Abstract
The American bullfrog (Aquarana catesbeiana) is both an economically important aquaculture species and a globally distributed invasive organism with high environmental adaptability. In this study, we present a high-quality chromosome-level genome assembly for the species, comprising 13 chromosomes with a total length of 6.32 Gb and a scaffold N50 of 691.8 Mb. Genome completeness was evaluated at 95.5% using BUSCO and 99.9% using Merqury. Repetitive sequences accounted for 79.51% of the genome. Through a combination of RNA-seq, Ab initio and homology-based gene prediction, we identified 32,382 protein-coding genes, with 98.96% of these genes functionally annotated. This chromosome-level assembly provides an important resource for future studies on the evolution, functional genomics and molecular breeding of the American bullfrog.
Similar content being viewed by others
Background & summary
Aquarana catesbeiana, commonly known as bullfrog, belongs to the class Amphibia, order Anura and family Ranidae. Native to North America, this species has undergone significant evolutionary adaptations, allowing it to thrive across diverse landforms and climatic conditions1. It has become a highly successful invasive species, expanding to over, 40 countries across four continents and contributing to the decline of indigenous species across multiple taxonomic categories2. Furthermore, bullfrog meat is considered a delicacy in many regions, driving increased global consumption3,4,5,6,7.
As a representative species of the order Anuran, the American bullfrog plays important roles in various fields of study, from developmental biology and physiology to ecology and evolution8,9. Previous studies utilised anuran species (frogs) to exhibit a wide range of sex chromosome differentiation stages as they exhibit diverse sex-determination systems and varying stages of sex chromosome differentiation, both between and within species10,11,12,13. Therefore, they serve as suitable candidates for investigating sex chromosome evolution and diversity13. Moreover, frogs display significant variation in genome size14,15, with sequenced species ranging from Platyplectrum ornatum (1.06 Gb)16 to Bombina bombina (9.8GB)17, making them ideal models for studying genome size evolution15.
To facilitate ecological, evolutionary and functional genomic studies, high-quality genome assemblies are essential. In 2017, a draft genome of A. catesbeiana was published, featuring a scaffold N50 of 51.6 Kb and 45.3% complete BUSCOs8. While these data provided valuable insights, the poor quality of the assembly limited its broader applicability. In the present study, we generate a chromosome-level genome assembly for the American bullfrog using integrated datasets from Pacific Biosciences (PacBio) HiFi reads, MGI short reads and Hi-C reads. This high-quality assembly will significantly advance research on the unique characteristics of frogs.
Methods
Sample collection
Tissue samples were collected from a female American bullfrog (Fig. 1) bred at the Qingyuan Yangshan breeding base, Zhongyang Group, Guangdong Province. Approximately 10 g of muscle tissue was extracted from this individual for whole-genome sequencing, including short-read, long-read and Hi-C sequencing. Additionally, multiple tissues were collected for transcriptome sequencing. Samples included intestines, lungs, vas deferens, liver, muscles, stomach and eyes, with approximately 1 g of each tissue collected. All tissue samples were fragmented, rapidly cooled with liquid nitrogen and stored at −80 °C for future use. The sample collection protocol was approved by the Animal Ethics Committee of Zhongkai University of Agriculture and Engineering (Guangzhou, China).
DNA extraction and genome sequencing
Genomic DNA (gDNA) was extracted from muscle tissue using the Qiagen Blood & Cell Culture DNA Kit (Qiagen, USA). The gDNA was randomly fragmented, and 1.5 µg was used to construct a 350-bp insert-size library with the MGIEasy Universal DNA Library Prep Set (MGI, China). Sequencing was performed on the MGISEQ2000 platform (MGI, China), generating 322.27 Gb of paired-end raw reads (150 bp). After filtering the raw reads using SOAPnuke (v2.1.0)18 to remove adaptors and low-quality sequences, 324.64 Gb of clean reads were obtained and used for genome size estimation and assembly.
