Letters 205, 40–50, https://doi.org/10.1016/j.imlet.2018.11.007 (2019). Special issue: The immune side of ectoenzymes.
Bladé, J. et al. Extramedullary disease in multiple myeloma: a systematic literature review. Blood Cancer Journal 12, 45 (2022).
Gorczyca, W. Atlas of differential diagnosis in neoplastic hematopathology (CRC Press, 2021).
Andrade, C. et al. PCMMD: Plasma Cells for Multiple Myeloma Diagnosis, https://doi.org/10.17632/3v2nrxpr9s.1 (2024).
Andrade, C. L. et al. Enhancing diagnostic accuracy of multiple myeloma through ml-driven analysis of hematological slides: new dataset and identification model to support hematologists. Scientific Reports 14, 11176 (2024).
Shearer, C. The crisp-dm model: the new blueprint for data mining. Journal of data warehousing 5, 13–22 (2000).
Gehlot, S., Gupta, A. & Gupta, R. A cnn-based unified framework utilizing projection loss in unison with label noise handling for multiple myeloma cancer diagnosis. Medical Image Analysis 72, 102099 (2021).
Kumar, D. et al. Automatic detection of white blood cancer from bone marrow microscopic images using convolutional neural networks. IEEE Access 8, 142521–142531 (2020).
Afshin, B., Reza, A., Eman, S. & Alaa, S. Multi-scale regional attention deeplab3+: Multiple myeloma plasma cells segmentation in microscopic images. Proceedings of Machine Learning Research 156, 47–56 (2021).
Rasal, T., Veerakumar, T., Subudhi, B. N. & Esakkirajan, S. Segmentation and counting of multiple myeloma cells using iemd based deep neural network. Leukemia Research 122, 106950, https://doi.org/10.1016/j.leukres.2022.106950 (2022).
Tzutalin, D. Labelimg. GitHub repository https://github.com/heartexlabs/labelImg Last access: March 29, 2023 (2015).
Jocher, G., Chaurasia, A. & Qiu, J. Ultralytics yolov8 (2023).
Redmon, J., Divvala, S., Girshick, R. & Farhadi, A. You only look once: Unified, real-time object detection. Proceedings of the IEEE conference on computer vision and pattern recognition 779–788 (2016).
Wang, C.-Y. et al. Cspnet: A new backbone that can enhance learning capability of cnn. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 10962–10971 (2020).
Li, H. et al. E-elan: Extended efficient layer aggregation network for real-time semantic segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 14652–14661 (2021).
Lin, T.-Y. et al. Microsoft coco: Common objects in context. In Computer Vision–ECCV 2014: 13th European Conference, Zurich, Switzerland, September 6-12, 2014, Proceedings, Part V 13, 740–755 (Springer, 2014).
Acknowledgements
T.N.R. and R.A.R. were supported by a grant from the Terumo Life Science Foundation, CNPq (Brazilian National Council for Scientific and Technological Development) grants [406354/2023-5, 312755/2023-6, 313053/2023-5], Maria Emilia Foundation grant to 01/2023, and INCITE FAPESB grant TO PIE0002/2022. M.V.F., B.M.A., R.A.R., and T.N.R. were supported by a grant from CAPES (Coordination for the Improvement of Higher Education Personnel—Brazilian federal government agency) finance code 001. M.V.F. and B.M.A. were supported by FAPESB (Bahia Research Foundation) grant [1589/2021]. R.A.R. was supported by Google Research Awards for Latin America 2021. T.J.S.L. was supported by the Council for Science, Technology and Innovation (CSTI), Cross-ministerial Strategic Innovation Promotion Program (SIP), “Innovative AI Hospital System”, the National Institute of Biomedical Innovation, Health and Nutrition (NIBIOHN) [grant number SIPAIH20D01], JSPS KAKENHI [JP22K06119] and the National Center for Child Health and Development internal grant [2022B-2].
Author information
Authors and Affiliations
Contributions
C.L.B.A., S.M.F., A.S.S., T.N.R., and R.A.R. conceptualized the study. A.S.S., M.M.S., M.I.C.S.S., I.M.D.R.P.R, G.C.C., H.H.M.S., M.M.L.S., and R.M. organized the data. M.V.F., B.M.A., I.P.C.M., and R.A.R. implemented the algorithms and performed the analyses. C.L.B.A., S.M.F., T.J.S.L., T.N.R., and R.A.R. interpreted the results. M.V.F., B.M.A., I.P.C.M., J.L.S.B.F., M.A.G., T.N.R., and R.A.R. conducted the experiments. C.L.B.A., S.M.F., T.J.S.L., T.N.R., and R.A.R. wrote the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
L. B. Andrade, C., Ferreira, M.V., M. Alencar, B. et al. PCMMD: A Novel Dataset of Plasma Cells to Support the Diagnosis of Multiple Myeloma. Sci Data 12, 161 (2025). https://doi.org/10.1038/s41597-025-04459-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41597-025-04459-1