For long-read sequencing, 2 µg of gDNA was used to create libraries following PacBio’s HiFi sequencing protocol (Pacific Biosciences, USA) with the SMRTbell Express Template Prep Kit 2.0. Sequencing was carried out on the PacBio Sequel II System, generating approximately 10.32 million consensus reads (156.21 Gb) with an average read length of 15.128 kb, processed using CCS software (SMRT Link v9.0)19 (-min-passes 1–min-rq 0.99–min-length 100).
Hi-C sequencing was performed on muscle tissue (~1 g) from the same female individual. Libraries were constructed using the GrandOmics Hi-C Kit (GrandOmics, China) with DpnII as the restriction enzyme. Sequencing was carried out on the Illumina NovaSeq platform (paired-end reads, 150 bp), yielding 711.68 Gb of raw reads. After quality filtration using fastp20, eliminating low-quality reads (quality scores <20) and those shorter than 30 bp, 697.99 Gb of high-quality reads (98.76% of the initial reads) were obtained for constructing pseudo-chromosomes.
RNA extraction and transcriptome sequencing
RNA was extracted from tissue samples using the standard Trizol protocol, followed by purification with the Qiagen RNeasy Mini Kit (Qiagen, USA). cDNA libraries were constructed following Illumina’s guidelines, and sequencing on the HiSeq X Ten platform (Illumina, USA). Finally, approximately 52 Gb of transcriptome data were generated. These data were used to support genome assembly and annotation.
Genome size estimation
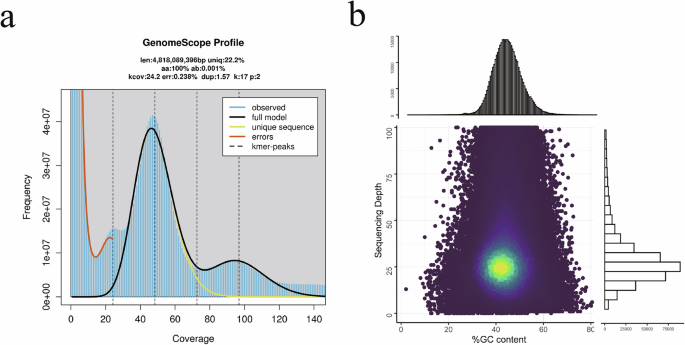
To genome size of the American bullfrog was estimated using a K-mer-based21 analysis method. The frequency of 17-mers was calculated using the GCE software22 (v1.0.2). The genome size (G) was determined by the formula G = K_num/K_depth, where K_depth represents K-mer depth and K_num is the total number of 17-mers. A BLASTN (v2.7.1)23 alignment against the NT database confirmed the absence of significant exogenous contamination in the library data. The estimated genome size was approximately 5.81 Gb, with an estimated genome heterozygosity of 0.51% (Fig. 2a).
De novo genome assembly
After obtaining subreads, we used the assembly software hifiasm (v0.16.1)24 to perform reference-free de novo assembly of long reads from the pacbio platform’s SMRT sequencing, using default parameters. Error correction was performed using racon (v1.4.3)25, gcpp (v2.0.2) (https://github.com/PacificBiosciences/gcpp) and pilon (v1.22)26, which utilised MGI short reads. The initial genome assembly size was 6.37 Gb, aligning with the previously estimated genome size. Various evaluation methods were applied to assess the quality of the assembly.
Pseudo-chromosome construction
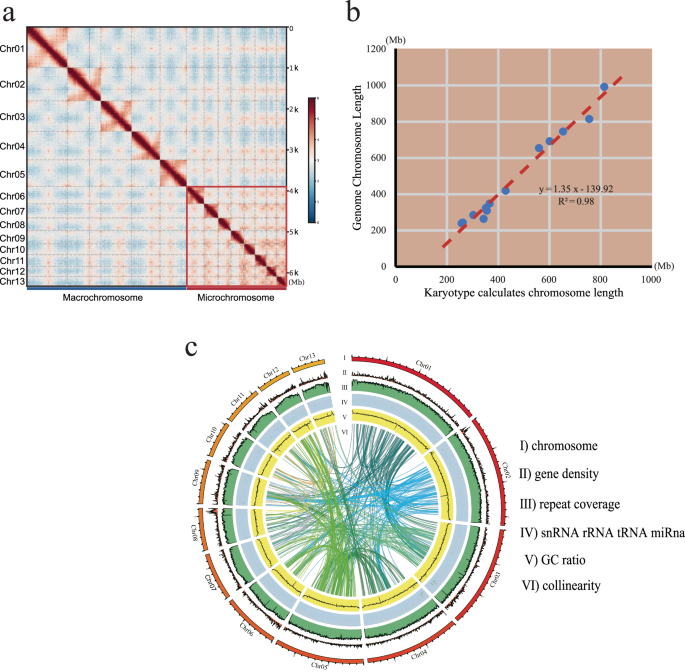
Hi-C technology was used to construct pseudo-chromosomes based on high-quality genome assembly. Trimmomatic software (LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:15)27 was employed to remove sequencing low-quality fragments and adapters. We aligned the retained data to the draft assembly using Juicer (baw: -SP5M)28, filtering out low-quality and redundant reads. Interaction maps were created using 3D-DNA software (Fig. 3a), and error correction was performed using JuiceBox software (v1.11.08)29. The final genome assembly reached 6.32 Gb, consisting of 13 pseudo-chromosomes (See details in Table 1), which covered 99.21% of the original genome. The overall assembly metrics included a scaffold N50 of 691.8 Mb.
(a) A total of 13 distinct blocks were visualised in the Hi-C contact matrixes. Chromosomes 1–6 are designated as macrochromosomes, while chromosomes 7–13 are classified as microchromosomes. (b) Pseudo-chromosome and Karyotype Correlation Analysis. The x-axis represents chromosome length measured based on existing karyotype data, assuming that each μm of chromosome contains 37.0 Mb. The y-axis represents the pseudo-chromosome cumulative length. (c) A Circos plot summarising the genome features. From outside to inside: (I) the lengths of 13 pseudo-chromosomes, (II) gene density, (III) repeat coverage, (IV) non-coding RNA content, (V) GC content and (VI) internal syntenic blocks.
Based on the existing karyotype data on American bullfrog30,31, we conducted a correlation analysis between the relative lengths obtained from the karyotype data and the cumulative length of pseudo-chromosomes. The analysis yielded an R-squared value above 0.99, confirming consistency with prior research. Using the karyotype image, we calculated physical chromosome lengths and determined a compression ratio of 37.0 Mb per μm (Table 1). The correlation analysis between the pseudo-chromosome lengths and karyotype measurements produced an R-squared value greater than 0.98 (Fig. 3b), demonstrating the accuracy of the pseudo-chromosome construction.
Gene annotation and functional assignment
Repetitive sequences were predicted using a Homolog-based approach, employing RepeatMasker and RepeatProteinMask software (v4.1.2)32 with the RepBase database. Additionally, an ab initio approach was employed, utilising RepeatModeler (v2.0.2a)32 and LTR-FINDER (v1.0.5)33 software to construct an ab initio repetitive sequence library, followed by prediction using RepeatMasker. Simultaneously, the TRF software (v4.09)34 was employed to identify Tandem Repeats within the genome. Finally, a total of 5.07 Gb of repetitive sequences were detected, accounting for 79.51% of the assembled genome (See details in Table 2).
Gene structure prediction involved three approaches: transcriptome-based, homology-based and de novo prediction. Exonerate (v2.4)35 was used to predict gene structures in homologous species, including Engystomops pustulosus, Bufo bufo, Rana temporaria and Xenopus laevis. De novo assembly of transcriptome reads was performed using Trinity (v2.13.2)36, followed by de novo prediction using PASA37 and Augustus (v3.3)38. A total of 32,382 genes were annotated, with an average gene length of 58,248.47 bp, an average CDS length of 1,264.88 bp and an average of 6.55 exons per gene (See details in Table 3).
Functional gene annotation was conducted through comparisons across various databases such as SwissProt39, NCBI NR, PFAM40, GO41, KEGG42, InterPro43 and TrEMBL44. Among these predicted genes, 98.96% were annotated in protein databases (See details in Table 3). Syntenic blocks were constructed using MCscan45, and a Circos46 plot (Fig. 3c) illustrated key features, including: (I) the lengths of 13 pseudo-chromosomes, (II) gene density, (III) repeat coverage, (IV) non-coding RNA content, (V) GC content and (VI) internal syntenic blocks.
Data Records
The genome assembly of the American bullfrog have been deposited in the GenBank database under the accession number GCA_042186555.147. The original genomic and transcriptomic data has been deposited in the China National GeneBank DataBase (CNGBdb) under project ID CNP0004806. Genome annotation files can be accessed publicly via the FigShare48.
Technical Validation
Quality assessment of the genome assembly
To evaluate the quality of the assembled genome, gDNA quality was assessed using both a Nanodrop spectrophotometer (Thermo Fisher Scientific, USA) and agarose gel electrophoresis. The OD260/280 values ranged between 1.8 and 2.0, while OD260/230 values were between 2.0 and 2.2, confirming sample quality. RNA extracted from the samples was analysed using a 2100 Bioanalyzer (Agilent Technologies, USA), yielding a 28S/18S ratio exceeding 1.0 and an RNA Integrity Number (RIN) greater than 7.0, further meeting quality requirements.
The minimap2 software (v2.12)49 was employed to map long reads to assembly data, using a 1000 bp sliding window, which confirmed that the genome assembly was free from contamination (Fig. 2b). Additionally, alignment of both long and short reads to the assembled genome indicated coverage of 99.13% and 99.96%, respectively, with average sequencing depth of 50.39x and 35.42x. The integrity of the genome assembly was further assessed using Benchmarking Universal Single Copy Orthologs (BUSCO, v5.2.2)50 with the vertebrata_odb10 database (Table 4). The results showed that the assembly and annotation contained 92.2% and 89.5% complete BUSCO genes, respectively, with single copy BUSCOs accounting for 90.1% and 86.9%, and duplicated BUSCOs representing 2.1% and 2.7%. Merqury (v1.3)51 analysis revealed a QV score and error rate of 38.924 and 1.28e-04, respectively. Gfastats (v1.3.1)52 was used to generate assembly summary statistics (Table 4).
Collinearity analysis
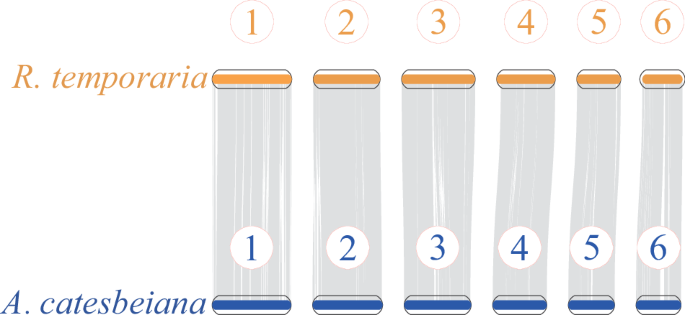
The JCVI package53 was used to perform whole-genome synteny analysis by aligning the chromosome-level genome data between the American bullfrog (this study) and its relative common frog (R. temporaria). Our results demonstrated strong one-to-one chromosomal correspondences between the two species (Fig. 4), highlighting the high-quality of the American Bullfrog genome assembly.
Code availability
The parameters and versions of the softwares used in the present study are described in the Methods section. Default parameters were used unless otherwise specified. No custom code was employed in the present study.
References
Halliday, T. The Book of Frogs. A Life-Size Guide to Six Hundred Species from around the World (University of Chicago Press, 2016).
Everts, T., Van Driessche, C., Neyrinck, S., Jacquemyn, H. & Brys, R. The American bullfrog exposed: distribution, invasion fronts, and spatial configuration of invasion hubs revealed by eDNA-based monitoring and environmental assessments. Manag. Biol. Invasions 14, 201–220 (2023).
Zhu, Y. et al. Comparison of the nutritional composition of bullfrog meat from different parts of the animal. Food Sci. Anim. Resour. 41, 1049–1059 (2021).
Zhang, C. et al. Apparent digestibility coef fi cients and amino acid availability of common protein ingredients in the diets of bullfrog, Rana (Lithobates) catesbeiana. Aquaculture 437, 38–45 (2015).
Wei, D. et al. Natural occurrences and characterization of Elizabethkingia miricola infection in cultured bullfrogs (Rana catesbeiana). Front. Cell. Infect. Microbiol. 13, 1–11 (2023).
Zhang, Cxiao et al. Effects of different lipid sources on growth performance, body composition and lipid metabolism of bullfrog Aquarana catesbeiana. Aquaculture 457, 104–108 (2016).
Trimpert, J. et al. Elizabethkingia miricola infection in multiple anuran species. Transbound. Emerg. Dis. 68, 931–940 (2021).
Hammond, S. A. et al. The North American bullfrog draft genome provides insight into hormonal regulation of long noncoding RNA. Nat. Commun. 8, 1–8 (2017).
Seidl, F. et al. Genome of spea multiplicata, a rapidly developing, phenotypically plastic, and desert-adapted spadefoot toad. G3 Genes, Genomes, Genet. 9, 3909–3919 (2019).
Malcom, J. W., Kudra, R. S. & Malone, J. H. The Sex Chromosomes of Frogs: Variability and Tolerance Offer Clues to Genome Evolution and Function. J. Genomics 2, 68–76 (2014).
Perrin, N. Sex-chromosome evolution in frogs: What role for sex-antagonistic genes? Philos. Trans. R. Soc. B Biol. Sci. 376, 20200049 (2021).
Eggert, C. Sex determination: the amphibian models. Reprod. Nutr. Dev. 44, 539–549 (2004).
Ma, W. J. & Veltsos, P. The diversity and evolution of sex chromosomes in frogs. Genes (Basel). 12, 483 (2021).
Olmo, E. & Morescalchi, A. Genome and cell sizes in frogs: A comparison with salamanders. Experientia 34, 44–46 (1978).
Zuo, B., Nneji, L. M. & Sun, Y.-B. Comparative genomics reveals insights into anuran genome size evolution. BMC Genomics 24, 379 (2023).
Lamichhaney, S. et al. A bird-like genome from a frog: Mechanisms of genome size reduction in the ornate burrowing frog, Platyplectrum ornatum. Proc. Natl. Acad. Sci. USA. 118, e2011649118 (2021).
Nürnberger, B. et al. A dense linkage map for a large repetitive genome: Discovery of the sex-determining region in hybridizing fire-bellied toads (Bombina bombina and Bombina variegata). G3 Genes, Genomes, Genet. 11, jkab286 (2021).
Chen, Y. et al. SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience 7, gix120 (2018).
Rhoads, A. & Au, K. F. PacBio Sequencing and Its Applications. Genomics. Proteomics Bioinformatics 13, 278–289 (2015).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Deorowicz, S., Kokot, M., Grabowski, S. & Debudaj-Grabysz, A. KMC 2: fast and resource-frugal k-mer counting. Bioinformatics 31, 1569–1576 (2015).
Sun, H., Ding, J., Piednoël, M. & Schneeberger, K. findGSE: estimating genome size variation within human and Arabidopsis using k-mer frequencies. Bioinformatics 34, 550–557 (2018).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Cheng, H., Concepcion, G. T., Feng, X., Zhang, H. & Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods 18, 170–175 (2021).
Abd-Alla, H. I., Souguir, D. & Radwan, M. O. Genus Sophora: a comprehensive review on secondary chemical metabolites and their biological aspects from past achievements to future perspectives. Arch. Pharm. Res. 44, 903–986 (2021).
Walker, B. J. et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS One 9, e112963 (2014).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Durand, N. C. et al. Juicer Provides a One-Click System for Analyzing Loop-Resolution Hi-C Experiments. Cell Syst. 3, 95–98 (2016).
Durand, N. C. et al. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst. 3, 99–101 (2016).
Vitelli, L., Batistoni, R., Andronico, F., Nardi, I. & Barsacchi-Pilone, G. Chromosomal localization of 18S + 28S and 5S ribosomal RNA genes in evolutionarily diverse anuran amphibians. Chromosoma 84, 475–491 (1982).
Reynhout, J. K. & Kimmel, D. L. Chromosome studies of the lethal hybrid Rana pipiens ♀ × Rana catesbeiana ♂. Dev. Biol. 20, 501–517 (1969).
Bedell, J. A., Korf, I. & Gish, W. MaskerAid: a performance enhancement to RepeatMasker. Bioinformatics 16, 1040–1041 (2000).
Xu, Z. & Wang, H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 35, W265–W268 (2007).
Benson, G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580 (1999).
Slater, G. S. C. & Birney, E. Automated generation of heuristics for biological sequence comparison. BMC bioinformatics. 6, 1–11 (2005).
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011).
Haas, B. J. et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31, 5654–5666 (2003).
Stanke, M. et al. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 34, W435–W439 (2006).
Bairoch, A. & Apweiler, R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 28, 45–48 (2000).
Mistry, J. et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 49, D412–D419 (2021).
Carbon, S. et al. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 47, D330–D338 (2019).
Ogata, H. et al. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 27, 29–34 (1999).
Jones, P. et al. InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240 (2014).
O’Donovan, C. et al. High-quality protein knowledge resource: SWISS-PROT and TrEMBL. Brief. Bioinform. 3, 275–284 (2002).
Wang, Y. et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49 (2012).
Krzywinski, M. et al. Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645 (2009).
NCBI GenBank https://identifiers.org/ncbi/insdc.gca:GCA_042186555.1 (2024).
Zhang, K. American bullfrog annotation files. figshare https://figshare.com/s/cd3056e928f4d91b3578 (2024).
Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018).
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015).
Rhie, A., Walenz, B. P., Koren, S. & Phillippy, A. M. Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol. 21, 1–27 (2020).
Formenti, G. et al. Gfastats: Conversion, evaluation and manipulation of genome sequences using assembly graphs. Bioinformatics 38, 4214–4216 (2022).
Tang, H. et al. Synteny and collinearity in plant genomes. Science 320, 486–488 (2008).
Acknowledgements
This project was supported by Development Project of "Biosafety Technology" of Guangdong Province (No. 2022B1111030001), Special Provincial Approval Project Funds for the Rural Revitalization Strategy of Department of Agriculture and Rural Affairs of Guangdong Province (No. KB23Y1101), and R & D Projects in Key Areas of Guangdong Province (No. 2021B0202030001).
Author information
Authors and Affiliations
Contributions
Lin Li, Chun Liu and Zhendong Qin conceived this study; Kai Zhang, Ye Tian and Yuxuan Zhang participated in the research and bioinformatics analysis; Kai Zhang and Chun Liu collected the samples; Bin Xu and Xiewu Jiang provided research advice; Kai Zhang, Ye Tian and Yuxuan Zhang wrote the original manuscript; Lin Li, Chun Liu and Zhendong Qin reviewed and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, K., Zhang, Y., Tian, Y. et al. A Chromosome-level genome assembly of the American bullfrog (Aquarana catesbeiana). Sci Data 12, 413 (2025). https://doi.org/10.1038/s41597-025-04697-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41597-025-04697-3